Predicting Adult Hospital Admission from Emergency Department Using Machine Learning: An Inclusive Gradient Boosting Model
Abstract
1. Introduction
2. Methods
2.1. Data Sources
2.2. Study Design
2.3. Study Population
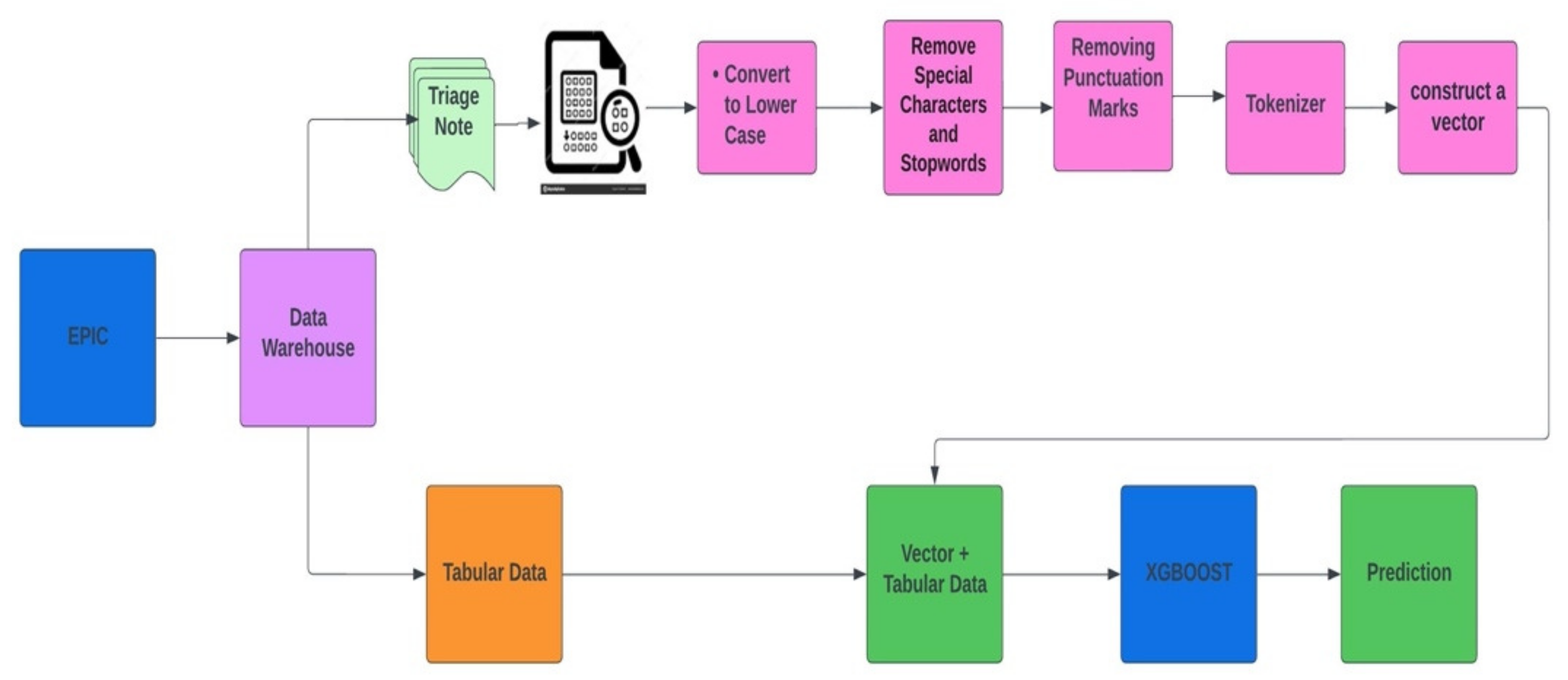
2.4. Study Data
2.5. Outcome Definition
2.6. Model Development
2.7. Models Training and Testing
2.8. Models Interpretation
2.9. Statistical Analysis
3. Results
3.1. Cohort Characteristics
3.2. Experimental Results
3.2.1. Experiment 1
3.2.2. Experiment 2
3.3. Single Feature Analysis
3.4. SHAP Plot Analysis:
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Chalfin, D.B.; Trzeciak, S.; Likourezos, A.; Baumann, B.M.; Dellinger, R.P. Impact of delayed transfer of critically ill patients from the emergency department to the intensive care unit. Crit. Care Med. 2007, 35, 1477–1483. [Google Scholar] [CrossRef]
- Rabin, E.; Kocher, K.; McClelland, M.; Pines, J.; Hwang, U.; Rathlev, N.; Asplin, B.; Trueger, N.S. Solutions to emergency department “boarding” and crowding are underused and may need to be legislated. Health Aff. 2012, 31, 1757–1766. [Google Scholar] [CrossRef]
- Forero, R.; McCarthy, S.; Hillman, K. Access block and emergency department overcrowding. Crit. Care 2011, 15, 216. [Google Scholar] [CrossRef]
- Sterling, N.W.; Patzer, R.E.; Di, M.; Schrager, J.D. Prediction of emergency department patient disposition based on natural language processing of triage notes. Int. J. Med. Inform. 2019, 129, 184–188. [Google Scholar] [CrossRef]
- King, Z.; Farrington, J.; Utley, M.; Kung, E.; Elkhodair, S.; Harris, S.; Sekula, R.; Gillham, J.; Li, K.; Crowe, S. Machine learning for real-time aggregated prediction of hospital admission for emergency patients. NPJ Digit. Med. 2022, 5, 104. [Google Scholar] [CrossRef]
- Yao, L.-H.; Leung, K.-C.; Hong, J.-H.; Tsai, C.-L.; Fu, L.-C. A System for Predicting Hospital Admission at Emergency Department Based on Electronic Health Record Using Convolution Neural Network. In Proceedings of the 2020 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Toronto, ON, Canada, 11–14 October 2020; pp. 546–551. [Google Scholar] [CrossRef]
- Pivovarov, R.; Elhadad, N. Automated methods for the summarization of electronic health records. J. Am. Med. Inform. Assoc. 2015, 22, 938–947. [Google Scholar] [CrossRef]
