Proteomics-Based Serum Alterations of the Human Protein Expression after Out-of-Hospital Cardiac Arrest: Pilot Study for Prognostication of Survivors vs. Non-Survivors at Day 1 after Return of Spontaneous Circulation (ROSC)
Abstract
:1. Introduction
2. Material and Methods
2.1. Study Design
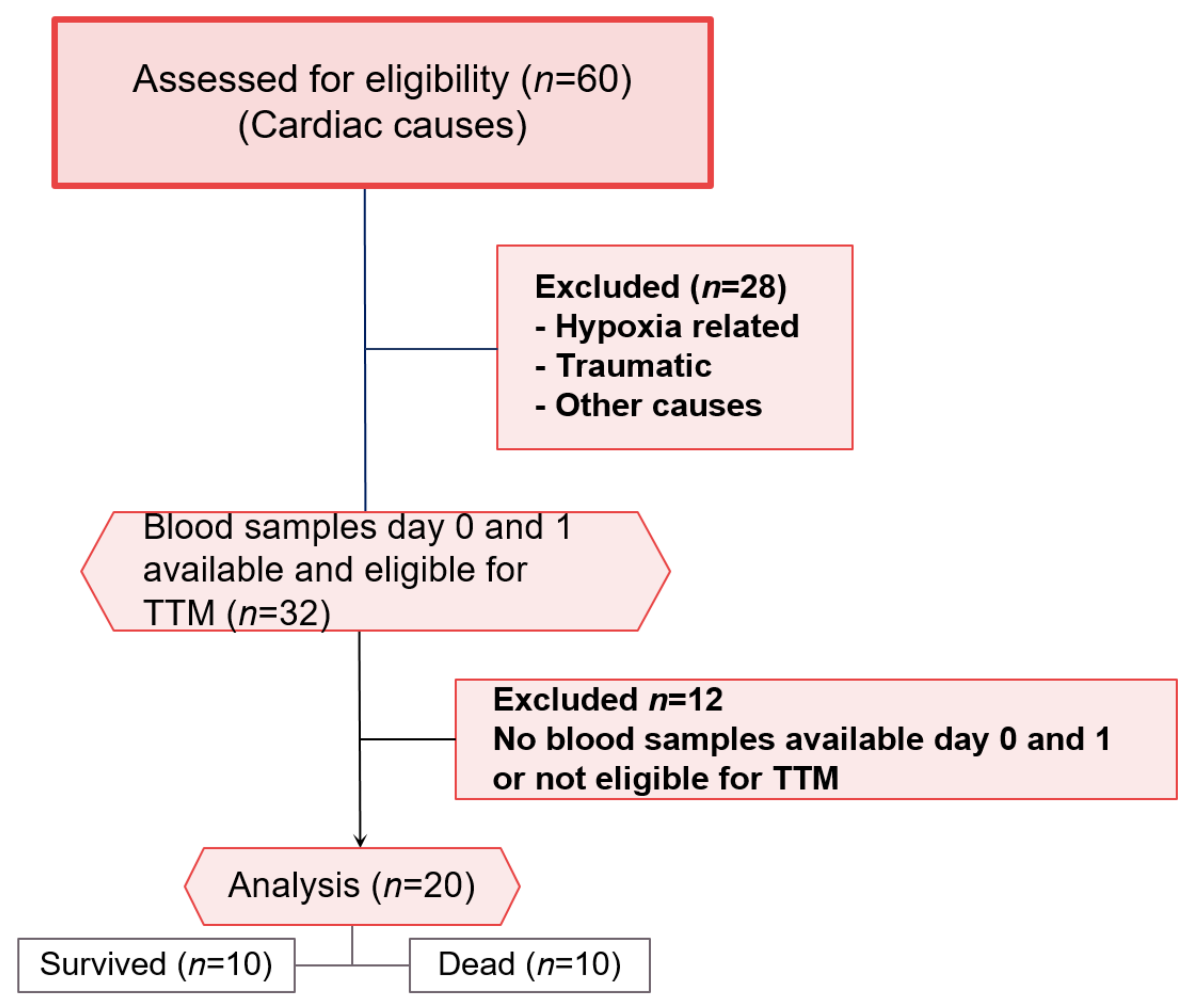
2.2. Patients
2.3. Sample Collection
2.4. TTM Therapy
2.5. Proteomic Analysis
2.6. Sample Preparation
2.7. OLINK Panels
- The inflammation panel covers a wide range of inflammation-related protein biomarkers, which enables the analysis of 92 biomarkers through a multiplex immunoassay. The panel is assembled to detect an assortment of traditional, as well as exploratory, biomarkers within the inflammation research field.
