Identification, Structure and Characterization of Bacillus tequilensis Biofilm with the Use of Electrophoresis and Complementary Approaches
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Molecular Identification of Bacterial Biofilm Formation
2.3. Capillary Zone Electrophoresis (CZE) Analysis
2.4. Zeta Potential Assay
2.5. MALDI-TOF MS Analysis
2.6. Fluorescence Microscopy Assay
2.7. Scanning Electron Microscopy Assay
3. Results
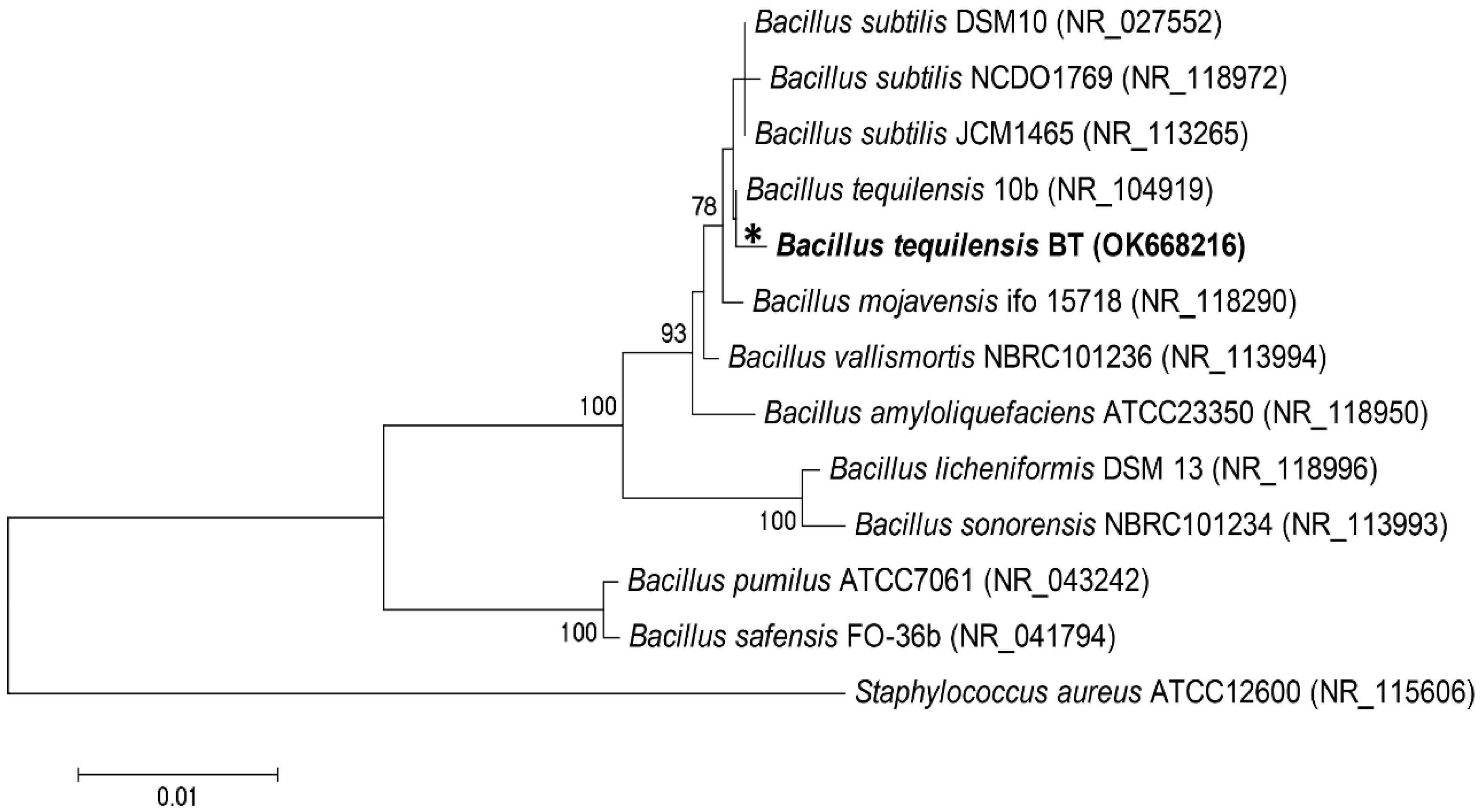
3.1. Molecular Identification of Bacteria Biofilm Formation
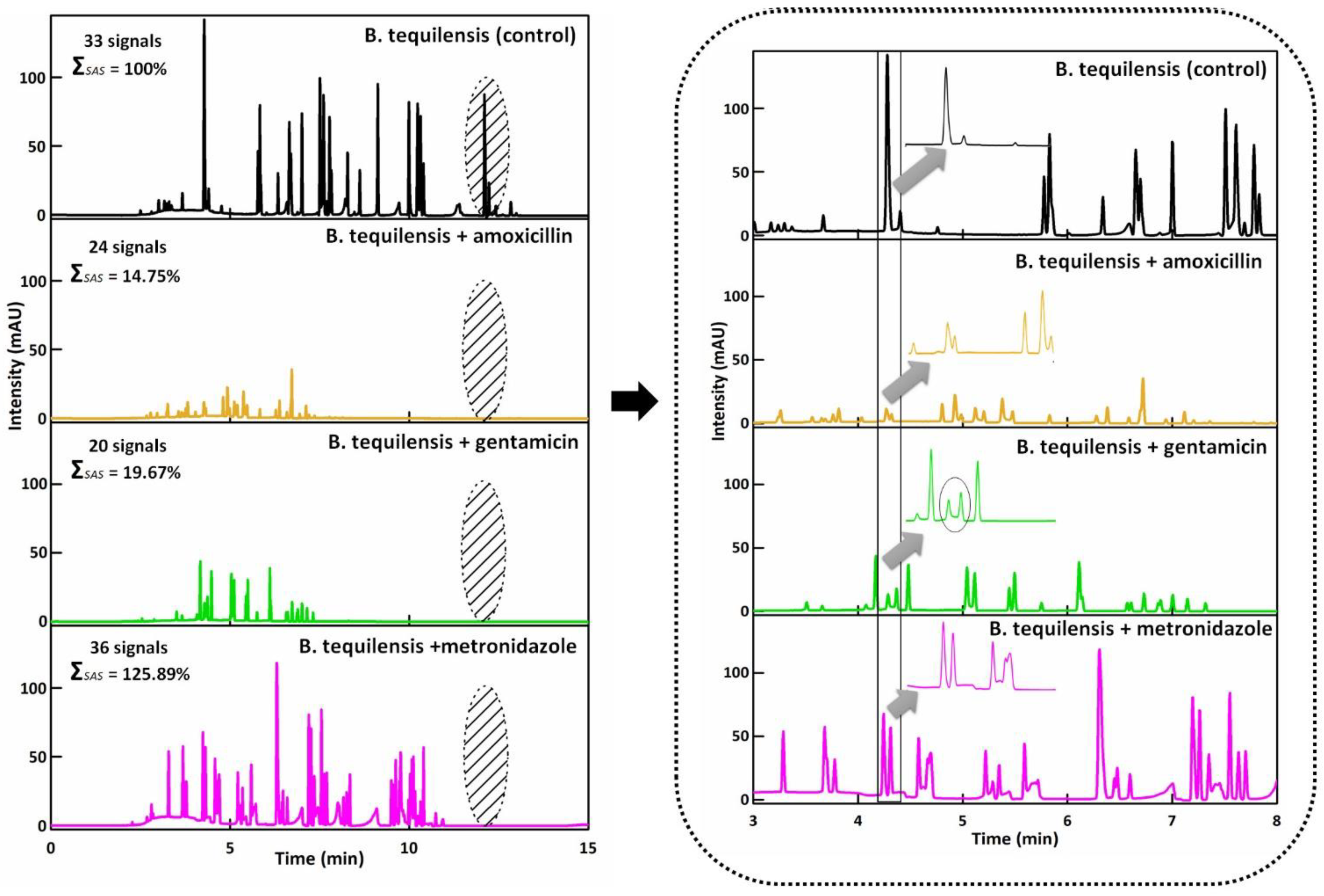
3.2. CZE Assay
3.3. Zeta Potential Analysis
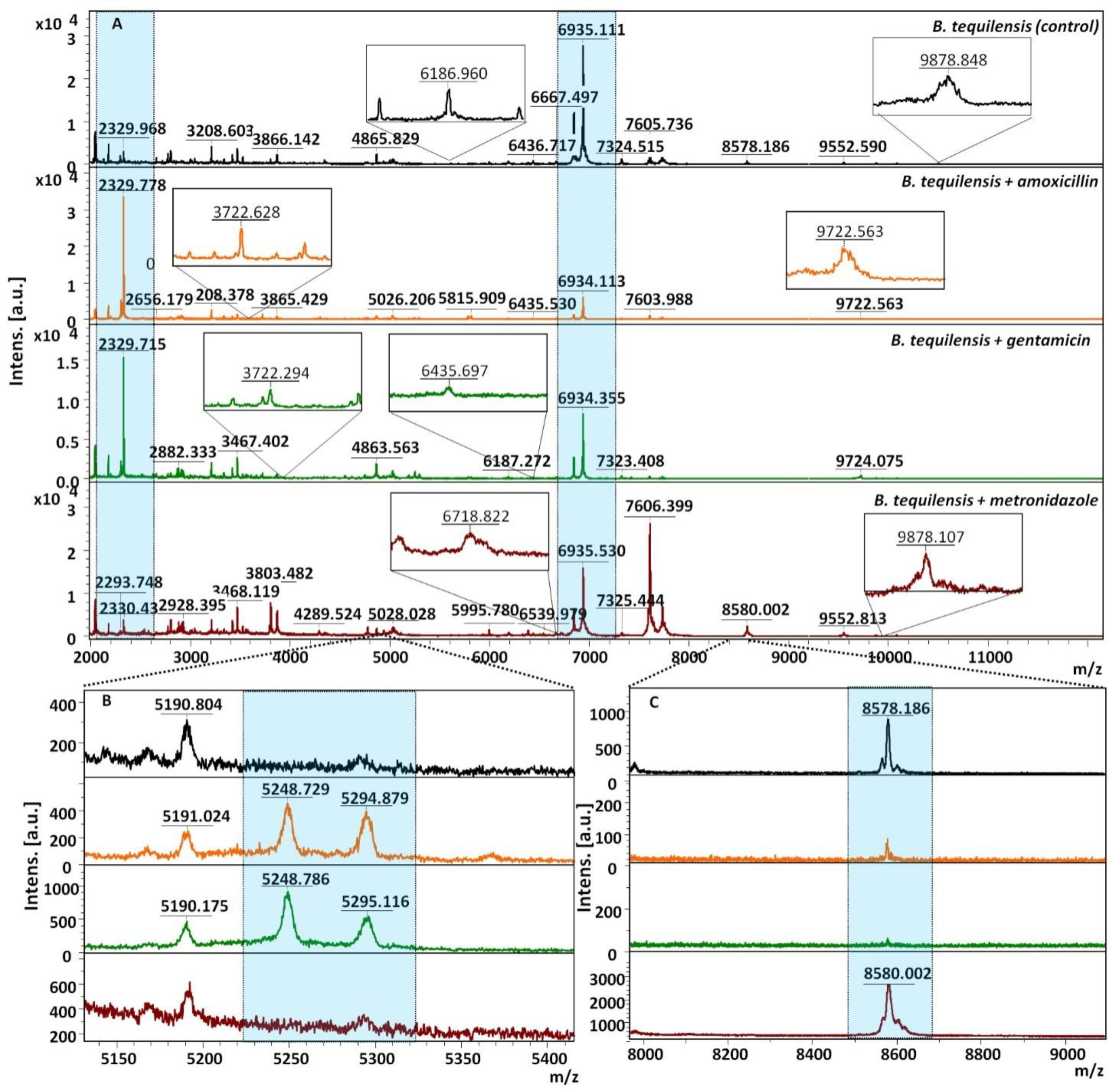
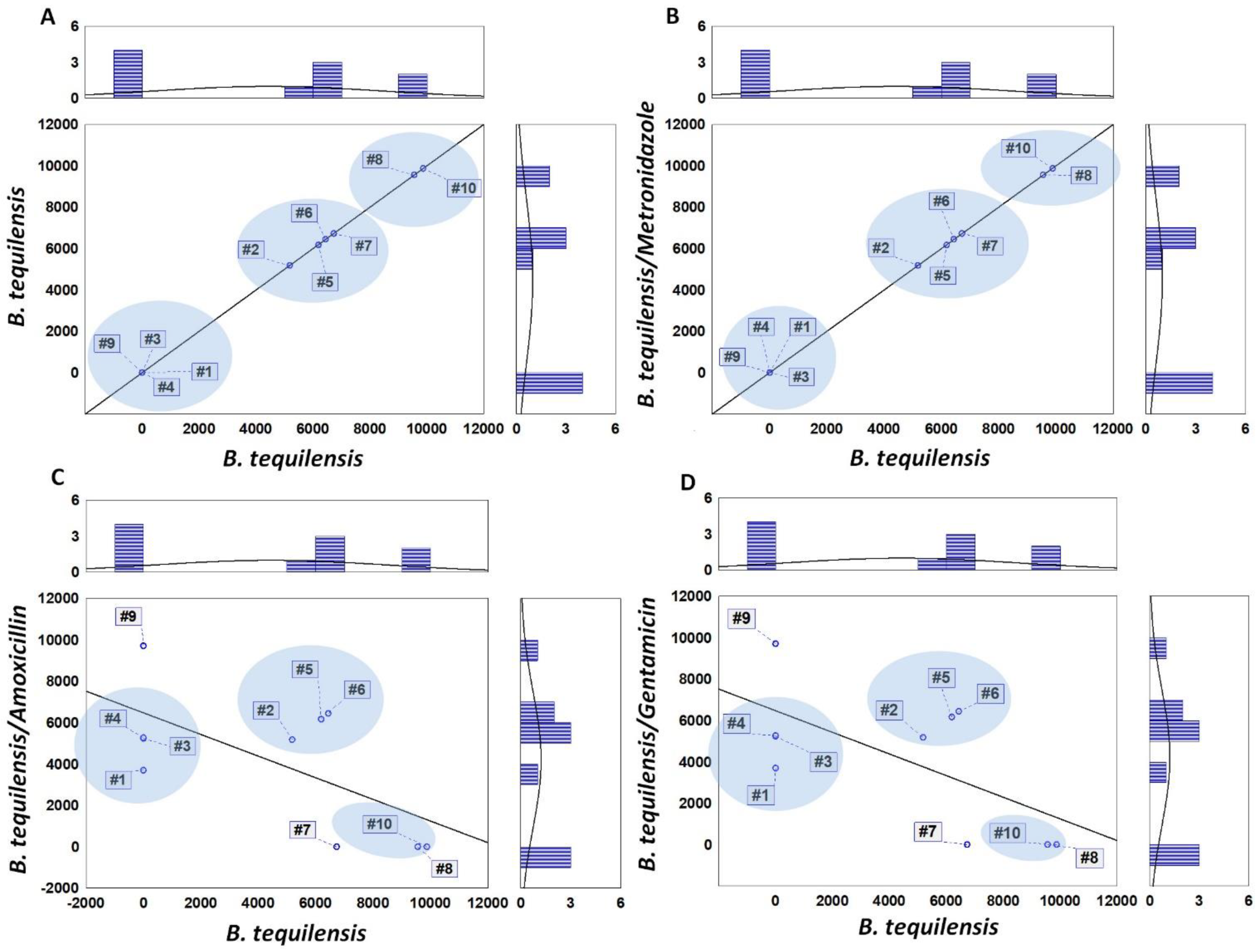
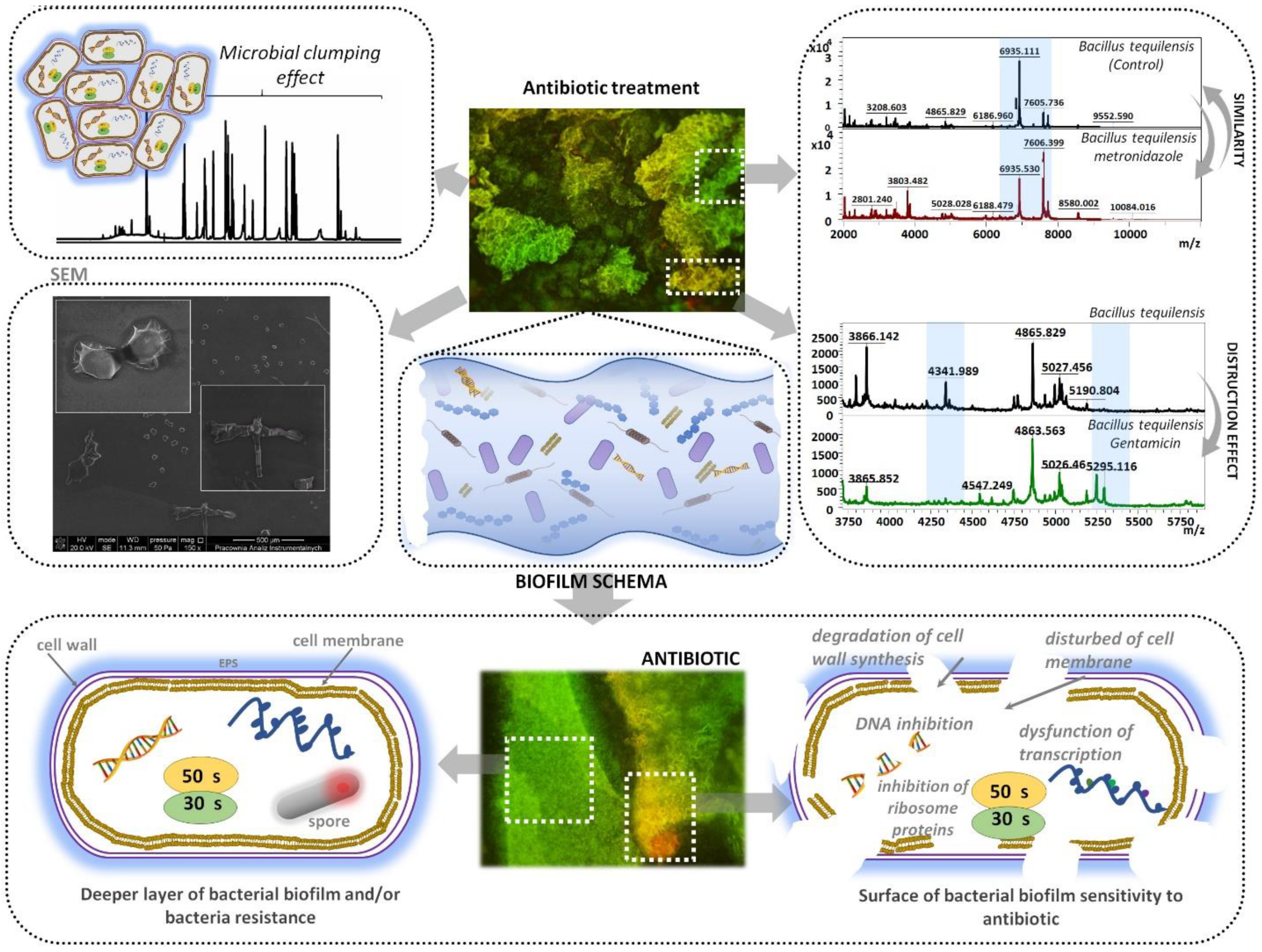
3.4. MALDI-TOF MS Analysis
3.5. Fluorescence Microscopy
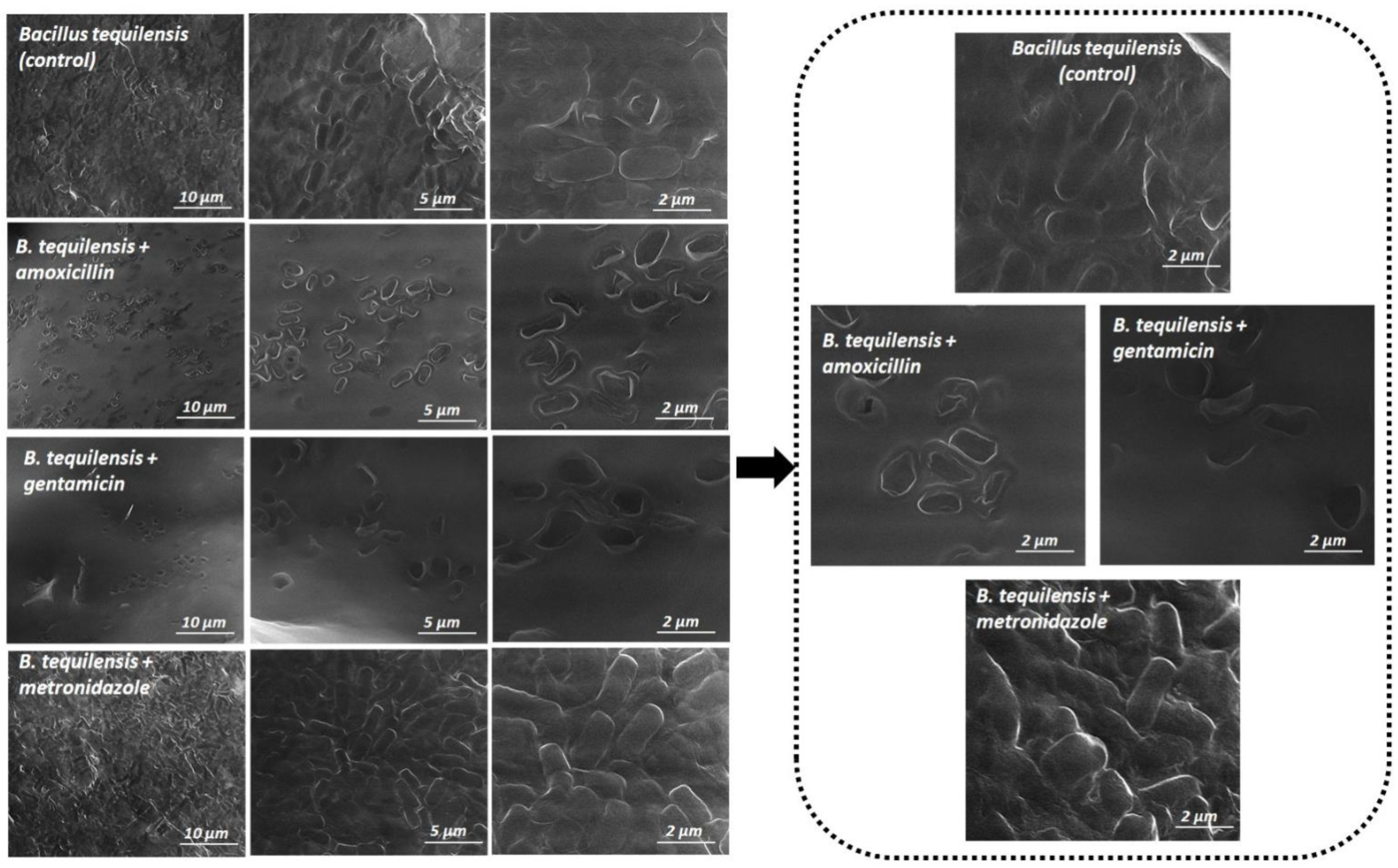
3.6. Scanning Electron Microscopy
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Van Merode, A.E.J.; Van Der Mei, H.C.; Busscher, H.J.; Krom, B.P. Influence of culture heterogeneity in cell surface charge on adhesion and biofilm formation by Enterococcus faecalis. J. Bacteriol. 2006, 188, 2421–2426. [Google Scholar] [CrossRef] [Green Version]
- Sharma, D.; Misba, L.; Khan, A. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 2019, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Flint, S.H.; Palmer, J.S. Bacillus cereus spores and toxins—The potential role of biofilms. Food Microbiol. 2020, 90, 103493. [Google Scholar] [CrossRef]