- Deo, R.C. Machine Learning in Medicine. Circulation 2015, 132, 1920–1930. [Google Scholar] [CrossRef]
- Klang, E.; Barash, Y.; Soffer, S.; Bechler, S.; Resheff, Y.S.; Granot, T.; Shahar, M.; Klug, M.; Guralnik, G.; Zimlichman, E.; et al. Promoting head CT exams in the emergency department triage using a machine learning model. Neuroradiology 2020, 62, 153–160. [Google Scholar] [CrossRef]
- Hong, W.S.; Haimovich, A.D.; Taylor, R.A. Predicting hospital admission at emergency department triage using machine learning. PLoS ONE 2018, 13, e0201016. [Google Scholar] [CrossRef]
- Dinh, M.M.; Russell, S.B.; Bein, K.J.; Rogers, K.; Muscatello, D.; Paoloni, R.; Hayman, J.; Chalkley, D.R.; Ivers, R. Te Sydney Triage to Admission Risk Tool (START) to predict Emergency Department Disposition: A derivation and internal validation study using retrospective state-wide data from New South Wales Australia. BMC Emerg. Med. 2016, 16, 46. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: San Francisco, CA, USA, 2016; pp. 785–794. [Google Scholar]
- Raita, Y.; Goto, T.; Faridi, M.K.; Brown, D.F.; Camargo, C.A.; Hasegawa, K. Emergency department triage prediction of clinical outcomes using machine learning models. Crit. Care 2019, 23, 64. [Google Scholar] [CrossRef]
- Lee, S.Y.; Chinnam, R.B.; Dalkiran, E.; Krupp, S.; Nauss, M. Prediction of emergency department patient disposition decision for proactive resource allocation for admission. Health Care Manag. Sci. 2020, 23, 339–359. [Google Scholar] [CrossRef]
- Barak-Corren, Y.; Israelit, S.H.; Reis, B.Y. Progressive prediction of hospitalisation in the emergency department: Uncovering hidden patterns to improve patient fow. Emerg. Med. J. 2017, 34, 308–314. [Google Scholar] [CrossRef]
- Lucini, F.R.; Fogliatto, F.S.; da Silveira, G.J.; Neyeloff, J.L.; Anzanello, M.J.; Kuchenbecker, R.S.; Schaan, B.D. Text mining approach to predict hospital admissions using early medical records from the emergency department. Int. J. Med. Inform. 2007, 100, 1–8. [Google Scholar] [CrossRef]
- Klang, E.; Kummer, B.R.; Dangayach, N.S.; Zhong, A.; Kia, M.A.; Timsina, P.; Cossentino, I.; Costa, A.B.; Levin, M.A.; Oermann, E.K. Predicting adult neuroscience intensive care unit admission from emergency department triage using a retrospective, tabular-free text machine learning approach. Sci. Rep. 2021, 11, 1381. [Google Scholar] [CrossRef]







