A Novel PiRNA Enhances CA19-9 Sensitivity for Pancreatic Cancer Identification by Liquid Biopsy
Abstract
:1. Introduction
2. Experimental Section
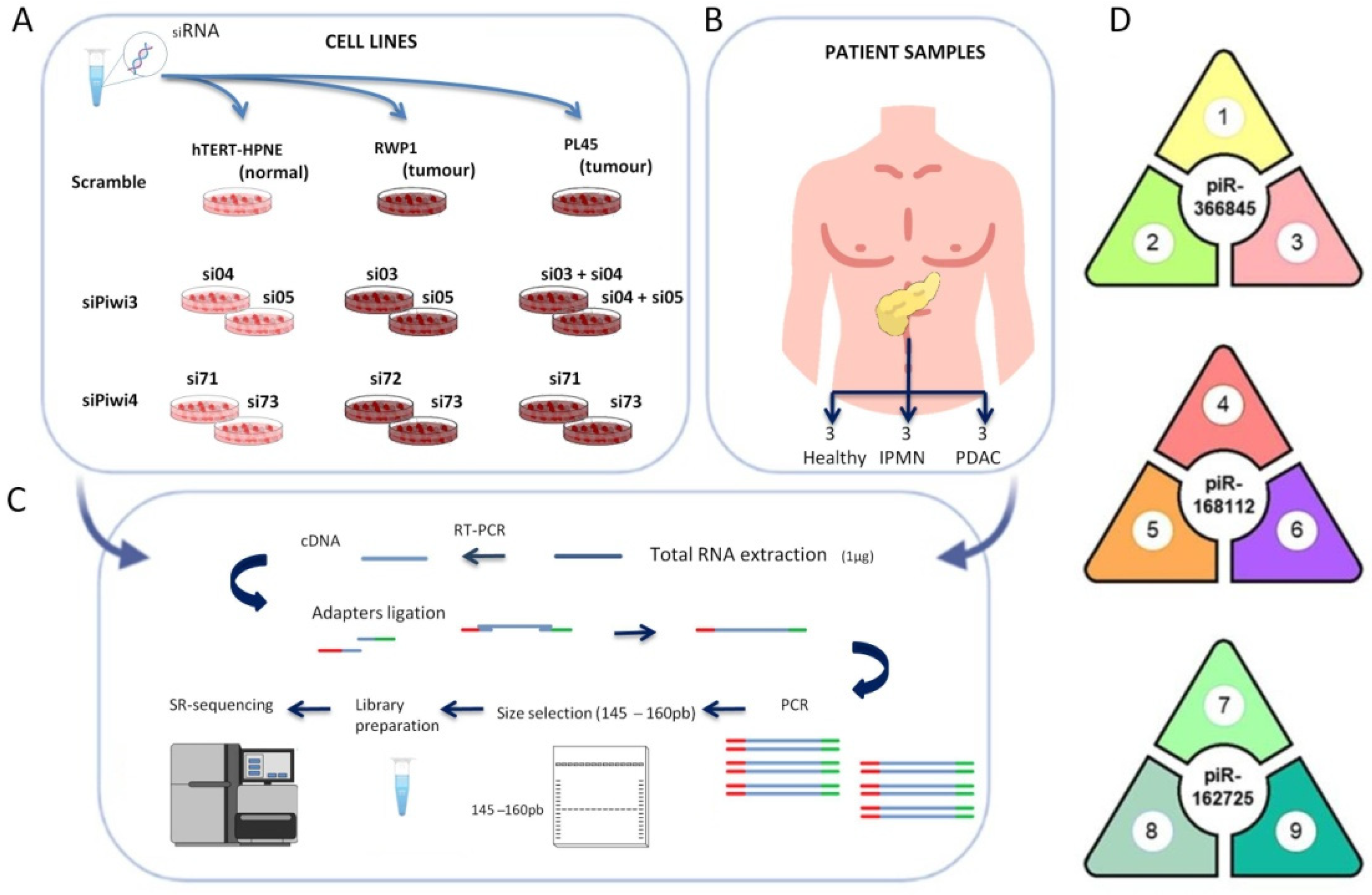
2.1. Cell Lines and Cell Culture
2.2. Patient Samples and Public Databases
2.3. RNA Interference, Western Blotting, and Immunocytochemistry
2.4. Small RNA Isolation, Library Preparation, Small RNA Sequencing, and Bioinformatic Analysis
2.5. Quantification of piRNAs in Blood Samples by Real-Time PCR
2.6. Statistical Analysis
3. Results
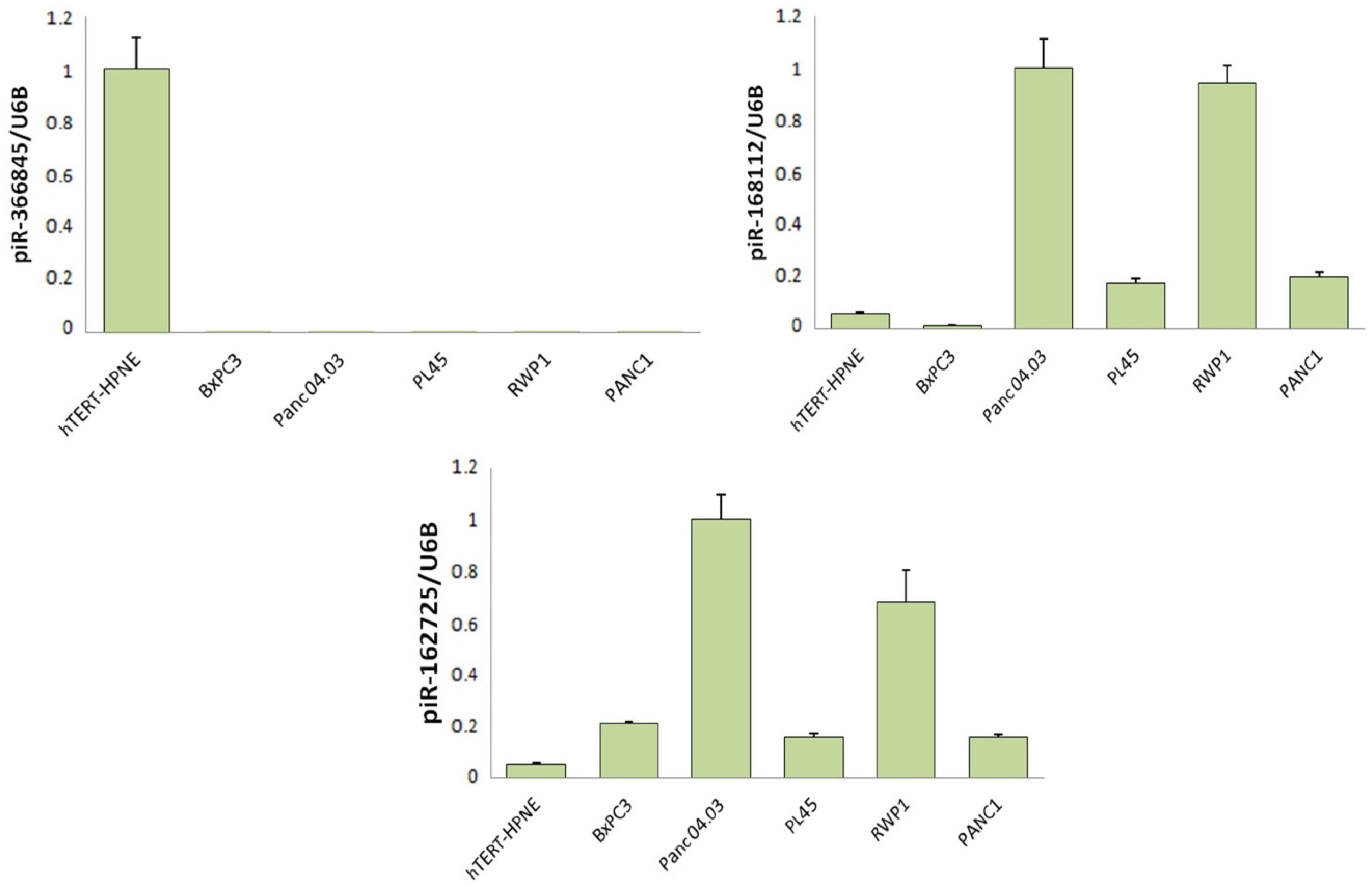
3.1. Differentially Expressed piRNAs Associated with PIWIL3 Exhibit a Transforming Effect, whereas a piRNA Associated with PIWIL4 Had Protective Effects
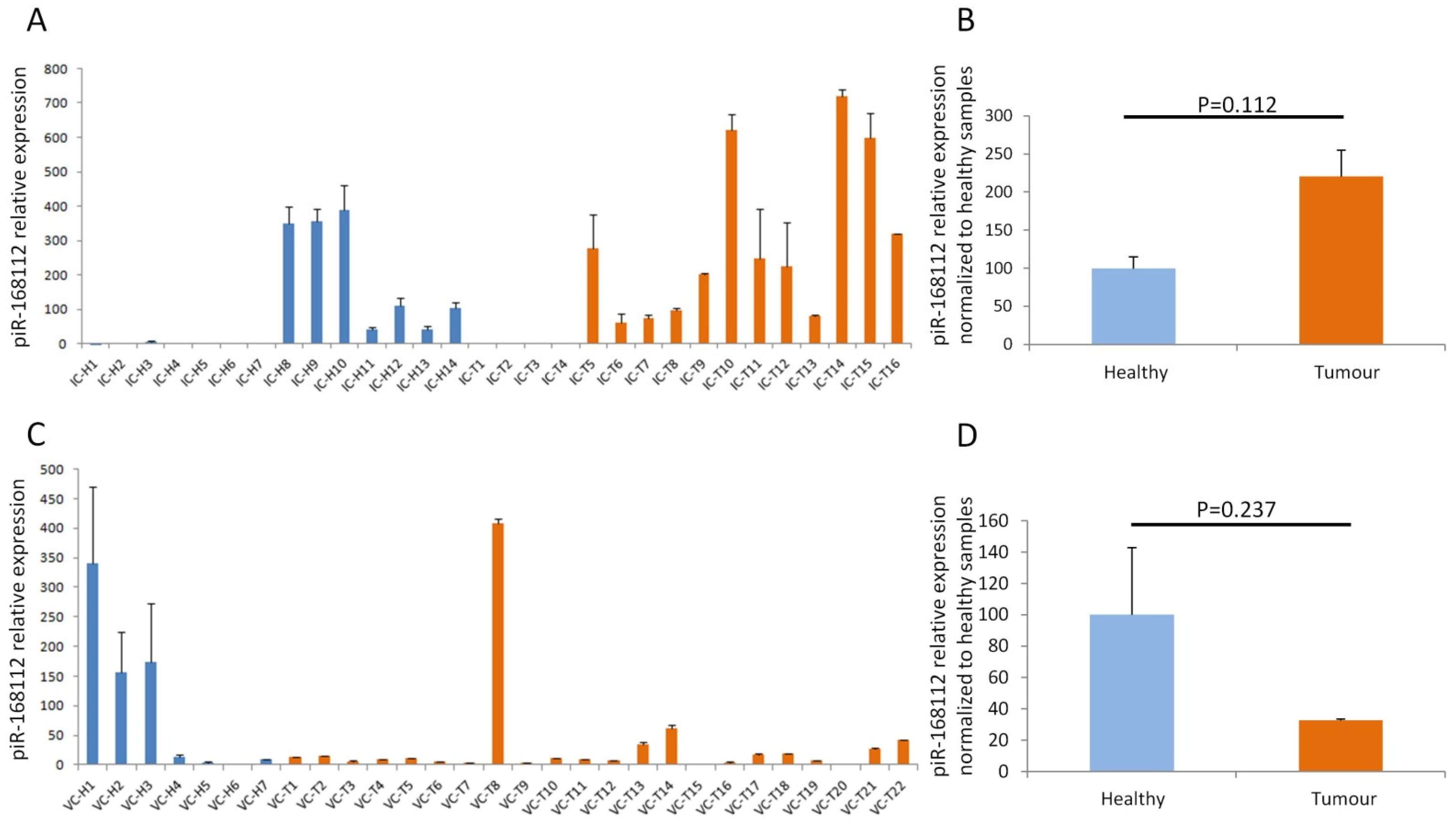
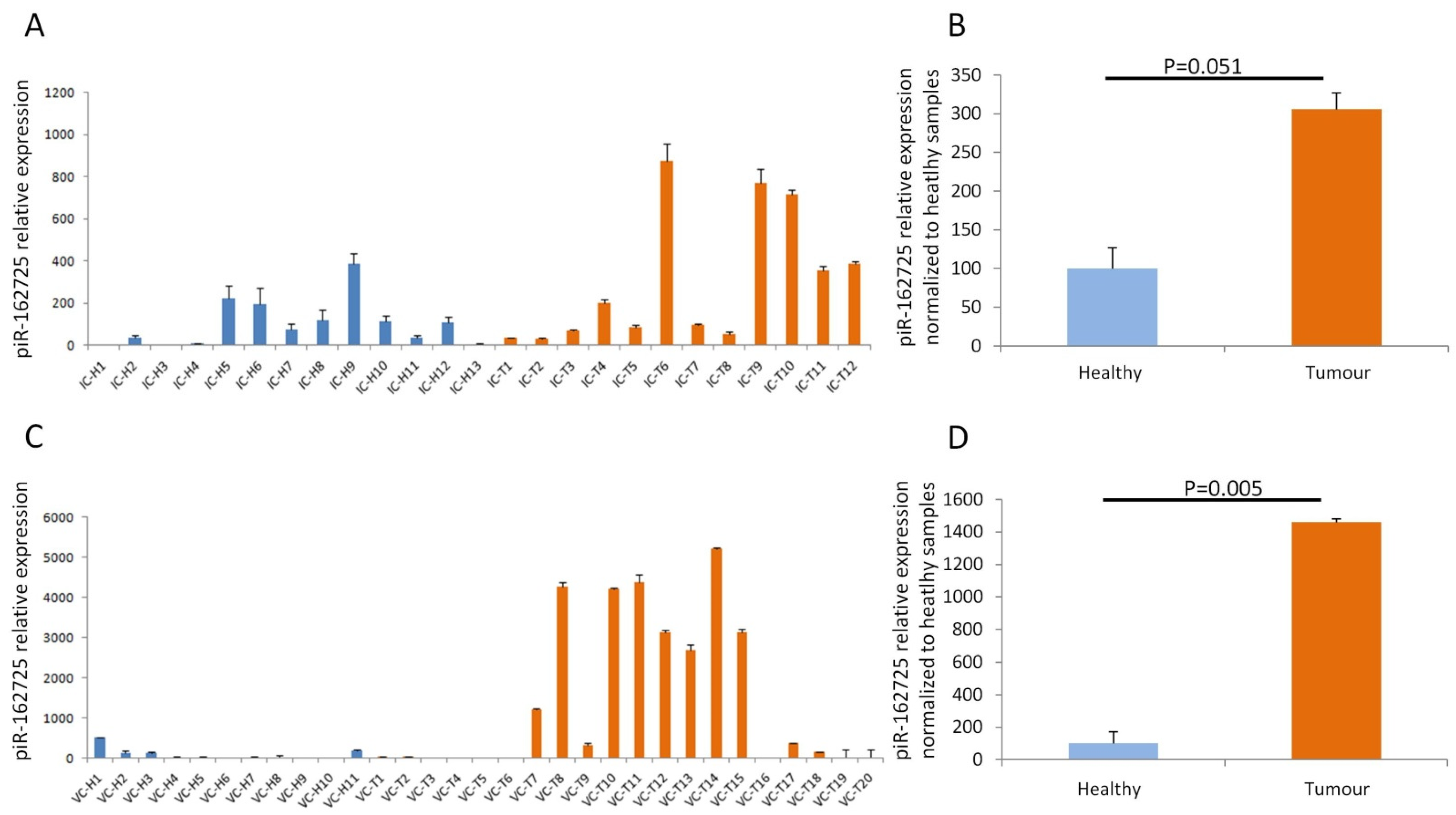
3.2. High Expression of piR-162725 in Human Plasma Samples Differentiates Tumour Samples from Healthy Samples
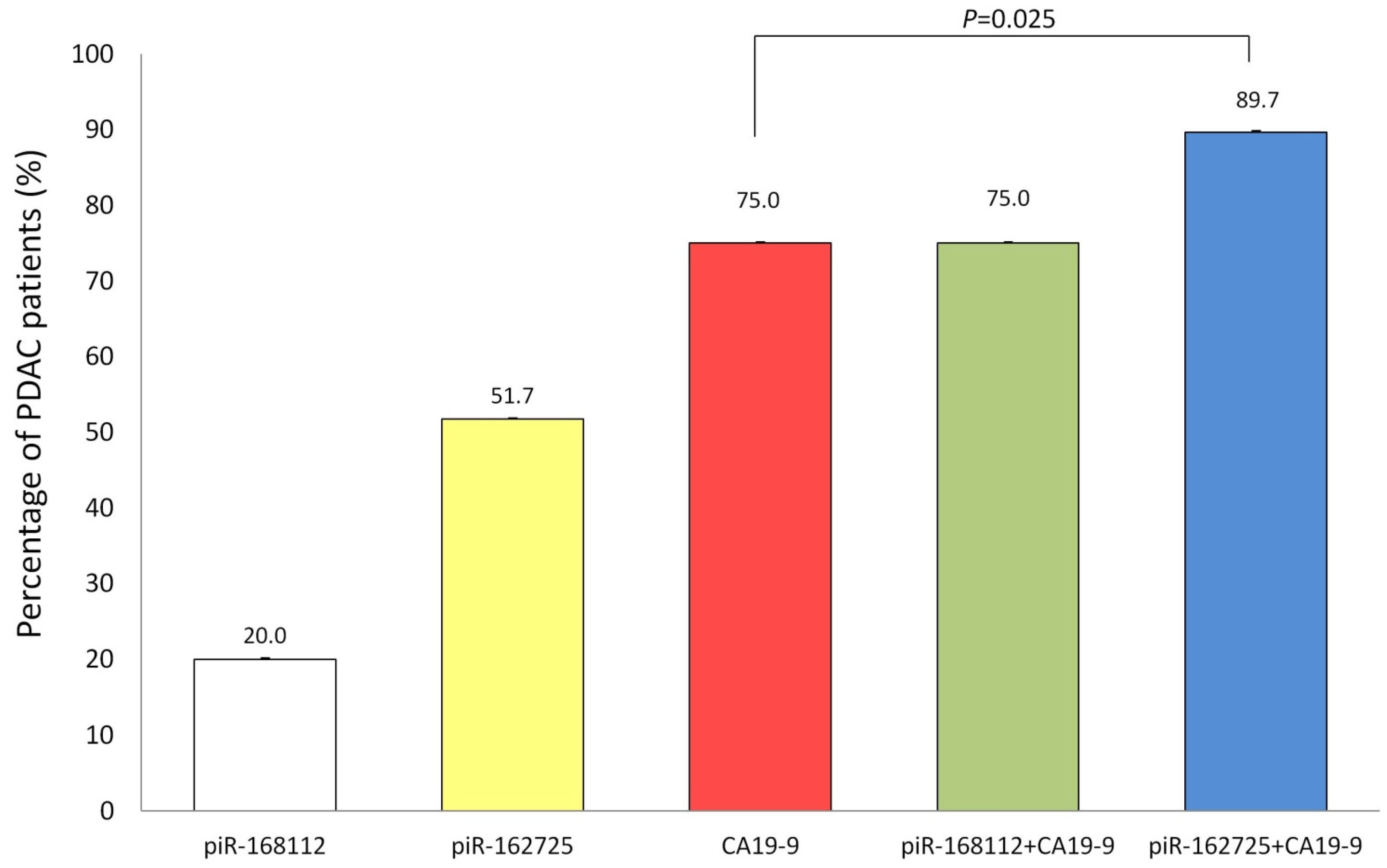
3.3. High Expression of piR-162725 in Human Plasma Samples Increases the Sensitivity of CA19-9 as a Liquid Biopsy Biomarker to Identify PDAC Patients
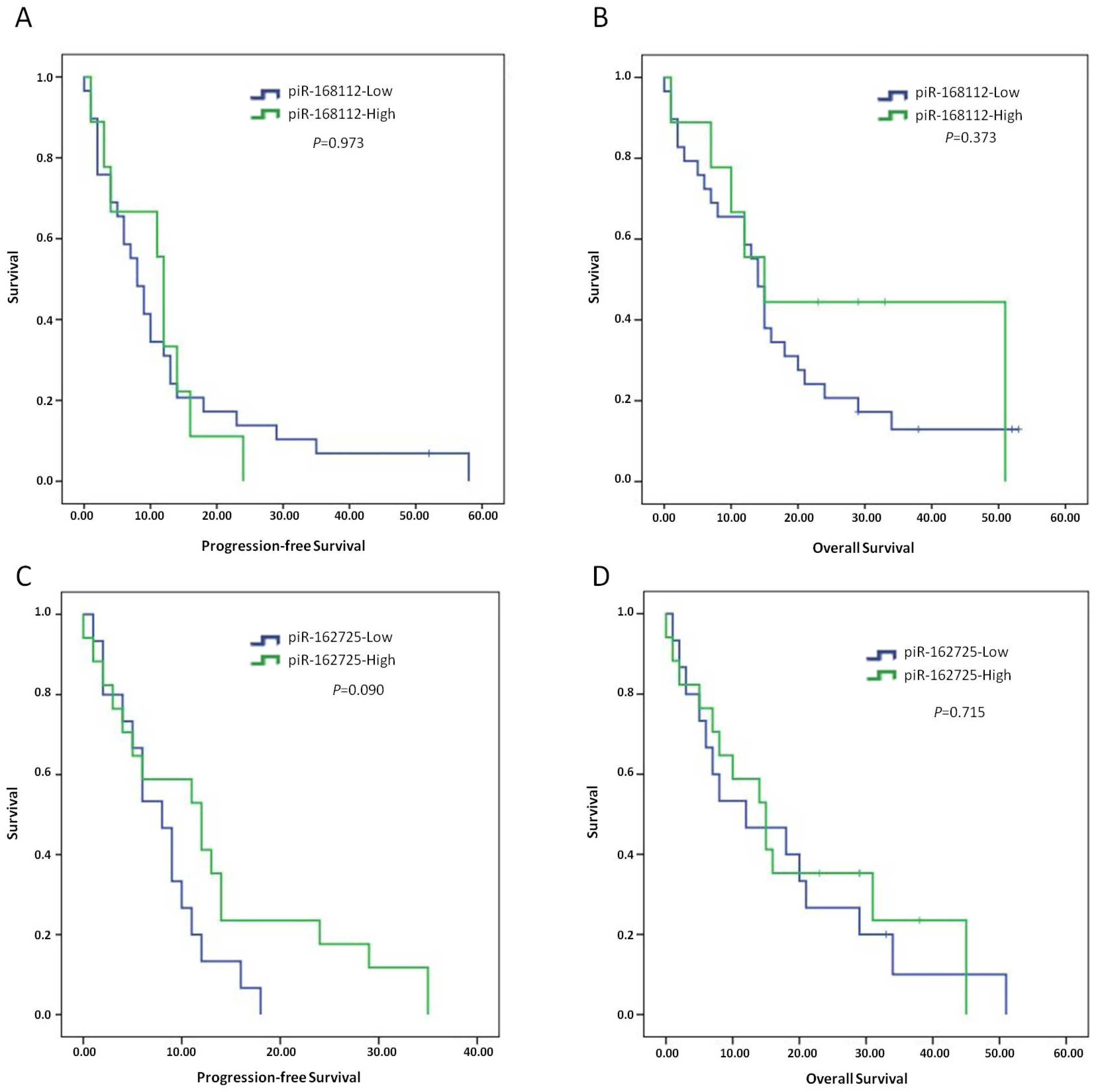
3.4. High Expression of piR-162725 or piR-168112 Is Not Associated with Patient Outcome
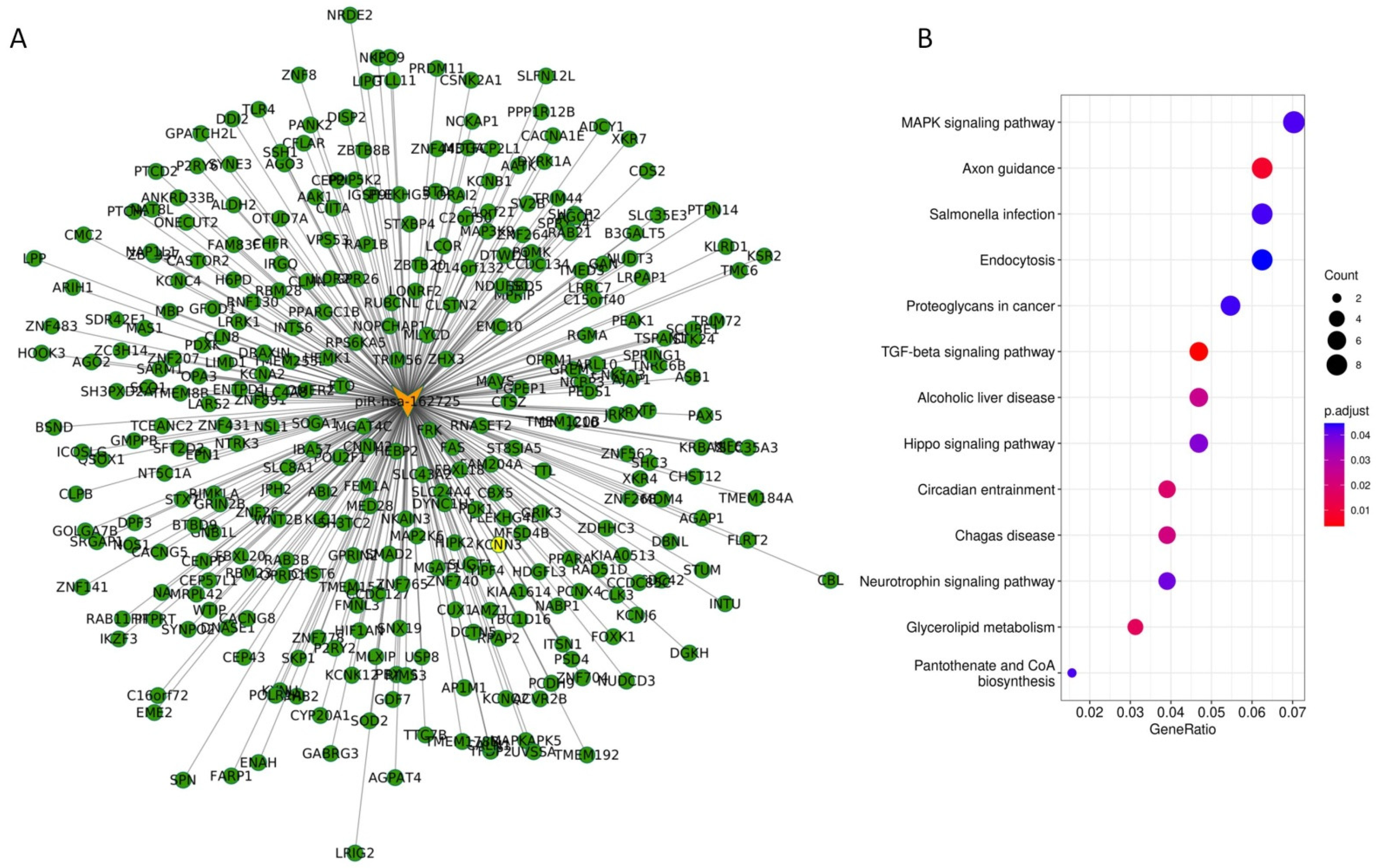
3.5. The Potential piR-162725/PIWIL3 Complex Is an Undercover Modulator of Tumourigenic Signalling Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PDAC | Pancreatic ductal adenocarcinoma |
| PIWI | P-element-induced wimpy testis |
| AGO | Argonaute protein family |
| piRNAs | PIWI-interacting RNAs |
| piRISC | piRNA-induced silencing complex |
| TERT | Telomerase reverse transcriptase |
| WB | Western blot |
| IHC | Immunohistochemistry |
| siRNA | Short interference RNA |
| EMT | Epithelial-to-mesenchymal transition |
| FOLFIRINOX | 5-fluorouracil, leucovorin, irinotecan, and oxaliplatin |
| Nab-Paclitaxel | Nano albumin-bound paclitaxel |
| TCGA | The Cancer Genome Atlas |
| FDA | Food and Drug Administration |
| KEGG | Kyoto Encyclopaedia of Genes and Genomes |