- Navaneethan, Y.; Effarizah, M.E. Prevalence, toxigenic profiles, multidrug resistance, and biofilm formation of Bacillus cereus isolated from ready-to eat cooked rice in Penang, Malaysia. Food Control 2021, 121, 107553. [Google Scholar] [CrossRef]
- Ruzicka, F.; Horka, M.; Hola, V. Extracellular Polysaccharides in Microbial Biofilm and Their Influence on the Electrophoretic Properties of Microbial Cells. In Capillary Electrophoresis of Carbohydrates; Volpi, N., Ed.; Humana Press: New York, NY, USA, 2011; pp. 105–126. [Google Scholar]
- Peng, N.; Cai, P.; Mortimer, M.; Wu, Y.; Gao, C.; Huang, Q. The exopolysaccharide-eDNA interaction modulates 3D architecture of Bacillus subtilis biofilm. BMC Microbiol. 2020, 20, 115. [Google Scholar] [CrossRef]
- Denkhaus, E.; Meisen, S.; Telgheder, U.; Wingender, J. Chemical and physical methods for characterisation of biofilms. Microchim. Acta 2007, 158, 1–27. [Google Scholar] [CrossRef]
- Wang, L.; Li, Y.; Wang, L.; Zhu, M.; Zhu, X.; Qian, C.; Li, W. Responses of biofilm microorganisms from moving bed biofilm reactor to antibiotics exposure: Protective role of extracellular polymeric substances. Bioresour. Technol. 2018, 254, 268–277. [Google Scholar] [CrossRef]