- The organ damage panel investigates 92 biomarkers from 1 µL of the biological sample. It provides the optimal dynamic range and focuses on proteins that are relevant for processes involved in the biological response to organ damage. The proteins analysed in this panel are important in processes of response to stress, regulation of cell proliferation, cell cycle, and cell death/apoptosis.
- The neurology panel consists of a proximity extension assay (PEA) technology, which tests 92 neurology-related protein biomarkers across 96 samples simultaneously without compromising on data quality.
2.8. Bioinformatic Analysis of Proteins
- Heatmapper (http://www.heatmapper.ca/, accessed on 14 December 2021) is an online server, which allows for the visualization of the results of gene expression profiling and cluster analysis in the form of heat maps through a graphical interface [12]. It allows for the accurate inspection of combinations of dataset characteristics to identify correlations and clustering results, as well as sample-related characteristics (e.g., survival time and gene expression levels). This approach allows for the visualization, as well as the accurate and rapid interpretation of the data obtained by large scale gene expression profiling [13]. By organizing complex data as matrix, the visualization of these data is improved. Heat mapping reorders rows and columns of the dataset to place the data with similar profiles, which are close to each other. In a second step, ranges of similar values are assigned to specific colour codes [14]. A heat map performs two actions on a matrix: First, it reorders the rows