The Correlation between Periodontal Parameters and Cell-Free DNA in the Gingival Crevicular Fluid, Saliva, and Plasma in Chinese Patients: A Cross-Sectional Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Selection
2.2. Sample Collection
2.3. Extraction and Quantification of cfDNA
2.4. Statistical Analysis
3. Results
3.1. Demographics and Clinical Parameters
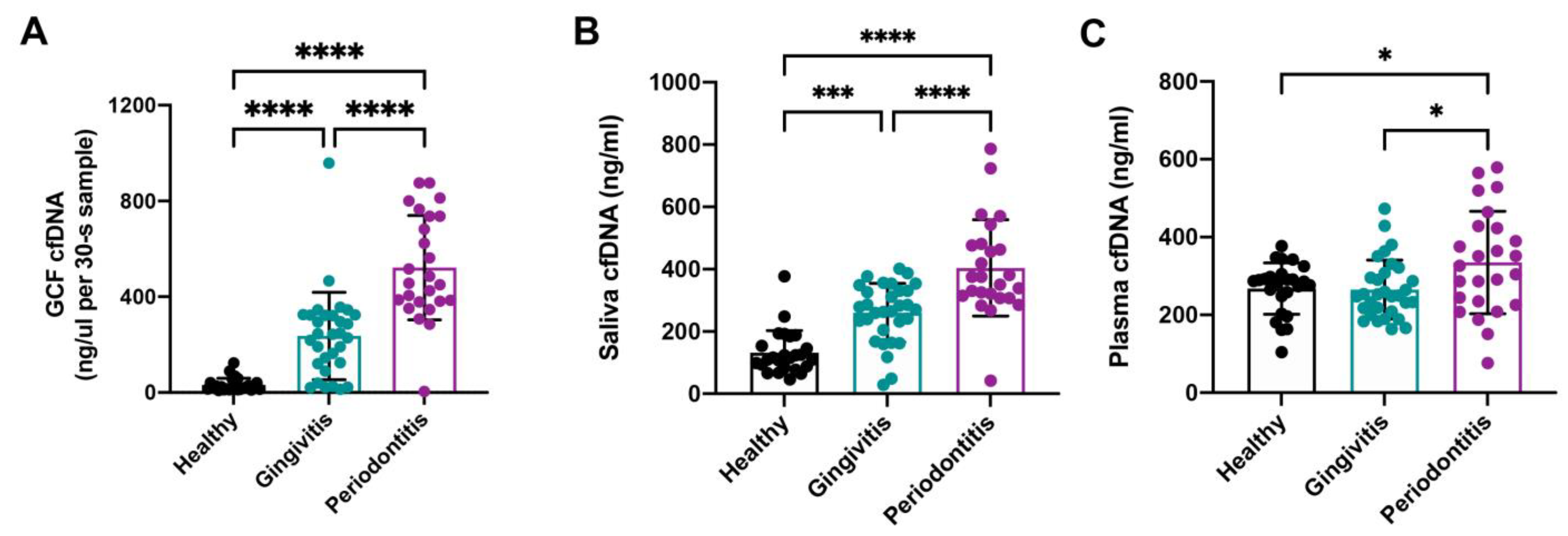
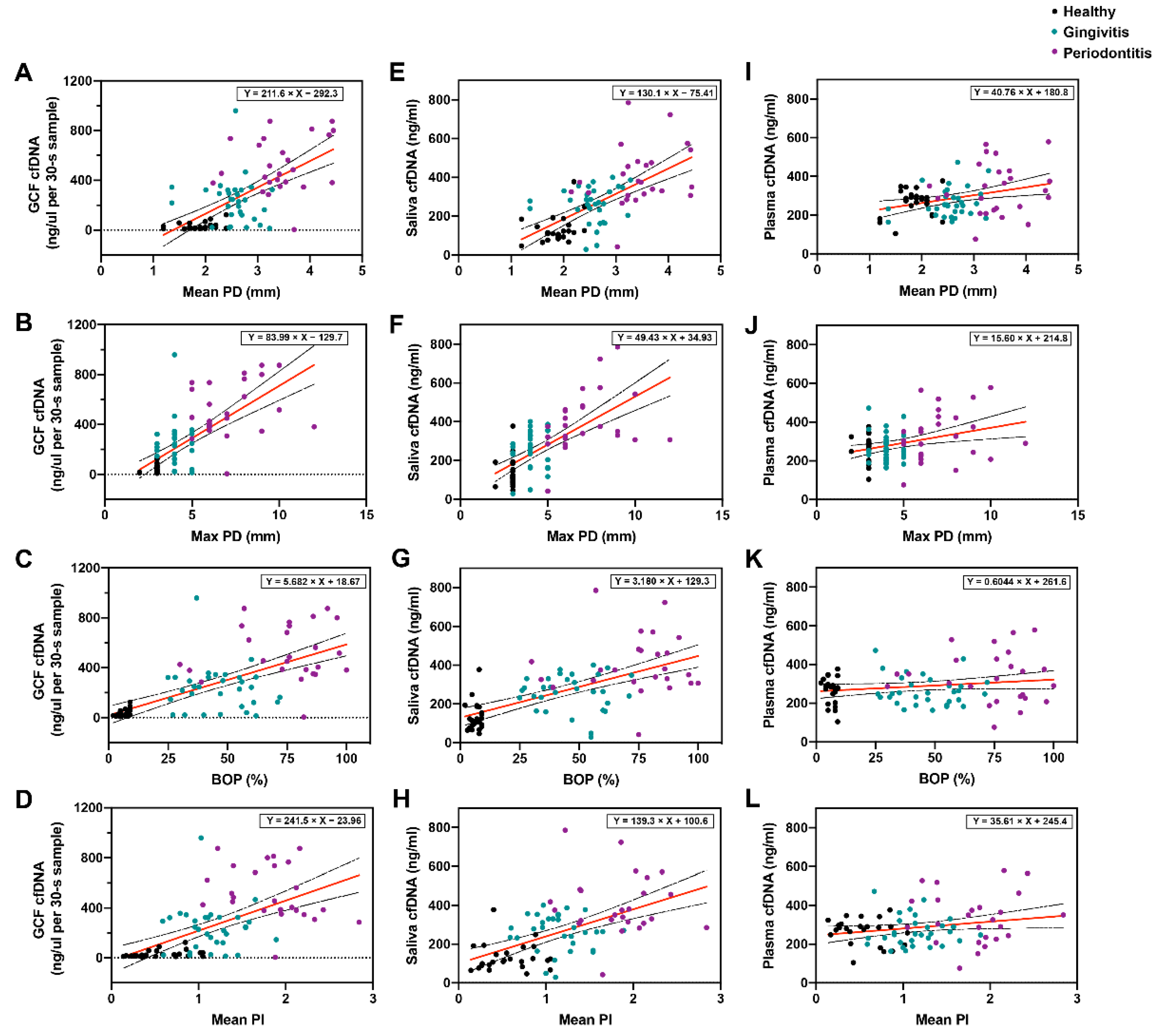
3.2. Comparison of GCF cfDNA Concentration in Relation to Periodontal Parameters
3.3. Comparison of Saliva cfDNA Concentration in Relation to Periodontal Parameters
3.4. Comparison of Plasma cfDNA Concentration in Relation to Periodontal Parameters
3.5. Multivariate Analysis of Age, Sex, and Clinical Parameters on cfDNA Concentrations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hajishengallis, G. Periodontitis: From microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 2015, 15, 30–44. [Google Scholar] [CrossRef] [Green Version]
- Thaiss, C.A.; Zmora, N.; Levy, M.; Elinav, E. The microbiome and innate immunity. Nature 2016, 535, 65–74. [Google Scholar] [CrossRef]
- Olive, C. Pattern recognition receptors: Sentinels in innate immunity and targets of new vaccine adjuvants. Expert Rev. Vaccines 2012, 11, 237–256. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swarup, V.; Rajeswari, M.R. Circulating (cell-free) nucleic acids--a promising, non-invasive tool for early detection of several human diseases. FEBS Lett. 2007, 581, 795–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar]
- Mortaz, E.; Adcock, I.M.; Abedini, A.; Kiani, A.; Kazempour-Dizaji, M.; Movassaghi, M.; Garssen, J. The role of pattern recognition receptors in lung sarcoidosis. Eur. J. Pharmacol. 2017, 808, 44–48. [Google Scholar] [CrossRef]
- Kim, Y.; Jo, A.R.; Jang, D.H.; Cho, Y.J.; Chun, J.; Min, B.M.; Choi, Y. Toll-like receptor 9 mediates oral bacteria-induced IL-8 expression in gingival epithelial cells. Immunol. Cell Biol. 2012, 90, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Sahingur, S.E.; Xia, X.J.; Schifferle, R.E. Oral bacterial DNA differ in their ability to induce inflammatory responses in human monocytic cell lines. J. Periodontol. 2012, 83, 1069–1077. [Google Scholar] [CrossRef] [PubMed]