- Ashrafudoulla, M.; Na, K.W.; Hossain, M.I.; Mizan, M.F.R.; Nahar, S.; Toushik, S.H.; Roy, P.K.; Park, S.H.; Ha, S. Do Molecular and pathogenic characterization of Vibrio parahaemolyticus isolated from seafood. Mar. Pollut. Bull. 2021, 172, 112927. [Google Scholar] [CrossRef]
- Roy, P.K.; Mizan, M.F.R.; Hossain, M.I.; Han, N.; Nahar, S.; Ashrafudoulla, M.; Toushik, S.H.; Shim, W.B.; Kim, Y.M.; Ha, S. Do Elimination of Vibrio parahaemolyticus biofilms on crab and shrimp surfaces using ultraviolet C irradiation coupled with sodium hypochlorite and slightly acidic electrolyzed water. Food Control 2021, 128, 108179. [Google Scholar] [CrossRef]
- Roy, P.K.; Ha, A.J.W.; Mizan, M.F.R.; Hossain, M.I.; Ashrafudoulla, M.; Toushik, S.H.; Nahar, S.; Kim, Y.K.; Ha, S. Do Effects of environmental conditions (temperature, pH, and glucose) on biofilm formation of Salmonella enterica serotype Kentucky and virulence gene expression. Poult. Sci. 2021, 100, 101209. [Google Scholar] [CrossRef] [PubMed]
- Relucenti, M.; Familiari, G.; Donfrancesco, O.; Taurino, M.; Li, X.; Chen, R.; Artini, M.; Papa, R.; Selan, L. Microscopy methods for biofilm imaging: Focus on sem and VP-SEM pros and cons. Biology 2021, 10, 51. [Google Scholar] [CrossRef] [PubMed]
- Dziubakiewicz, E.; Buszewski, B. Capillary electrophoresis of microbial aggregates. Electrophoresis 2014, 35, 1160–1164. [Google Scholar] [CrossRef] [PubMed]