and columns to ensure that rows (and columns) with similar profiles are closer to one another, causing these profiles to be more visible. Second, each entry in the data matrix is displayed as a colour, making it possible to view the patterns graphically [14].
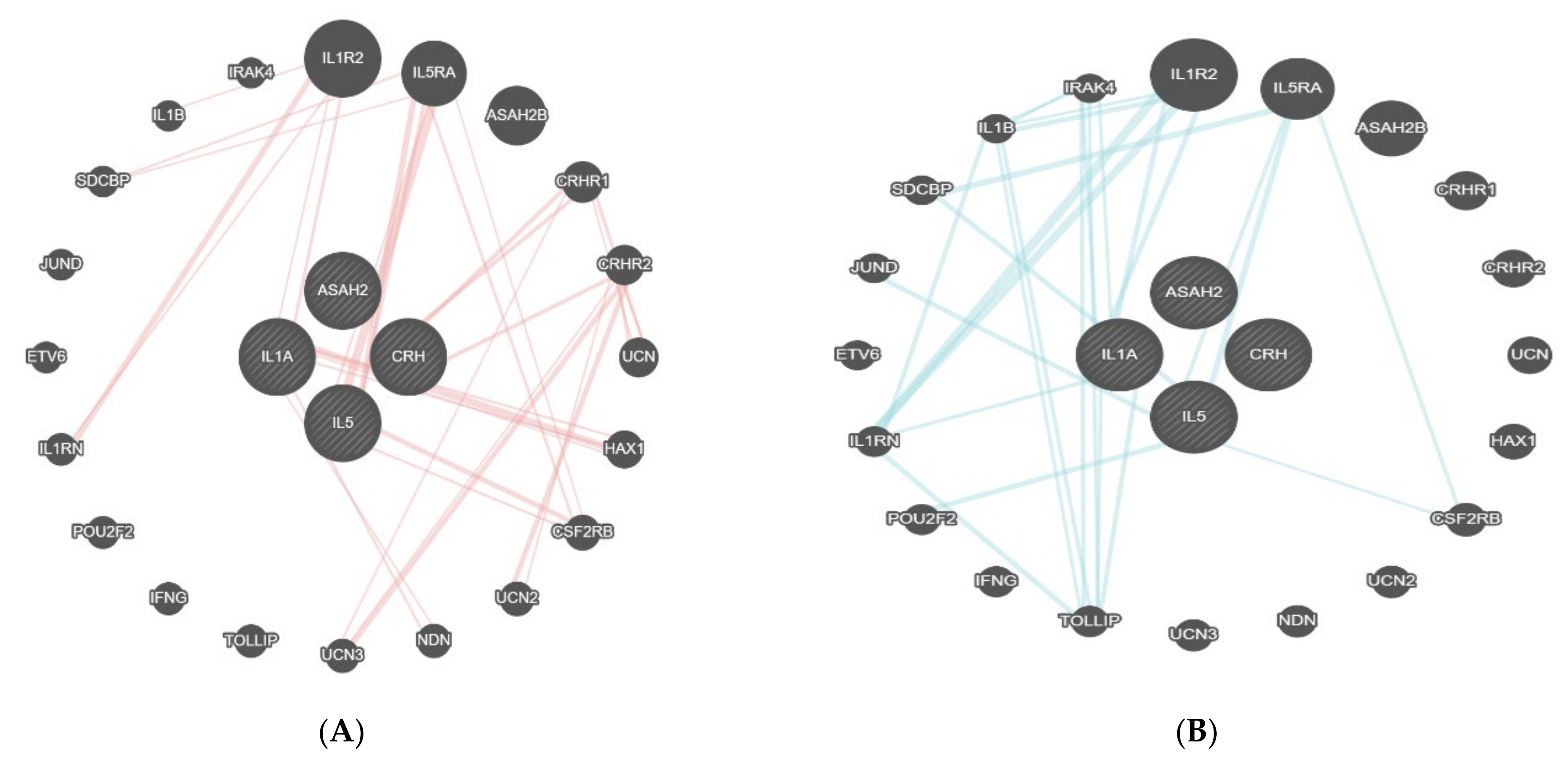
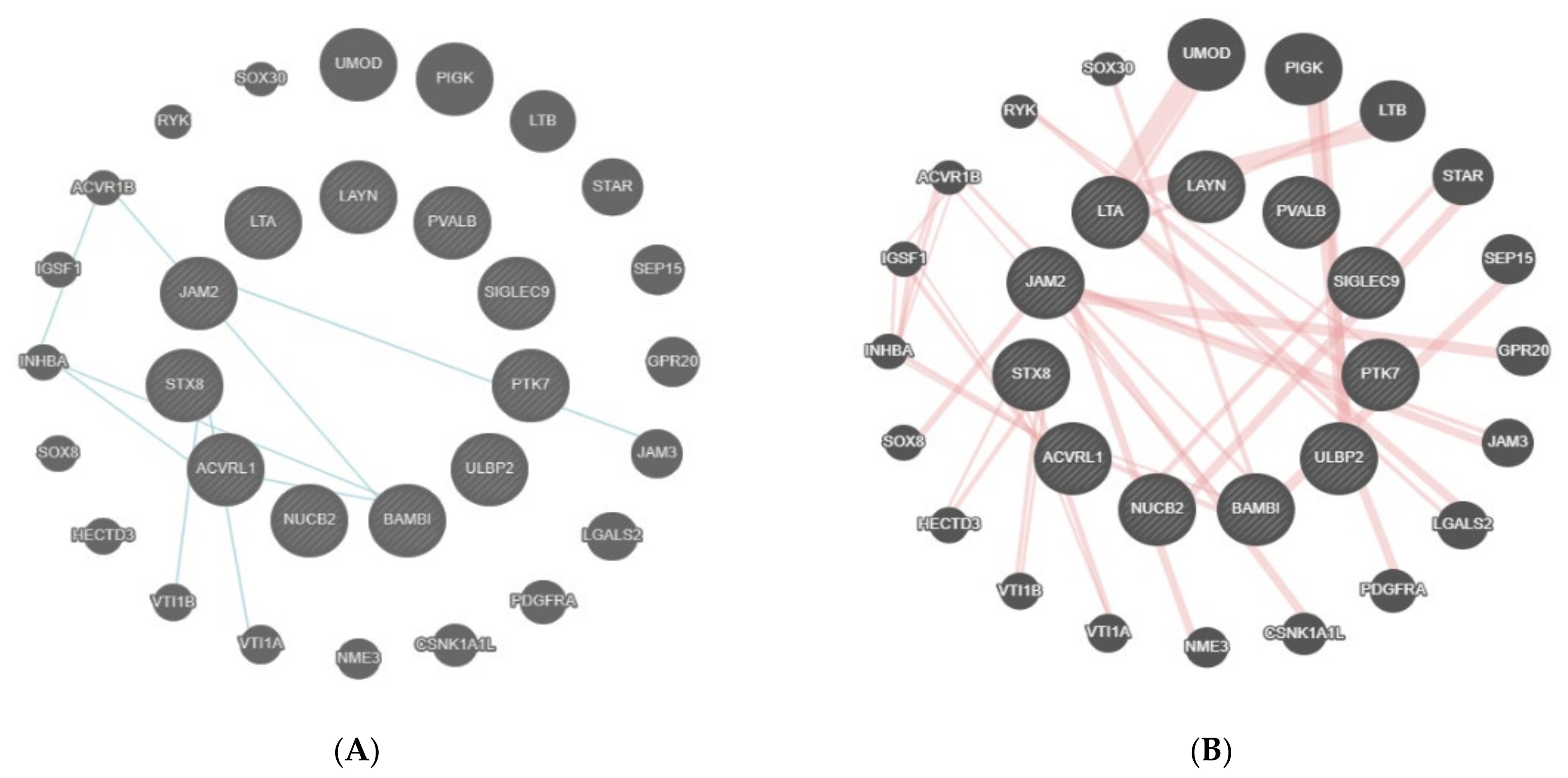
- GeneMANIA (http://www.genemania.org/, accessed on 14 December 2021) is a tool that helps in predicting the interactions and functions of a list of genes in a network form or when feasible, in pathways [15,16]. GeneMANIA provides the possibility of customizing the network, allowing for the choice of data sources or highlighting specific functions, with a more comfortable graphic experience [16]. GeneMANIA knowledge is based on data from large databases, which comprehend Gene Expression Omnibus, BioGRID, EMBL-EBI, Pfam, Ensembl, Mouse Genome Informatics, the National Center for Biotechnology Information, InParanoid, and Pathway Commons [15,16]. A network of interactions is created and the strength of the interaction is weighed. In the case of no interaction, an association weight of zero is assigned, while in the case of interaction, a positive value reflecting the strength of the interaction and the reliability of the finding, is assigned [17]. For example, the association of a pair of genes in a gene expression dataset is the Pearson correlation coefficient of their expression levels across multiple conditions in an experiment. The more the genes are co-expressed, the higher the weight they are linked by, ranging up to 1.0, indicating a perfectly correlated expression [15].