- Govindarajan, S.; Veeraraghavan, V.P.; Dillibabu, T.; Patil, S. Oral Cavity-A Resilient Source for DNA Sampling. J. Contemp. Dent. Pract. 2022, 23, 1–2. [Google Scholar] [PubMed]
- Silva, L.M.; Doyle, A.D.; Greenwell-Wild, T.; Dutzan, N.; Tran, C.L.; Abusleme, L.; Juang, L.J.; Leung, J.; Chun, E.M.; Lum, A.G.; et al. Fibrin is a critical regulator of neutrophil effector function at the oral mucosal barrier. Science 2021, 374, eabl5450. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Kawai, T.; Akira, S. Microbial sensing by Toll-like receptors and intracellular nucleic acid sensors. Cold Spring Harb. Perspect. Biol. 2014, 7, a016246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Chen, Z.J. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014, 32, 461–488. [Google Scholar] [CrossRef]
- Kaneko, C.; Kobayashi, T.; Ito, S.; Sugita, N.; Murasawa, A.; Nakazono, K.; Yoshie, H. Circulating levels of carbamylated protein and neutrophil extracellular traps are associated with periodontitis severity in patients with rheumatoid arthritis: A pilot case-control study. PLoS ONE 2018, 13, e0192365. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Martinez, R.E.; Abud-Mendoza, C.; Patino-Marin, N.; Rizo-Rodriguez, J.C.; Little, J.W.; Loyola-Rodriguez, J.P. Detection of periodontal bacterial DNA in serum and synovial fluid in refractory rheumatoid arthritis patients. J. Clin. Periodontol. 2009, 36, 1004–1010. [Google Scholar] [CrossRef]
- Rao, A.; D'Souza, C.; Subramanyam, K.; Rai, P.; Thomas, B.; Gopalakrishnan, M.; Karunasagar, I.; Kumar, B.K. Molecular analysis shows the presence of periodontal bacterial DNA in atherosclerotic plaques from patients with coronary artery disease. Indian Heart J. 2021, 73, 218–220. [Google Scholar] [CrossRef]
- Papapanou, P.N.; Sanz, M.; Buduneli, N.; Dietrich, T.; Feres, M.; Fine, D.H.; Flemmig, T.F.; Garcia, R.; Giannobile, W.V.; Graziani, F.; et al. Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 2018, 89 (Suppl. S1), S173–S182. [Google Scholar] [CrossRef] [Green Version]
- Joss, A.; Adler, R.; Lang, N.P. Bleeding on probing. A parameter for monitoring periodontal conditions in clinical practice. J. Clin. Periodontol. 1994, 21, 402–408. [Google Scholar] [CrossRef]
- Fischman, S.L. Clinical index systems used to assess the efficacy of mouthrinses on plaque and gingivitis. J. Clin. Periodontol. 1988, 15, 506–510. [Google Scholar] [CrossRef]