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting Cancer Incidence and Deaths to 2030: The Unexpected Burden of Thyroid, Liver, and Pancreas Cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamm, E.P.; Bhosale, P.R.; Vikram, R.; de Almeida Marcal, L.P.; Balachandran, A. Imaging of Pancreatic Ductal Adenocarcinoma: State of the Art. World J. Radiol 2013, 5, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Useros, J.; Martin-Galan, M.; Garcia-Foncillas, J. The Match between Molecular Subtypes, Histology and Microenvironment of Pancreatic Cancer and Its Relevance for Chemoresistance. Cancers 2021, 13, 322. [Google Scholar] [CrossRef] [PubMed]
- Chari, S.T.; Leibson, C.L.; Rabe, K.G.; Timmons, L.J.; Ransom, J.; de Andrade, M.; Petersen, G.M. Pancreatic Cancer-Associated Diabetes Mellitus: Prevalence and Temporal Association with Diagnosis of Cancer. Gastroenterology 2008, 134, 95–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mavros, M.N.; Moris, D.; Karanicolas, P.J.; Katz, M.H.G.; O’Reilly, E.M.; Pawlik, T.M. Clinical Trials of Systemic Chemotherapy for Resectable Pancreatic Cancer: A Review. JAMA Surg. 2021, 156, 663–672. [Google Scholar] [CrossRef]
- Tempero, M.A.; Reni, M.; Riess, H.; Pelzer, U.; O’Reilly, E.M.; Winter, J.M.; Oh, D.-Y.; Li, C.-P.; Tortora, G.; Chang, H.-M.; et al. APACT: Phase III, Multicenter, International, Open-Label, Randomized Trial of Adjuvant Nab-Paclitaxel plus Gemcitabine (Nab-P/G) vs Gemcitabine (G) for Surgically Resected Pancreatic Adenocarcinoma. JCO 2019, 37, 4000. [Google Scholar] [CrossRef]
- Conroy, T.; Hammel, P.; Hebbar, M.; Ben Abdelghani, M.; Wei, A.C.; Raoul, J.-L.; Choné, L.; Francois, E.; Artru, P.; Biagi, J.J.; et al. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. N. Engl. J. Med. 2018, 379, 2395–2406. [Google Scholar] [CrossRef]
- Zeng, S.; Pöttler, M.; Lan, B.; Grützmann, R.; Pilarsky, C.; Yang, H. Chemoresistance in Pancreatic Cancer. Int. J. Mol. Sci. 2019, 20, 4504. [Google Scholar] [CrossRef] [Green Version]