- WebGestalt is a tool to interpret the lists of genes from large scale x-OMICS (proteomics, genomics) studies [18]. The proteins of interest were uploaded to the tool where user IDs are unambiguously mapped to unique Entrez gene IDs, and all of them are mapped from a selected platform genome. Through the GoSlim classification plot, it is possible to examine the distribution of the genes of interest across the major branches of the gene ontology (GO) biological process, cellular component, and molecular function ontologies [19]. Each biological process, cellular component, and molecular function category is represented by a red, blue, and green bar, respectively.
2.9. Statistics
2.10. Ethical Registration
3. Results
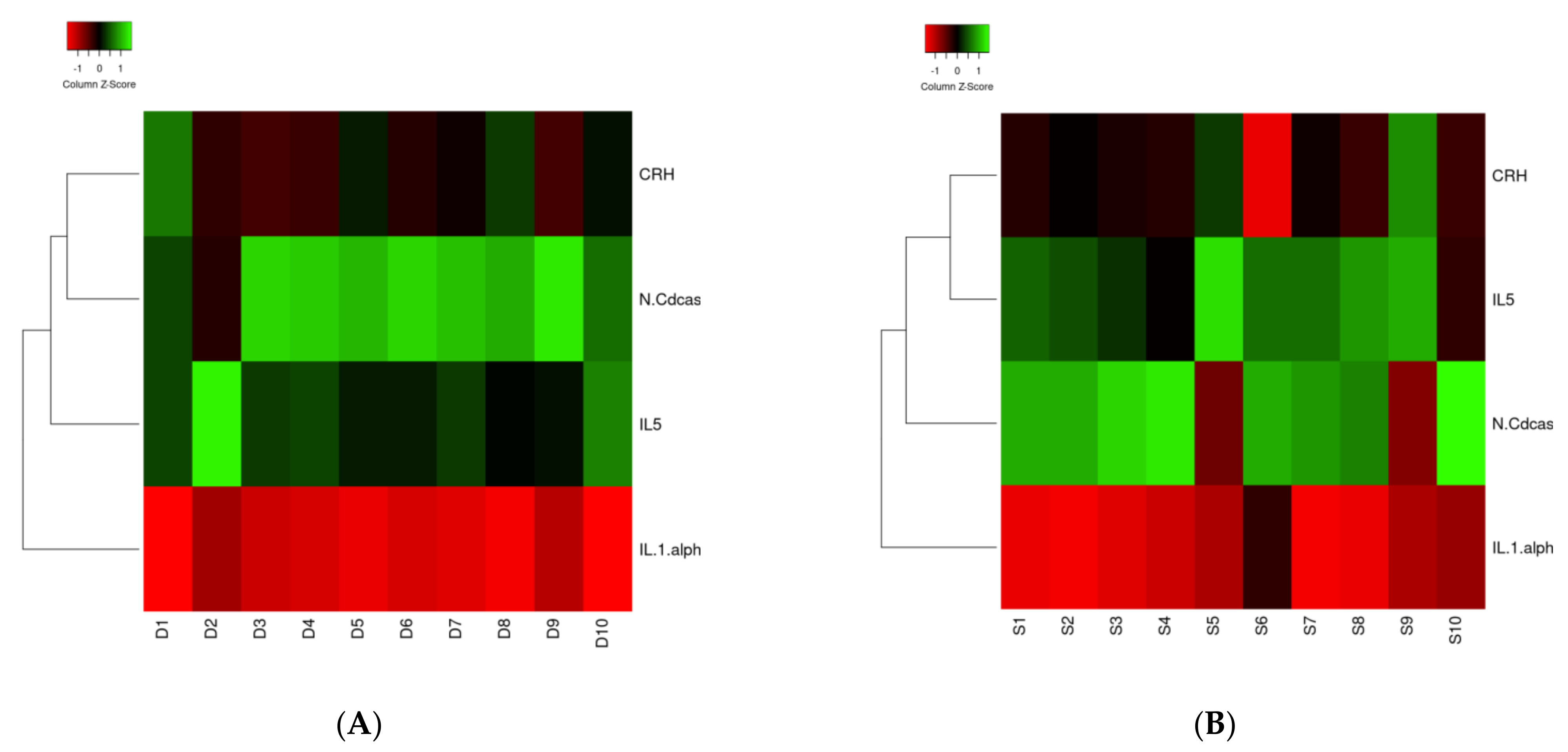
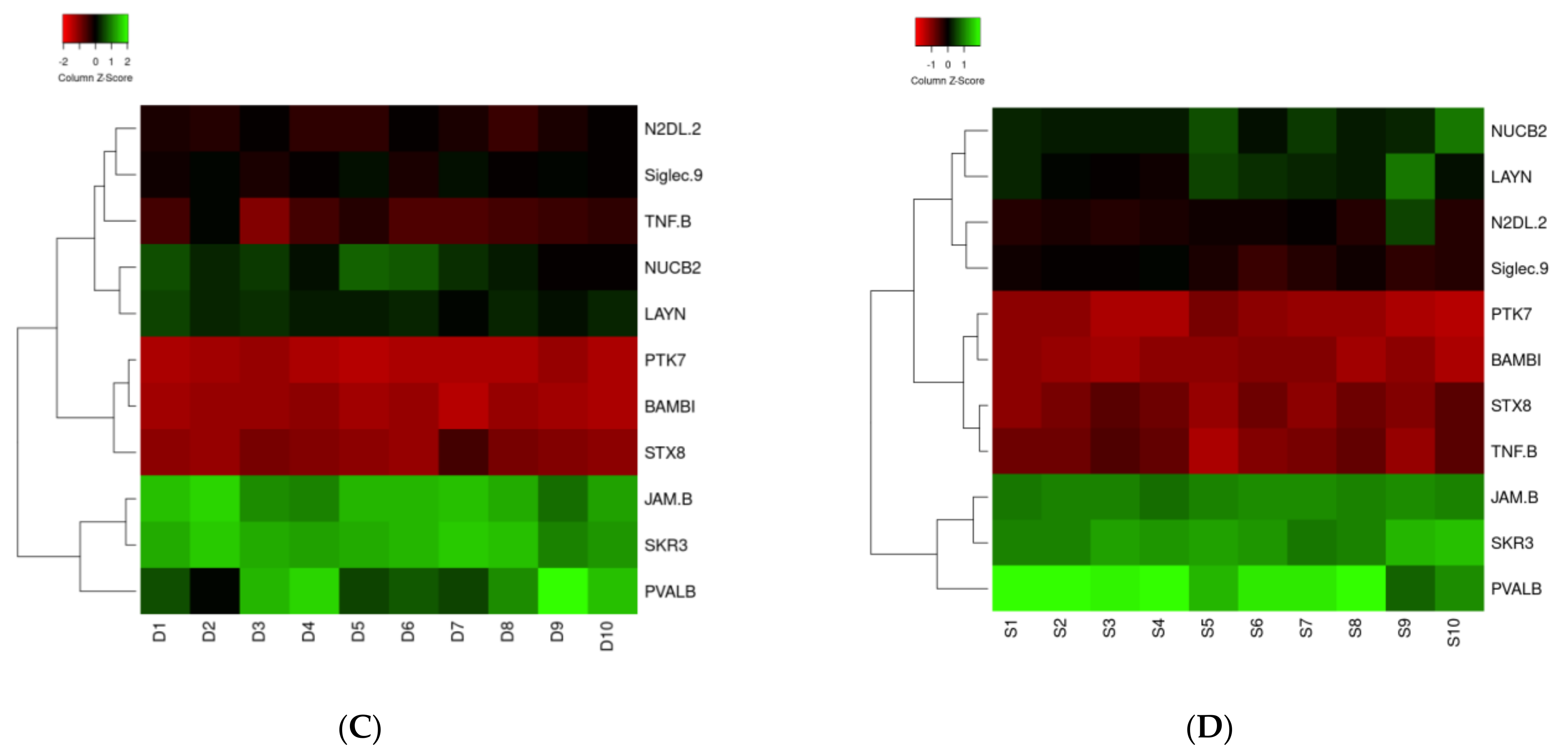
3.1. Proteomic Analysis
3.2. Bioinformatic Analysis
4. Discussion
4.1. Patient Population
4.2. Protein Identification
4.3. Biological Processes and Cascades
4.4. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| ACVR1B | Activin receptor type-1B |
| ASAH2 | N-acylsphingosine amidohydrolase-2 |
| BAMBI | Activin membrane-bound inhibitor homolog |
| CCT | Cranial computer tomography |
| CPR | Cardiopulmonary resuscitation |
| CRH | Corticotropin-releasing hormone |
| CRHR1 | Corticotropin-releasing hormone receptor-1 |
| CRHR2 | Corticotropin-releasing hormone receptor-2 |
| CSF2RB | Cytokine receptor common subunit beta |
| DNA | Desoxyribonucleid acid |
| EEG | Electroencephalography |
| ERC | European resuscitation council |
| FC | Fold change |
| GO | Gene ontology |
| HIBI | Hypoxic−ischemic brain injury |
| ICU | Intensive care unit |
| IL-1A | Interleukin-1-alpha |
| IL1R2 | Interleukin-1 receptor type 2 |
| IL5 | Interleukin-5 |
| IL5RA | Interleukin-5 receptor subunit alpha |
| INHBA | Inhibin subunit beta A |
| IRAK4 | Interleukin-1 receptor-associated kinase 4 |
| JAM-B | Junctional adhesion molecule B |
| JAM3 | Junctional adhesion molecule 3 |
| LAYN | Layilin |
| LOD | Lower detection limit |
| NPX | Normalized protein expression |
| NSE | Enolase-2 |
| NUCB2 | Nucleobindin-2 |
| N2DL-2 | NKG2D ligand-2 |
| n-CDase | Neutral ceramidase |
| OHCA | Out-of-hospital cardiac arrest |
| PCR | Polymerase chain reaction |
| PEA | Proximity extension assay |
| PIGK | Phosphatidylinositol glycan anchor biosynthesis class K LTB |