- Suwannagindra, S.; Thaweboo, B.; Kerdvongbundit, V. Correlation between cell free DNA in gingival crevicular fluid and clinical periodontal parameters by using two collection techniques. M. Dent. J. 2020, 40, 165–174. [Google Scholar]
- Wassall, R.R.; Preshaw, P.M. Clinical and technical considerations in the analysis of gingival crevicular fluid. Periodontol 2016, 70, 65–79. [Google Scholar] [CrossRef]
- Henson, B.S.; Wong, D.T. Collection, storage, and processing of saliva samples for downstream molecular applications. Methods Mol. Biol. 2010, 666, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Green, E.A.; Li, R.; Albiges, L.; Choueiri, T.K.; Freedman, M.; Pal, S.; Dyrskjot, L.; Kamat, A.M. Clinical Utility of Cell-free and Circulating Tumor DNA in Kidney and Bladder Cancer: A Critical Review of Current Literature. Eur. Urol. Oncol. 2021, 4, 893–903. [Google Scholar] [CrossRef]
- Corcoran, R.B.; Chabner, B.A. Application of Cell-free DNA Analysis to Cancer Treatment. N. Engl. J. Med. 2018, 379, 1754–1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Han, S.; Zhang, M.; Wang, Y. Non-invasive prenatal paternity testing using cell-free fetal DNA from maternal plasma: DNA isolation and genetic marker studies. Leg. Med. 2018, 32, 98–103. [Google Scholar] [CrossRef]
- Shimony, A.; Zahger, D.; Gilutz, H.; Goldstein, H.; Orlov, G.; Merkin, M.; Shalev, A.; Ilia, R.; Douvdevani, A. Cell free DNA detected by a novel method in acute ST-elevation myocardial infarction patients. Acute Card. Care 2010, 12, 109–111. [Google Scholar] [CrossRef]
- Duvvuri, B.; Lood, C. Cell-Free DNA as a Biomarker in Autoimmune Rheumatic Diseases. Front. Immunol. 2019, 10, 502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hajishengallis, G.; Sahingur, S.E. Novel inflammatory pathways in periodontitis. Adv. Dent. Res. 2014, 26, 23–29. [Google Scholar] [CrossRef]
- Sahingur, S.E.; Xia, X.J.; Voth, S.C.; Yeudall, W.A.; Gunsolley, J.C. Increased nucleic Acid receptor expression in chronic periodontitis. J. Periodontol. 2013, 84, e48–e57. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Liu, C.; Wang, Z. The induction of inflammation by the cGAS-STING pathway in human dental pulp cells: A laboratory investigation. Int. Endod. J. 2022, 55, 54–63. [Google Scholar] [CrossRef]