- Rogowska, A.; Pomastowski, P.; Złoch, M.; Railean-Plugaru, V.; Król, A.; Rafińska, K.; Szultka-Młyńska, M.; Buszewski, B. The influence of different pH on the electrophoretic behaviour of Saccharomyces cerevisiae modified by calcium ions. Sci. Rep. 2018, 8, 2–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruzicka, F.; Horka, M.; Hola, V.; Votava, M. Capillary Isoelectric Focusing—Useful tool for detection of the biofilm formation in Staphylococcus epidermidis. J. Microbiol. Methods 2007, 68, 530–535. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Comi, T.J.; Si, T.; Dunham, S.J.B.; Sweedler, J.V. A one-step matrix application method for MALDI mass spectrometry imaging of bacterial colony biofilms. J. Mass Spectrom. 2016, 51, 1030–1035. [Google Scholar] [CrossRef] [PubMed]
- Pomastowski, P.; Złoch, M.; Rodzik, A.; Ligor, M.; Kostrzewa, M.; Buszewski, B. Analysis of bacteria associated with honeys of different geographical and botanical origin using two different identification approaches: MALDI-TOF MS and 16S rDNA PCR technique. PLoS ONE 2019, 14, e0217078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gatson, J.W.; Benz, B.F.; Chandrasekaran, C.; Satomi, M.; Venkateswaran, K.; Hart, M.E. Bacillus tequilensis sp. nov., isolated from a 2000-year-old Mexican shaft-tomb, is closely related to Bacillus subtilis. Int. J. Syst. Evol. Microbiol. 2006, 56, 1475–1484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters—2021, Version 11.0; 2021. Available online: http://www.eucast.org (accessed on 9 December 2021).