| PT | Prothrombin time |
| PTK7 | Tyrosin protein kinase 7 |
| PTT | Partial thromboplastin time |
| QC | Quality control |
| ROSC | Return of spontaneous circulation |
| SEP | Somatosensory-evocable potentials |
| Siglec-9 | Sialic acid-binding immunoglobulin-like lectin 9 |
| SOP | Standard operating procedure |
| SOX30 | SRY-box transcription factor 30 |
| STX8 | Syntaxin 8 |
| TNF-B | Tumour necrosis factor |
| TTM | Targeted temperature management |
| UMOD | Uromodulin |
| VTI1A | Vesicle transport through interaction with T-SNAREs 1A |
| VTI1B | Vesicle transport through interaction with T-SNAREs 1B |
References
- Hayashi, M.; Shimizu, W.; Albert, C.M. The spectrum of epidemiology underlying sudden cardiac death. Circ. Res. 2015, 116, 1887–1906. [Google Scholar] [CrossRef] [PubMed]
- The Hypothermia after Cardiac Arrest Study Group. Mild therapeutic hypothermia to improve the neurologic outcome after cardiac arrest. N. Engl. J. Med. 2002, 346, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Peberdy, M.A.; Callaway, C.W.; Neumar, R.W.; Geocadin, R.G.; Zimmerman, J.L.; Donnino, M.; Gabrielli, A.; Silvers, S.M.; Zaritsky, A.L.; Merchant, A.; et al. Part 9: Post-cardiac arrest care: 2010 American Heart Association guidelines for cardiopulmonary resuscitation and emergency cardiovascular care. Circulation 2010, 122, S768–S786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vane, M.F.; Carmona, M.J.C.; Pereira, S.M.; Kern, K.B.; Timerman, S.; Perez, G.; Vane, L.A.; Otsuki, D.A.; Auler, J.O., Jr. Predictors and their prognostic value for no ROSC and mortality after a non-cardiac surgery intraoperative cardiac arrest: A retrospective cohort study. Sci. Rep. 2019, 9, 14975–14979. [Google Scholar] [CrossRef] [PubMed]
- Hawkes, M.A.; Rabinstein, A.A. Neurological prognostication after cardiac arrest in the era of target temperature management. Curr. Neurol. Neurosci. Rep. 2019, 19, 10. [Google Scholar] [CrossRef]
- Callaway, C.W.; Donnino, M.W.; Fink, E.L.; Geocadin, R.G.; Golan, E.; Kern, K.B.; Leary, M.; Meurer, W.J.; Peberdy, M.A.; Thompson, T.M.; et al. Part 8: Post–cardiac arrest care: 2015 American Heart Association guidelines update for cardiopulmonary resuscitation and emergency cardiovascular care. Circulation 2015, 132, S465–S482. [Google Scholar] [CrossRef] [Green Version]