- Barros, S.P.; Williams, R.; Offenbacher, S.; Morelli, T. Gingival crevicular fluid as a source of biomarkers for periodontitis. Periodontol 2016, 70, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Sandquist, M.; Wong, H.R. Biomarkers of sepsis and their potential value in diagnosis, prognosis and treatment. Expert Rev. Clin. Immunol. 2014, 10, 1349–1356. [Google Scholar] [CrossRef] [PubMed]
- Trombelli, L.; Scapoli, C.; Carrieri, A.; Giovannini, G.; Calura, G.; Farina, R. Interleukin-1 beta levels in gingival crevicular fluid and serum under naturally occurring and experimentally induced gingivitis. J. Clin. Periodontol. 2010, 37, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.H.; Joo, J.Y.; Lee, Y.J.; Koh, J.K.; Choi, J.H.; Shin, Y.; Cho, J.; Park, E.; Kang, J.; Lee, K.; et al. Grading system for periodontitis by analyzing levels of periodontal pathogens in saliva. PLoS ONE 2018, 13, e0200900. [Google Scholar] [CrossRef]
- Shakeeb, N.; Varkey, P.; Ajit, A. Human Saliva as a Diagnostic Specimen for Early Detection of Inflammatory Biomarkers by Real-Time RT-PCR. Inflammation 2021, 44, 1713–1723. [Google Scholar] [CrossRef]
- Zhu, X.; Huang, H.; Zhao, L. PAMPs and DAMPs as the Bridge Between Periodontitis and Atherosclerosis: The Potential Therapeutic Targets. Front. Cell Dev. Biol. 2022, 10, 856118. [Google Scholar] [CrossRef]
- Hegde, R.; Awan, K.H. Effects of periodontal disease on systemic health. Dis. Mon. 2019, 65, 185–192. [Google Scholar] [CrossRef]
- Listyarifah, D.; Al-Samadi, A.; Salem, A.; Syaify, A.; Salo, T.; Tervahartiala, T.; Grenier, D.; Nordstrom, D.C.; Sorsa, T.; Ainola, M. Infection and apoptosis associated with inflammation in periodontitis: An immunohistologic study. Oral Dis. 2017, 23, 1144–1154. [Google Scholar] [CrossRef] [Green Version]
- Sordi, M.B.; Magini, R.S.; Panahipour, L.; Gruber, R. Pyroptosis-Mediated Periodontal Disease. Int. J. Mol. Sci. 2021, 23, 372. [Google Scholar] [CrossRef]
- Ribeiro, C.C.C.; Carmo, C.D.S.; Benatti, B.B.; Casarin, R.V.C.; Alves, C.M.C.; Nascimento, G.G.; Moreira, A.R.O. Systemic circulating inflammatory burden and periodontitis in adolescents. Clin. Oral Investig. 2021, 25, 5855–5865. [Google Scholar] [CrossRef]