- Buszewski, B.; Rogowska, A.; Railean-Plugaru, V.; Złoch, M.; Walczak-Skierska, J.; Pomastowski, P. The influence of different forms of silver on selected pathogenic bacteria. Materials 2020, 13, 2403. [Google Scholar] [CrossRef]
- Buszewski, B.; Król, A.; Pomastowski, P.; Railean-Plugaru, V.; Szultka-Młyńska, M. Electrophoretic Determination of Lactococcus lactis Modified by Zinc Ions. Chromatographia 2019, 82, 347–355. [Google Scholar] [CrossRef] [Green Version]
- Saitou, N.; Nei, M. The Neighbor-joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Lemon, K.P.; Earl, A.M.; Vlamakis, H.C.; Aguilar, C.; Kolter, R. Biofilm development with emphasis on Bacillus subtilis. In Current Topics in Microbial Immunity; Romeo, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; Volume 322, pp. 1–16. [Google Scholar]
- Kłodzińska, E.; Jaworski, M.; Kupczyk, W.; Jackowski, M.; Buszewski, B. A study of interactions between bacteria and antibiotics by capillary electrophoresis. Electrophoresis 2012, 33, 3095–3100. [Google Scholar] [CrossRef] [PubMed]
- Kłodzińska, E.; Szumski, M.; Dziubakiewicz, E.; Hrynkiewicz, K.; Skwarek, E.; Janusz, W.; Buszewski, B. Effect of zeta potential value on bacterial behavior during electrophoretic separation. Electrophoresis 2010, 31, 1590–1596. [Google Scholar] [CrossRef]