- Dragancea, I.; Rundgren, M.; Englund, E.; Friberg, H.; Cronberg, T. The influence of induced hypothermia and delayed prognostication on the mode of death after cardiac arrest. Resuscitation 2013, 84, 337–342. [Google Scholar] [CrossRef]
- Sandroni, C.; D’Arrigo, S.; Nolan, J.P. Prognostication after cardiac arrest. Crit. Care 2018, 22, 150. [Google Scholar] [CrossRef] [Green Version]
- Tiainen, M.; Roine, R.O.; Pettilä, V.; Takkunen, O. Serum neuron-specific enolase and S-100B protein in cardiac arrest patients treated with hypothermia. Stroke 2003, 34, 2881–2886. [Google Scholar] [CrossRef] [Green Version]
- Nolan, J.P.; Soar, J.; Cariou, A.; Cronberg, T.; Moulaert, V.R.M.; Deakin, C.D.; Bottiger, B.W.; Friberg, H.; Sunde, K.; Sandroni, C. European Resuscitation Council and European Society of Intensive Care Medicine guidelines for post-resuscitation care 2015. Resuscitation 2015, 95, 202–222. [Google Scholar] [CrossRef]
- Petrera, A.; von Toerne, C.; Behler, J.; Huth, C.; Thorand, B.; Hilgendorff, A.; Hauck, S.M. Multi-platforms approach for serum proteomics: Complementarity of Olink PEA technology to mass spectrometry-based protein profiling. J. Proteome Res. 2021, 20, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
- Verhaak, R.G.W.; Sanders, M.A.; Bijl, M.A.; Delwel, R.; Horsman, S.; Moorhouse, M.J.; Van Der Spek, P.J.; Löwenberg, B.; Valk, P.J.M. HeatMapper: Powerful combined visualization of gene expression profile correlations, genotypes, phenotypes and sample characteristics. BMC Bioinform. 2006, 7, 337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Key, M. A tutorial in displaying mass spectrometry-based proteomic data using heat maps. BMC Bioinform. 2012, 13, S10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mostafavi, S.; Ray, D.; Warde-Farley, D.; Grouios, C.; Morris, Q. GeneMANIA: A real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008, 9, S4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Franz, M.; Rodriguez, H.; Lopes, C.; Zuberi, K.; Montojo, J.; Bader, G.D.; Morris, Q. GeneMANIA update 2018. Nucleic Acids Res. 2018, 46, W60–W64. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Vasaikar, S.; Shi, Z.; Greer, M.; Zhang, B. WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017, 45, W130–W137. [Google Scholar] [CrossRef]