- Bergquist, J.R.; Ivanics, T.; Shubert, C.R.; Habermann, E.B.; Smoot, R.L.; Kendrick, M.L.; Nagorney, D.M.; Farnell, M.B.; Truty, M.J. Type of Resection (Whipple vs. Distal) Does Not Affect the National Failure to Provide Post-Resection Adjuvant Chemotherapy in Localized Pancreatic Cancer. Ann. Surg. Oncol. 2017, 24, 1731–1738. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugarbaker, P.H.; Stuart, O.A. Intraperitoneal Gemcitabine Chemotherapy Is Safe for Patients with Resected Pancreatic Cancer: Final Clinical and Pharmacologic Data from a Phase II Protocol and Recommended Future Directions. J. Gastrointest. Oncol. 2021, 12, S99–S109. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.-H.; Jang, J.-Y.; Kang, J.S.; Kim, J.R.; Han, Y.; Kim, E.; Kwon, W.; Kim, S.-W. Recent Treatment Patterns and Survival Outcomes in Pancreatic Cancer According to Clinical Stage Based on Single-Center Large-Cohort Data. Ann. Hepatobiliary Pancreat. Surg. 2018, 22, 386–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, C.M.; Kim, J.Y.; Choi, G.H.; Kim, K.S.; Choi, J.S.; Lee, W.J.; Kim, B.R. The Use of Adjusted Preoperative CA 19-9 to Predict the Recurrence of Resectable Pancreatic Cancer. J. Surg. Res. 2007, 140, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Maithel, S.K.; Maloney, S.; Winston, C.; Gönen, M.; D’Angelica, M.I.; Dematteo, R.P.; Jarnagin, W.R.; Brennan, M.F.; Allen, P.J. Preoperative CA 19-9 and the Yield of Staging Laparoscopy in Patients with Radiographically Resectable Pancreatic Adenocarcinoma. Ann. Surg. Oncol. 2008, 15, 3512–3520. [Google Scholar] [CrossRef]
- Kim, J.-E.; Lee, K.T.; Lee, J.K.; Paik, S.W.; Rhee, J.C.; Choi, K.W. Clinical Usefulness of Carbohydrate Antigen 19-9 as a Screening Test for Pancreatic Cancer in an Asymptomatic Population. J. Gastroenterol. Hepatol. 2004, 19, 182–186. [Google Scholar] [CrossRef]
- Kawai, S.; Suzuki, K.; Nishio, K.; Ishida, Y.; Okada, R.; Goto, Y.; Naito, M.; Wakai, K.; Ito, Y.; Hamajima, N. Smoking and Serum CA19-9 Levels According to Lewis and Secretor Genotypes. Int. J. Cancer 2008, 123, 2880–2884. [Google Scholar] [CrossRef]