- Poortinga, A.; Bos, R.; Norde, W.; Busscher, H. Electric double layer interactions in bacterial adhesion to surfaces. Surf. Sci. Rep. 2002, 47, 1–32. [Google Scholar] [CrossRef]
- Buszewski, B.; Maślak, E.; Złoch, M.; Railean-Plugaru, V.; Kłodzińska, E.; Pomastowski, P. A new approach to identifying pathogens, with particular regard to viruses, based on capillary electrophoresis and other analytical techniques. TrAC-Trends Anal. Chem. 2021, 139, 116250. [Google Scholar] [CrossRef]
- Thornsberry, C.; Anhalt, J.P.; Washington, J.A. Clinical laboratory evaluation of the Abbott MS-2 Automated Antimicrobial susceptibility testing system: Report of a collaborative study. J. Clin. Microbiol. 1980, 12, 375–390. [Google Scholar] [CrossRef] [Green Version]
- Luis, D.; Guadix, J.A.; Mar, A.; Doan, H.K.; Leveau, J.H.J.; Romero, D. Sporulation is dispensable for the vegetable-associated life cycle of the human pathogen Bacillus cereus. Microb. Biotechnol. 2021, 14, 1550–1565. [Google Scholar] [CrossRef]
- Kruft, V.; Kapp, U.; Wittmann-Liebold, B. Max-Pianck-institut Characterization and primary structure of proteins L28, L33 and L34 from Bacillus stearothermophilus ribosomes. Biochimie 1991, 73, 855–860. [Google Scholar] [CrossRef]
- Abee, T.; Groot, M.N.; Tempelaars, M.; Zwietering, M.; Moezelaar, R.; Voort, M. van der Germination and outgrowth of spores of Bacillus cereus group members: Diversity and role of germinant receptors. Food Microbiol. 2011, 28, 199–208. [Google Scholar] [CrossRef]