- Jacobs, I.; Nadkarni, V.; Bahr, J.; Berg, R.A.; Billi, J.E.; Bossaert, L.; Cassan, P.; Coovadia, A.; D’Este, K.; Finn, J.; et al. Cardiac arrest and cardiopulmonary resuscitation outcome reports: Update and simplification of the Utstein templates for resuscitation registries: A statement for healthcare professionals from a task force of the International Liaison Committee on Resuscitation (American Heart Association, European Resuscitation Council, Australian Resuscitation Council, New Zealand Resuscitation Council, Heart and Stroke Foundation of Canada, InterAmerican Heart Foundation, Resuscitation Councils of Southern Africa). Circulation 2004, 110, 3385–3397. [Google Scholar]
- Franek, O. “Survival incidence” should be an important parameter of cardiac arrest survival chance, beside to “survival rate”. Resuscitation 2015, 96, 79. [Google Scholar] [CrossRef]
- Andersen, L.W.; Holmberg, M.J.; Berg, K.M.; Donnino, M.W.; Granfeldt, A. In-hospital cardiac arrest: A review. JAMA 2019, 321, 1200. [Google Scholar] [CrossRef] [PubMed]
- Zelfani, S.; Manai, H.; Riahi, Y.; Daghfous, M. Out of hospital cardiac arrest: When to resuscitate. Pan Afr. Med. J. 2019, 33, 289. [Google Scholar] [CrossRef] [PubMed]
- Safdar, B.; Stolz, U.; Stiell, I.; Cone, D.C.; Bobrow, B.J.; Deboehr, M.; Dreyer, J.; Maloney, J.; Spaite, D. Differential survival for men and women from out-of-hospital cardiac arrest varies by age: Results from the OPALS study. Acad. Emerg. Med. 2014, 21, 1503–1511. [Google Scholar] [CrossRef] [PubMed]
- Cakmak, S.; Sogut, O.; Albayrak, L.; Yildiz, A. Serum copeptin levels predict the return of spontaneous circulation and the short-term prognosis of patients with out-of-hospital cardiac arrest: A randomized control study. Prehosp. Disaster Med. 2020, 35, 120–127. [Google Scholar] [CrossRef] [PubMed]

| Total | Survived | Dead | p-Value | |
|---|---|---|---|---|
| Number of patients | n = 20 | n = 10 | n =10 | |
| Age | 69.9 (9.5) | 70.8 (3.8) | 69.2 (12.3) | 0.697 |
| Gender | <0.001 | |||
| Female | 1 (5%) | 0 (0%) | 1 (10%) | |
| Male | 19 (95%) | 10 (100%) | 9 (90%) | |
| Cause of cardiac arrest | ||||
| Myocardial infarction–no. (%) | 14 (70) | 8 (80) | 6 (60) | 0.620 |
| Primary arrhythmia–no. (%) | 6 (30) | 2 (20) | 4 (40) | 0.620 |
| Cardiac arrest characteristics | ||||
| No-flow time (min) | 4.2 ± 2.9 | 3.0 ± 1.7 | 5.4 ± 3.6 | 0.030 |
| Witnessed arrest by bystander–no. (%) | 14 (70) | 9 (90) | 5 (50) | 0.140 |