- Furutama, D.; Matsuda, S.; Yamawaki, Y.; Hatano, S.; Okanobu, A.; Memida, T.; Oue, H.; Fujita, T.; Ouhara, K.; Kajiya, M.; et al. IL-6 Induced by Periodontal Inflammation Causes Neuroinflammation and Disrupts the Blood-Brain Barrier. Brain Sci. 2020, 10, 679. [Google Scholar] [CrossRef] [PubMed]
- Padilla-Martinez, F.; Wojciechowska, G.; Szczerbinski, L.; Kretowski, A. Circulating Nucleic Acid-Based Biomarkers of Type 2 Diabetes. Int. J. Mol. Sci. 2021, 23, 295. [Google Scholar] [CrossRef] [PubMed]
- Cerne, D.; Bajalo, J.L. Cell-free nucleic acids as a non-invasive route for investigating atherosclerosis. Curr. Pharm. Des. 2014, 20, 5004–5009. [Google Scholar] [CrossRef]
- Shi, C.; Dawulieti, J.; Shi, F.; Yang, C.; Qin, Q.; Shi, T.; Wang, L.; Hu, H.; Sun, M.; Ren, L.; et al. A nanoparticulate dual scavenger for targeted therapy of inflammatory bowel disease. Sci. Adv. 2022, 8, eabj2372. [Google Scholar] [CrossRef] [PubMed]
- Dawulieti, J.; Sun, M.; Zhao, Y.; Shao, D.; Yan, H.; Lao, Y.H.; Hu, H.; Cui, L.; Lv, X.; Liu, F.; et al. Treatment of severe sepsis with nanoparticulate cell-free DNA scavengers. Sci. Adv. 2020, 6, eaay7148. [Google Scholar] [CrossRef]
- Deng, K.; Pelekos, G.; Jin, L.; Tonetti, M.S. Gingival bleeding on brushing as a sentinel sign of gingival inflammation: A diagnostic accuracy trial for the discrimination of periodontal health and disease. J. Clin. Periodontol. 2021, 48, 1537–1548. [Google Scholar] [CrossRef]
- Kim, P.D.; Xia-Juan, X.; Crump, K.E.; Abe, T.; Hajishengallis, G.; Sahingur, S.E. Toll-Like Receptor 9-Mediated Inflammation Triggers Alveolar Bone Loss in Experimental Murine Periodontitis. Infect. Immun. 2015, 83, 2992–3002. [Google Scholar] [CrossRef] [Green Version]
- Liang, H.; Peng, B.; Dong, C.; Liu, L.; Mao, J.; Wei, S.; Wang, X.; Xu, H.; Shen, J.; Mao, H.Q.; et al. Cationic nanoparticle as an inhibitor of cell-free DNA-induced inflammation. Nat. Commun. 2018, 9, 4291. [Google Scholar] [CrossRef] [Green Version]
- Peng, B.; Liang, H.; Li, Y.; Dong, C.; Shen, J.; Mao, H.Q.; Leong, K.W.; Chen, Y.; Liu, L. Tuned Cationic Dendronized Polymer: Molecular Scavenger for Rheumatoid Arthritis Treatment. Angew. Chem. Int. Ed. Engl. 2019, 58, 4254–4258. [Google Scholar] [CrossRef]