- Martinez-Useros, J.; Garcia-Foncillas, J. Can Molecular Biomarkers Change the Paradigm of Pancreatic Cancer Prognosis? Biomed. Res. Int. 2016, 2016, 4873089. [Google Scholar] [CrossRef] [Green Version]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. PIWI-Interacting Small RNAs: The Vanguard of Genome Defence. Nat. Rev. Mol. Cell Biol 2011, 12, 246–258. [Google Scholar] [CrossRef]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef]
- Tan, L.; Mai, D.; Zhang, B.; Jiang, X.; Zhang, J.; Bai, R.; Ye, Y.; Li, M.; Pan, L.; Su, J.; et al. PIWI-Interacting RNA-36712 Restrains Breast Cancer Progression and Chemoresistance by Interaction with SEPW1 Pseudogene SEPW1P RNA. Mol. Cancer 2019, 18, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, J.; Deng, H.; Xiao, B.; Zhou, H.; Zhou, F.; Shen, Z.; Guo, J. PiR-823, a Novel Non-Coding Small RNA, Demonstrates in Vitro and in Vivo Tumor Suppressive Activity in Human Gastric Cancer Cells. Cancer Lett. 2012, 315, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Jiang, X.-Y.; Qi, W.; Ji, C.-G.; Xie, X.-L.; Zhang, D.-X.; Cui, Z.-J.; Wang, C.-K.; Bai, Y.; Wang, J.; et al. PiR-823 Contributes to Colorectal Tumorigenesis by Enhancing the Transcriptional Activity of HSF1. Cancer Sci. 2017, 108, 1746–1756. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Luo, Y.; Gao, Y.; Yang, Y.; Wang, Y.; Xu, Y.; Tan, S.; Zhang, Y.; Duan, J.; Yang, Y. PiR-651 Promotes Tumor Formation in Non-Small Cell Lung Carcinoma through the Upregulation of Cyclin D1 and CDK4. Int. J. Mol. Med. 2016, 38, 927–936. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, T.; Shiohama, A.; Minoshima, S.; Shimizu, N. Identification of Eight Members of the Argonaute Family in the Human Genome. Genomics 2003, 82, 323–330. [Google Scholar] [CrossRef]
- Tan, Y.; Liu, L.; Liao, M.; Zhang, C.; Hu, S.; Zou, M.; Gu, M.; Li, X. Emerging Roles for PIWI Proteins in Cancer. Acta Biochim. Biophys. Sin. 2015, 47, 315–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.-N.; Li, Y.; Xia, S.-Q.; Zhang, Y.-Y.; Zheng, J.-H.; Li, W. PIWI Proteins and PIWI-Interacting RNA: Emerging Roles in Cancer. Cell. Physiol. Biochem. 2017, 44, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Martinez-Useros, J.; Garcia-Carbonero, N.; Fernandez-Aceñero, M.J.; Ortega-Medina, L.; Garcia-Botella, S.; Perez-Aguirre, E.; Diez-Valladares, L.; Garcia-Foncillas, J. The Prognosis Value of PIWIL1 and PIWIL2 Expression in Pancreatic Cancer. JCM 2019, 8, 1275. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Martinez-Useros, J.; Garcia-Carbonero, N.; Fernandez-Aceñero, M.J.; Orta, A.; Ortega-Medina, L.; Garcia-Botella, S.; Perez-Aguirre, E.; Diez-Valladares, L.; Celdran, A.; et al. The Clinical Significance of PIWIL3 and PIWIL4 Expression in Pancreatic Cancer. J. Clin. Med. 2020, 9, 1252. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the CBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The CBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Kim, T.-K.; Baxter, D.; Scherler, K.; Gordon, A.; Fong, O.; Etheridge, A.; Galas, D.J.; Wang, K. SRNAnalyzer-a Flexible and Customizable Small RNA Sequencing Data Analysis Pipeline. Nucleic Acids Res. 2017, 45, 12140–12151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, P3. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R Package for Comparing Biological Themes among Gene Clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Ku, H.-Y.; Lin, H. PIWI Proteins and Their Interactors in PiRNA Biogenesis, Germline Development and Gene Expression. Natl. Sci. Rev. 2014, 1, 205–218. [Google Scholar] [CrossRef]
- Kishikawa, T.; Otsuka, M.; Ohno, M.; Yoshikawa, T.; Takata, A.; Koike, K. Circulating RNAs as New Biomarkers for Detecting Pancreatic Cancer. World J. Gastroenterol. 2015, 21, 8527–8540. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA Therapeutics—Challenges and Potential Solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Sole, C.; Arnaiz, E.; Manterola, L.; Otaegui, D.; Lawrie, C.H. The Circulating Transcriptome as a Source of Cancer Liquid Biopsy Biomarkers. Semin. Cancer Biol. 2019, 58, 100–108. [Google Scholar] [CrossRef]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of Human Plasma-Derived Exosomal RNAs by Deep Sequencing. BMC Genom. 2013, 14, 319. [Google Scholar] [CrossRef]