| BLS provided by bystander–no. (%) | 15 (75) | 8 (80) | 7 (70) | 0.990 |
| Dose of epinephrine during CPR (mg) | 5.5 ± 4.2 | 4.8 ± 4.1 | 6.0 ± 4.2 | 0.060 |
| Number of shocks | 5.0 ± 4.2 | 4.7 ± 4.1 | 5.3 ± 4.3 | 0.730 |
| Time to ROSC (min) | 17 ± 11 | 15 ± 12 | 23 ± 15 | 0.080 |
| ICU treatment | ||||
| Period of ICU hospitalization (days) | 12 ± 12 | 14 ± 9 | 7 ± 6 | 0.04 |
| Ventilation time (days) | 11 ± 10 | 10 ± 12 | 7 ± 5 | 0.07 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hinkelbein, J.; Kolaparambil Varghese Johnson, L.; Kiselev, N.; Schmitz, J.; Hellmich, M.; Drinhaus, H.; Lichtenstein, T.; Storm, C.; Adler, C. Proteomics-Based Serum Alterations of the Human Protein Expression after Out-of-Hospital Cardiac Arrest: Pilot Study for Prognostication of Survivors vs. Non-Survivors at Day 1 after Return of Spontaneous Circulation (ROSC). J. Clin. Med. 2022, 11, 996. https://doi.org/10.3390/jcm11040996
Hinkelbein J, Kolaparambil Varghese Johnson L, Kiselev N, Schmitz J, Hellmich M, Drinhaus H, Lichtenstein T, Storm C, Adler C. Proteomics-Based Serum Alterations of the Human Protein Expression after Out-of-Hospital Cardiac Arrest: Pilot Study for Prognostication of Survivors vs. Non-Survivors at Day 1 after Return of Spontaneous Circulation (ROSC). Journal of Clinical Medicine. 2022; 11(4):996. https://doi.org/10.3390/jcm11040996
Chicago/Turabian StyleHinkelbein, Jochen, Lydia Kolaparambil Varghese Johnson, Nikolai Kiselev, Jan Schmitz, Martin Hellmich, Hendrik Drinhaus, Theresa Lichtenstein, Christian Storm, and Christoph Adler. 2022. "Proteomics-Based Serum Alterations of the Human Protein Expression after Out-of-Hospital Cardiac Arrest: Pilot Study for Prognostication of Survivors vs. Non-Survivors at Day 1 after Return of Spontaneous Circulation (ROSC)" Journal of Clinical Medicine 11, no. 4: 996. https://doi.org/10.3390/jcm11040996
APA StyleHinkelbein, J., Kolaparambil Varghese Johnson, L., Kiselev, N., Schmitz, J., Hellmich, M., Drinhaus, H., Lichtenstein, T., Storm, C., & Adler, C. (2022). Proteomics-Based Serum Alterations of the Human Protein Expression after Out-of-Hospital Cardiac Arrest: Pilot Study for Prognostication of Survivors vs. Non-Survivors at Day 1 after Return of Spontaneous Circulation (ROSC). Journal of Clinical Medicine, 11(4), 996. https://doi.org/10.3390/jcm11040996