- Pan, W.; Yin, W.; Yang, L.; Xue, L.; Ren, J.; Wei, W.; Lu, Q.; Ding, H.; Liu, Z.; Nabar, N.R.; et al. Inhibition of Ctsk alleviates periodontitis and comorbid rheumatoid arthritis via downregulation of the TLR9 signalling pathway. J. Clin. Periodontol. 2019, 46, 286–296. [Google Scholar] [CrossRef]
- Michalowicz, B.S.; Hodges, J.S.; Pihlstrom, B.L. Is change in probing depth a reliable predictor of change in clinical attachment loss? J. Am. Dent. Assoc. 2013, 144, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Fleszar, T.J.; Knowles, J.W.; Morrison, E.C.; Burgett, F.G.; Nissle, R.R.; Ramfjord, S.P. Tooth mobility and periodontal therapy. J. Clin. Periodontol. 1980, 7, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Jing, W.; Si, Y.; Feng, X.; Tai, B.; Hu, D.; Lin, H.; Wang, B.; Wang, C.; Zheng, S.; et al. The prevalence and severity of periodontal disease in Mainland China: Data from the Fourth National Oral Health Survey (2015–2016). J. Clin. Periodontol. 2021, 48, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Meddeb, R.; Dache, Z.A.A.; Thezenas, S.; Otandault, A.; Tanos, R.; Pastor, B.; Sanchez, C.; Azzi, J.; Tousch, G.; Azan, S.; et al. Quantifying circulating cell-free DNA in humans. Sci. Rep. 2019, 9, 5220. [Google Scholar] [CrossRef] [Green Version]
- Laine, M.L.; Crielaard, W.; Loos, B.G. Genetic susceptibility to periodontitis. Periodontol 2012, 58, 37–68. [Google Scholar] [CrossRef]
| Characteristics | Groups | ||
|---|---|---|---|
| Healthy | Gingivitis | Periodontitis | |
| Number of subjects (n) | 25 | 31 | 25 |
| Male/female | 16/9 | 13/18 | 11/24 |
| Cigarette (Y/N) | 0/25 | 0/31 | 0/25 |
| Mean ± SD | |||
| Age range (years) | 25.08 ± 1.96 (range 23–32) | 26.16 ± 4.12 (range 21–33) | 33.52 ± 11.31 (range 22–60) |
| Clinical parameters | |||
| Mean PD (mm) | 1.87 ± 0.31 | 2.55 ± 0.43 | 3.44 ± 0.61 |
| Max PD (mm) | 2.92 ± 0.28 | 4.03 ± 0.66 | 7.16 ± 1.84 |
| BOP (%) | 6.56 ± 2.04 | 46.35 ± 13.51 | 74.24 ± 19.06 |
| Mean PI | 0.57 ± 0.29 | 1.15 ± 0.31 | 1.83 ± 0.43 |
| Correlation Coefficient (r Value) a | |||||
|---|---|---|---|---|---|
| Clinical Parameters | |||||
| Mean PD (mm) | Max PD (mm) | BOP (%) | Mean PI | ||
| 0.644 | 0.680 | 0.670 | 0.576 | ||
| (p < 0.0001) | (p < 0.0001) | (p < 0.0001) | (p < 0.0001) | ||
| Regression analyses (R2 value) b | |||||
| Mean PD (mm) | Max PD (mm) | BOP (%) | Mean PI | ||