- Pereira, F.D.E.S.; Bonatto, C.C.; Lopes, C.A.P.; Pereira, A.L.; Silva, L.P. Use of MALDI-TOF mass spectrometry to analyze the molecular profile of Pseudomonas aeruginosa biofilms grown on glass and plastic surfaces. Microb. Pathog. 2015, 86, 32–37. [Google Scholar] [CrossRef] [Green Version]
- Si, T.; Li, B.; Zhang, K.; Xu, Y.; Zhao, H.; Sweedler, J.V. Characterization of bacillus subtilis colony biofilms via mass spectrometry and fluorescence imaging. J. Proteome Res. 2016, 15, 1955–1962. [Google Scholar] [CrossRef] [PubMed]
- Tseng, B.S.; Zhang, W.; Harrison, J.J.; Quach, T.P.; Song, J.L.; Penterman, J.; Singh, P.K.; Chopp, D.L.; Packman, A.I.; Parsek, M.R. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ. Microbiol. 2013, 15, 2865–2878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daddi Oubekka, S.; Briandet, R.; Fontaine-Aupart, M.P.; Steenkeste, K. Correlative time-resolved fluorescence microscopy to assess antibiotic diffusion-reaction in biofilms. Antimicrob. Agents Chemother. 2012, 56, 3349–3358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, G.A.; Chesnel, L.; Boegli, L.; de Lancey Pulcini, E.; Fisher, S.; Stewart, P.S. Analysis of Clostridium difficile biofilms: Imaging and antimicrobial treatment. J. Antimicrob. Chemother. 2018, 73, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Anderl, J.N.; Franklin, M.J.; Stewart, P.S. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 2000, 44, 1818–1824. [Google Scholar] [CrossRef] [Green Version]