- Yuan, T.; Huang, X.; Woodcock, M.; Du, M.; Dittmar, R.; Wang, Y.; Tsai, S.; Kohli, M.; Boardman, L.; Patel, T.; et al. Plasma Extracellular RNA Profiles in Healthy and Cancer Patients. Sci. Rep. 2016, 6, 19413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.R.; Kimchi, E.T.; Manjunath, Y.; Gajagowni, S.; Stuckel, A.J.; Kaifi, J.T. Author Correction: RNA Cargos in Extracellular Vesicles Derived from Blood Serum in Pancreas Associated Conditions. Sci. Rep. 2020, 10, 9981. [Google Scholar] [CrossRef] [PubMed]
- Müller, S.; Raulefs, S.; Bruns, P.; Afonso-Grunz, F.; Plötner, A.; Thermann, R.; Jäger, C.; Schlitter, A.M.; Kong, B.; Regel, I.; et al. Erratum to: Next-Generation Sequencing Reveals Novel Differentially Regulated MRNAs, LncRNAs, MiRNAs, SdRNAs and a PiRNA in Pancreatic Cancer. Mol. Cancer 2015, 14, 144. [Google Scholar] [CrossRef] [Green Version]
- Xie, J.; Xing, S.; Shen, B.-Y.; Chen, H.-T.; Sun, B.; Wang, Z.-T.; Wang, J.-W.; Lu, X.-X. PIWIL1 Interacting RNA PiR-017061 Inhibits Pancreatic Cancer Growth via Regulating EFNA5. Hum. Cell 2021, 34, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Xin, J.; Du, M.; Jiang, X.; Wu, Y.; Ben, S.; Zheng, R.; Chu, H.; Li, S.; Zhang, Z.; Wang, M. Systematic Evaluation of the Effects of Genetic Variants on PIWI-Interacting RNA Expression across 33 Cancer Types. Nucleic Acids Res. 2021, 49, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Goonetilleke, K.S.; Siriwardena, A.K. Systematic Review of Carbohydrate Antigen (CA 19-9) as a Biochemical Marker in the Diagnosis of Pancreatic Cancer. Eur. J. Surg. Oncol. 2007, 33, 266–270. [Google Scholar] [CrossRef]
- Satake, K.; Chung, Y.S.; Umeyama, K.; Takeuchi, T.; Kim, Y.S. The Possibility of Diagnosing Small Pancreatic Cancer (Less than 4.0 Cm) by Measuring Various Serum Tumor Markers. A Retrospective Study. Cancer 1991, 68, 149–152. [Google Scholar] [CrossRef]
- Miyata, T.; Hayashi, H.; Yamashita, Y.-I.; Matsumura, K.; Nakao, Y.; Itoyama, R.; Yamao, T.; Tsukamoto, M.; Okabe, H.; Imai, K.; et al. Prognostic Value of the Preoperative Tumor Marker Index in Resected Pancreatic Ductal Adenocarcinoma: A Retrospective Single-Institution Study. Ann. Surg. Oncol. 2021, 28, 1572–1580. [Google Scholar] [CrossRef]