| Mean GCF cfDNA conc. | Model | R2 = 0.415 | R2 = 0.463 | R2 = 0.449 | R2 = 0.331 |
| SE | 196,025.625 | 187,864.649 | 190,166.355 | 209,571.977 | |
| p value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| Correlation Coefficient (r Value) a | |||||
|---|---|---|---|---|---|
| Clinical Parameters | |||||
| Mean PD (mm) | Max PD (mm) | BOP (%) | Mean PI | ||
| 0.657 | 0.664 | 0.622 | 0.550 | ||
| (p < 0.0001) | (p < 0.0001) | (p < 0.0001) | (p < 0.0001) | ||
| Regression analyses (R2 value) b | |||||
| Mean PD (mm) | Max PD (mm) | BOP (%) | Mean PI | ||
| Mean saliva cfDNA conc. | Model | R2 = 0.432 | R2 = 0.441 | R2 = 0.387 | R2 = 0.303 |
| SE | 116.411 | 115.485 | 120.905 | 128.942 | |
| p value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| Correlation Coefficient (r Value) a | |||||
|---|---|---|---|---|---|
| Clinical Parameters | |||||
| Mean PD (mm) | Max PD (mm) | BOP (%) | Mean PI | ||
| 0.321 | 0.327 | 0.185 | 0.220 | ||
| (p = 0.003) | (p = 0.003) | (p = 0.099) | (p = 0.049) | ||
| Regression analyses (R2 value) b | |||||
| Mean PD (mm) | Max PD (mm) | BOP (%) | Mean PI | ||
| Mean plasma cfDNA conc. | Model | R2 = 0.103 | R2 = 0.107 | R2 = 0.034 | R2 = 0.048 |
| SE | 93.723 | 93.526 | 97.270 | 96.552 | |
| p value | 0.003 | 0.003 | 0.099 | 0.049 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, X.; Chu, C.-J.; Pan, W.; Li, Y.; Huang, H.; Zhao, L. The Correlation between Periodontal Parameters and Cell-Free DNA in the Gingival Crevicular Fluid, Saliva, and Plasma in Chinese Patients: A Cross-Sectional Study. J. Clin. Med. 2022, 11, 6902. https://doi.org/10.3390/jcm11236902
Zhu X, Chu C-J, Pan W, Li Y, Huang H, Zhao L. The Correlation between Periodontal Parameters and Cell-Free DNA in the Gingival Crevicular Fluid, Saliva, and Plasma in Chinese Patients: A Cross-Sectional Study. Journal of Clinical Medicine. 2022; 11(23):6902. https://doi.org/10.3390/jcm11236902
Chicago/Turabian StyleZhu, Xuanzhi, Chao-Jung Chu, Weiyi Pan, Yan Li, Hanyao Huang, and Lei Zhao. 2022. "The Correlation between Periodontal Parameters and Cell-Free DNA in the Gingival Crevicular Fluid, Saliva, and Plasma in Chinese Patients: A Cross-Sectional Study" Journal of Clinical Medicine 11, no. 23: 6902. https://doi.org/10.3390/jcm11236902