| Demographics and Presentation | Vital Signs | Laboratory |
|---|---|---|
| Sex | Temperature | WBC |
| Age | Respirations | HGB |
| ESI | PainScale | PLT |
| Month | PulseOximetry | NEUT |
| Year | Pulse | LYMPH |
| Day | SystolicBP | CR |
| Hour | DiastolicBP | BUN |
| DayOfWeek | GLC | |
| NA | ||
| K | ||
| CHLORIDE | ||
| CALCIUM | ||
| LACTATE | ||
| ALT | ||
| AST | ||
| BILIRUBIN | ||
| ALK PHOS | ||
| ALBUMIN | ||
| CRP | ||
| D-DIMER | ||
| BNP | ||
| CPK | ||
| TROPONIN |
| Discharged (n = 841,825, 80.7%) | Hospitalized (n = 201,520, 19.3%) | p Value | |
|---|---|---|---|
| Age, median (IQR), y | 43.0 (29.0–59.0) | 64.0 (49.0–77.0) | <0.001 |
| Female, N. (%) | 465,178 (55.3) | 103,030 (51.1) | <0.001 |
| SBP, median (IQR), mmHg | 130.0 (118.0–145.0) | 134.0 (117.0–153.0) | <0.001 |
| DBP, median (IQR), mmHg | 76.0 (68.0–85.0) | 74.0 (65.0–85.0) | <0.001 |
| Heart rate, median (IQR), beats/min | 82.0 (73.0–93.0) | 87.0 (75.0–101.0) | <0.001 |
| Temperature, median (IQR), Celsius | 36.7 (36.4–36.9) | 36.7 (36.4–37.0) | <0.001 |
| Respirations, median (IQR), breaths/min | 18.0 (17.0–19.0) | 18.0 (18.0–20.0) | <0.001 |
| O2 saturation, median (IQR)% | 98.0 (97.0–99.0) | 98.0 (96.0–99.0) | <0.001 |
| WBC, median (IQR), ×103/uL | 7.8 (6.1–9.9) | 8.8 (6.6–11.9) | <0.001 |
| NEUT, median (IQR), ×103/uL | 5.0 (3.6–7.0) | 6.2 (4.2–9.3) | <0.001 |
| HGB, median (IQR), g/dL | 13.1 (11.9–14.3) | 12.3 (10.5–13.8) | <0.001 |
| BUN, median (IQR), mg/dL | 13.0 (10.0–17.0) | 17.0 (12.0–27.0) | <0.001 |
| Cr, median (IQR), mg/dL | 0.8 (0.7–1.0) | 0.9 (0.7–1.4) | <0.001 |
| Minutes | True Positive | False Positive | True Negative | False Negative | Sensitivity | Specificity | Precision (PPV) |
|---|---|---|---|---|---|---|---|
| 30 | 21,221 | 9224 | 255,897 | 39,829 | 0.348 | 0.965 | 0.697 |
| 60 | 21,887 | 9252 | 255,869 | 39,163 | 0.359 | 0.965 | 0.703 |
| 90 | 23,247 | 9575 | 255,546 | 37,803 | 0.381 | 0.964 | 0.708 |
| 120 | 25,172 | 9731 | 255,390 | 35,878 | 0.412 | 0.963 | 0.721 |
| 150 | 27,473 | 10,367 | 254,754 | 33,577 | 0.450 | 0.961 | 0.726 |
| Minutes | True Positive | False Positive | True Negative | False Negative | Sensitivity | Specificity | Precision (PPV) |
|---|---|---|---|---|---|---|---|
| 30 | 19,352 | 7810 | 257,311 | 41,698 | 0.317 | 0.971 | 0.712 |
| 60 | 21,342 | 8850 | 256,271 | 39,708 | 0.350 | 0.967 | 0.707 |