- Ballehaninna, U.K.; Chamberlain, R.S. The Clinical Utility of Serum CA 19-9 in the Diagnosis, Prognosis and Management of Pancreatic Adenocarcinoma: An Evidence Based Appraisal. J. Gastrointest. Oncol. 2012, 3, 105–119. [Google Scholar] [CrossRef]
- Ferri, M.J.; Saez, M.; Figueras, J.; Fort, E.; Sabat, M.; López-Ben, S.; de Llorens, R.; Aleixandre, R.N.; Peracaula, R. Improved Pancreatic Adenocarcinoma Diagnosis in Jaundiced and Non-Jaundiced Pancreatic Adenocarcinoma Patients through the Combination of Routine Clinical Markers Associated to Pancreatic Adenocarcinoma Pathophysiology. PLoS ONE 2016, 11, e0147214. [Google Scholar] [CrossRef]
- Hu, L.-P.; Zhang, X.-X.; Jiang, S.-H.; Tao, L.-Y.; Li, Q.; Zhu, L.-L.; Yang, M.-W.; Huo, Y.-M.; Jiang, Y.-S.; Tian, G.-A.; et al. Targeting Purinergic Receptor P2Y2 Prevents the Growth of Pancreatic Ductal Adenocarcinoma by Inhibiting Cancer Cell Glycolysis. Clin. Cancer Res. 2019, 25, 1318–1330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Misir, S.; Hepokur, C.; Aliyazicioglu, Y.; Enguita, F.J. Biomarker Potentials of MiRNA-Associated CircRNAs in Breast Cancer (MCF-7) Cells: An in Vitro and in Silico Study. Mol. Biol. Rep. 2021, 48, 2463–2471. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Song, X.; Wang, Y.; Fu, S.; Han, P. Genome-Wide Analysis of the Expression of Circular RNA Full-Length Transcripts and Construction of the CircRNA-MiRNA-MRNA Network in Cervical Cancer. Front. Cell Dev. Biol. 2020, 8, 603516. [Google Scholar] [CrossRef] [PubMed]
| Univariate PFS (95% CI) | Univariate OS (95% CI) | |||||||
|---|---|---|---|---|---|---|---|---|
| HR | Lower | Upper | p | HR | Lower | Upper | p | |
| Gender (Female vs. Male) | 1.252 | 0.637 | 2.460 | 0.514 | 1.376 | 0.656 | 2.889 | 0.398 |
| Stage (I/II vs. III/IV) | 2.467 | 1.137 | 5.353 | 0.022 | 5.545 | 1.909 | 16.109 | 0.002 |
| pT (0–I vs. II–III) | 1.417 | 0.641 | 3.131 | 0.389 | 3.302 | 1.134 | 9.616 | 0.029 |
| pN (N0 vs. N1/N2/N3) | 1.498 | 0.612 | 3.668 | 0.377 | 1.151 | 0.462 | 2.867 | 0.763 |
| pM (M0 vs. M1) | 2.062 | 1.047 | 4.061 | 0.036 | 5.840 | 2.386 | 14.298 | 0.001 |
| Alcohol (No vs. Yes) | 1.331 | 0.621 | 2.851 | 0.462 | 1.519 | 0.650 | 3.553 | 0.334 |