- Dincer, S.; Masume Uslu, F.; Delik, A. Antibiotic resistance in biofilm. In Bacterial Biofilms; Dincer, S., Sümengen Özdenefe, M., Arkut, A., Eds.; IntechOpen: London, UK, 2020. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pauter, K.; Railean-Plugaru, V.; Złoch, M.; Pomastowski, P.; Szultka-Młyńska, M.; Buszewski, B. Identification, Structure and Characterization of Bacillus tequilensis Biofilm with the Use of Electrophoresis and Complementary Approaches. J. Clin. Med. 2022, 11, 722. https://doi.org/10.3390/jcm11030722
Pauter K, Railean-Plugaru V, Złoch M, Pomastowski P, Szultka-Młyńska M, Buszewski B. Identification, Structure and Characterization of Bacillus tequilensis Biofilm with the Use of Electrophoresis and Complementary Approaches. Journal of Clinical Medicine. 2022; 11(3):722. https://doi.org/10.3390/jcm11030722
Chicago/Turabian StylePauter, Katarzyna, Viorica Railean-Plugaru, Michał Złoch, Paweł Pomastowski, Małgorzata Szultka-Młyńska, and Bogusław Buszewski. 2022. "Identification, Structure and Characterization of Bacillus tequilensis Biofilm with the Use of Electrophoresis and Complementary Approaches" Journal of Clinical Medicine 11, no. 3: 722. https://doi.org/10.3390/jcm11030722