| 90 | 23,000 | 9345 | 255,776 | 38,050 | 0.377 | 0.965 | 0.711 |
| 120 | 25,147 | 9991 | 255,130 | 35,903 | 0.412 | 0.962 | 0.716 |
| 150 | 27,708 | 11,197 | 253,924 | 33,342 | 0.454 | 0.958 | 0.712 |
| Full Model AUC | Single Models AUC | |
|---|---|---|
| 30 min | 0.845 (95% CI: 0.843–0.847) | 0.847 (95% CI:0.845–0.849) |
| 60 min | 0.849 (95% CI: 0.848–0.851) | 0.850 (95% CI:0.849–0.852) |
| 90 min | 0.856 (95% CI: 0.855–0.858) | 0.857 (95% CI:0.855–0.858) |
| 120 min | 0.866 (95% CI: 0.864–0.867) | 0.867 (95% CI:0.865–0.868) |
| 150 min | 0.875 (95% CI: 0.874–0.877) | 0.877 (95% CI:0.876–0.879) |
| Feature | AUC |
|---|---|
| Age | 0.726 |
| ESI | 0.722 |
| Na+ | 0.621 |
| Ca++ | 0.619 |
| K+ | 0.618 |
| Hgb | 0.611 |
| Glucose | 0.603 |
| Lactate | 0.603 |
| Cl- | 0.602 |
| White Blood Cell Count | 0.595 |
| Neutrophil Count | 0.595 |
| Platelet Count | 0.594 |
| Lymphocyte Count | 0.591 |
| Pulse Rate | 0.591 |
| Sytolic BP | 0.581 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patel, D.; Cheetirala, S.N.; Raut, G.; Tamegue, J.; Kia, A.; Glicksberg, B.; Freeman, R.; Levin, M.A.; Timsina, P.; Klang, E. Predicting Adult Hospital Admission from Emergency Department Using Machine Learning: An Inclusive Gradient Boosting Model. J. Clin. Med. 2022, 11, 6888. https://doi.org/10.3390/jcm11236888
Patel D, Cheetirala SN, Raut G, Tamegue J, Kia A, Glicksberg B, Freeman R, Levin MA, Timsina P, Klang E. Predicting Adult Hospital Admission from Emergency Department Using Machine Learning: An Inclusive Gradient Boosting Model. Journal of Clinical Medicine. 2022; 11(23):6888. https://doi.org/10.3390/jcm11236888
Chicago/Turabian StylePatel, Dhavalkumar, Satya Narayan Cheetirala, Ganesh Raut, Jules Tamegue, Arash Kia, Benjamin Glicksberg, Robert Freeman, Matthew A. Levin, Prem Timsina, and Eyal Klang. 2022. "Predicting Adult Hospital Admission from Emergency Department Using Machine Learning: An Inclusive Gradient Boosting Model" Journal of Clinical Medicine 11, no. 23: 6888. https://doi.org/10.3390/jcm11236888
APA StylePatel, D., Cheetirala, S. N., Raut, G., Tamegue, J., Kia, A., Glicksberg, B., Freeman, R., Levin, M. A., Timsina, P., & Klang, E. (2022). Predicting Adult Hospital Admission from Emergency Department Using Machine Learning: An Inclusive Gradient Boosting Model. Journal of Clinical Medicine, 11(23), 6888. https://doi.org/10.3390/jcm11236888