| Smoker (Yes vs. No) | 1.371 | 0.702 | 2.678 | 0.335 | 1.782 | 0.860 | 3.693 | 0.120 |
| Diabetes (No vs. Yes) | 1.278 | 0.638 | 2.560 | 0.489 | 1.343 | 0.637 | 2.830 | 0.439 |
| AHT (No vs. Yes) | 1.292 | 0.444 | 3.759 | 0.638 | 1.314 | 0.338 | 5.111 | 0.694 |
| Multivariate PFS (95% CI) | Multivariate OS (95% CI) | |||||||
| Stage (I/II vs. III/IV) | 1.943 | 0.658 | 5.738 | 0.229 | 10.512 | 0.373 | 296.493 | 0.167 |
| pM (M0 vs. M1) | 1.322 | 0.515 | 3.393 | 0.562 | 3.391 | 1.189 | 9.673 | 0.022 |
| pT (0–I vs. II–III) | - | - | - | - | 4.995 | 0.191 | 130.586 | 0.334 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Gonzalez-Gonzalez, M.; Sanz-Criado, L.; Garcia-Carbonero, N.; Celdran, A.; Villarejo-Campos, P.; Minguez, P.; Pazo-Cid, R.; Garcia-Jimenez, C.; Orta-Ruiz, A.; et al. A Novel PiRNA Enhances CA19-9 Sensitivity for Pancreatic Cancer Identification by Liquid Biopsy. J. Clin. Med. 2022, 11, 7310. https://doi.org/10.3390/jcm11247310
Li W, Gonzalez-Gonzalez M, Sanz-Criado L, Garcia-Carbonero N, Celdran A, Villarejo-Campos P, Minguez P, Pazo-Cid R, Garcia-Jimenez C, Orta-Ruiz A, et al. A Novel PiRNA Enhances CA19-9 Sensitivity for Pancreatic Cancer Identification by Liquid Biopsy. Journal of Clinical Medicine. 2022; 11(24):7310. https://doi.org/10.3390/jcm11247310
Chicago/Turabian StyleLi, Weiyao, Miguel Gonzalez-Gonzalez, Lara Sanz-Criado, Nuria Garcia-Carbonero, Angel Celdran, Pedro Villarejo-Campos, Pablo Minguez, Roberto Pazo-Cid, Custodia Garcia-Jimenez, Alberto Orta-Ruiz, and et al. 2022. "A Novel PiRNA Enhances CA19-9 Sensitivity for Pancreatic Cancer Identification by Liquid Biopsy" Journal of Clinical Medicine 11, no. 24: 7310. https://doi.org/10.3390/jcm11247310
APA StyleLi, W., Gonzalez-Gonzalez, M., Sanz-Criado, L., Garcia-Carbonero, N., Celdran, A., Villarejo-Campos, P., Minguez, P., Pazo-Cid, R., Garcia-Jimenez, C., Orta-Ruiz, A., Garcia-Foncillas, J., & Martinez-Useros, J. (2022). A Novel PiRNA Enhances CA19-9 Sensitivity for Pancreatic Cancer Identification by Liquid Biopsy. Journal of Clinical Medicine, 11(24), 7310. https://doi.org/10.3390/jcm11247310









