A Tailored Lipid Supplement Restored Membrane Fatty Acid Composition and Ameliorates In Vitro Biological Features of Human Amniotic Epithelial Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Isolation of Human Amniotic Epithelial Cells (hAECs)
2.3. Immunophenotypic Analysis of hAECs
2.4. hAEC Membrane Isolation and Fatty Acid Composition Analysis
2.5. Refeed® Supplement
2.6. hAEC Culture and Refeed® Lipid Supplementation
2.7. hAEC Viability and Proliferation Analysis
2.8. Immunofluorescence Analysis of hAECs
2.9. Morphological and Dimensional Analysis of hAECs
2.10. Celector® Technology: Fractionation Principle, Procedure, and Optical Analysis
2.11. Senescence Analysis of hAECs by SA-β-Galactosidase Assay
2.12. RNA Extraction and cDNA Synthesis
2.13. Real-Time PCR
2.14. Analysis of hAEC Migratory Potential by Scratch Wound Assay
2.15. Isolation of Peripheral Blood Mononuclear Cells (PBMCs)
2.16. Analysis of BrdU Incorporation in PBMCs Co-Cultured with hAECs
2.17. Statistical Analysis
3. Results
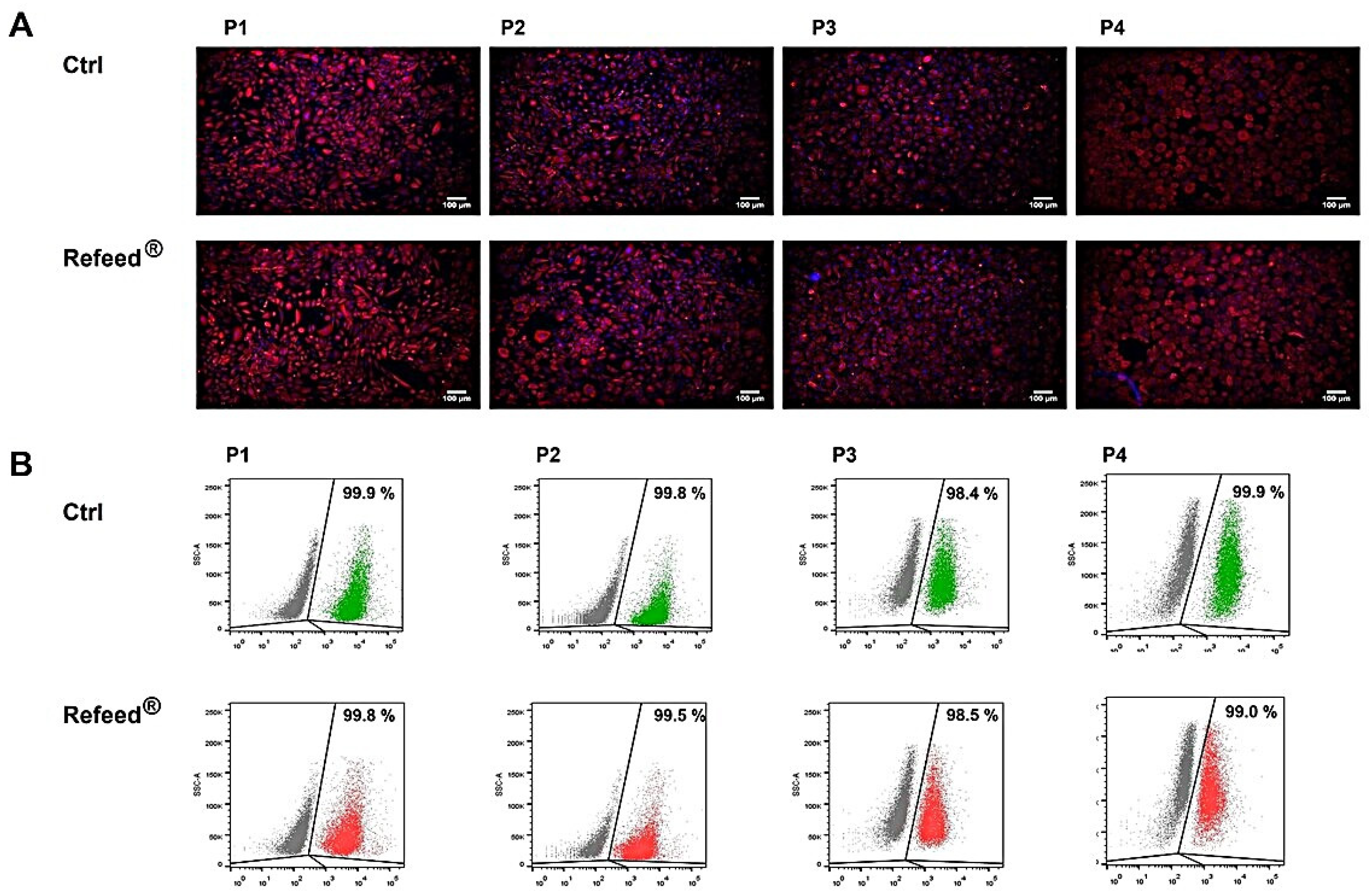
3.1. Immunophenotypic Characterization of Isolated hAECs
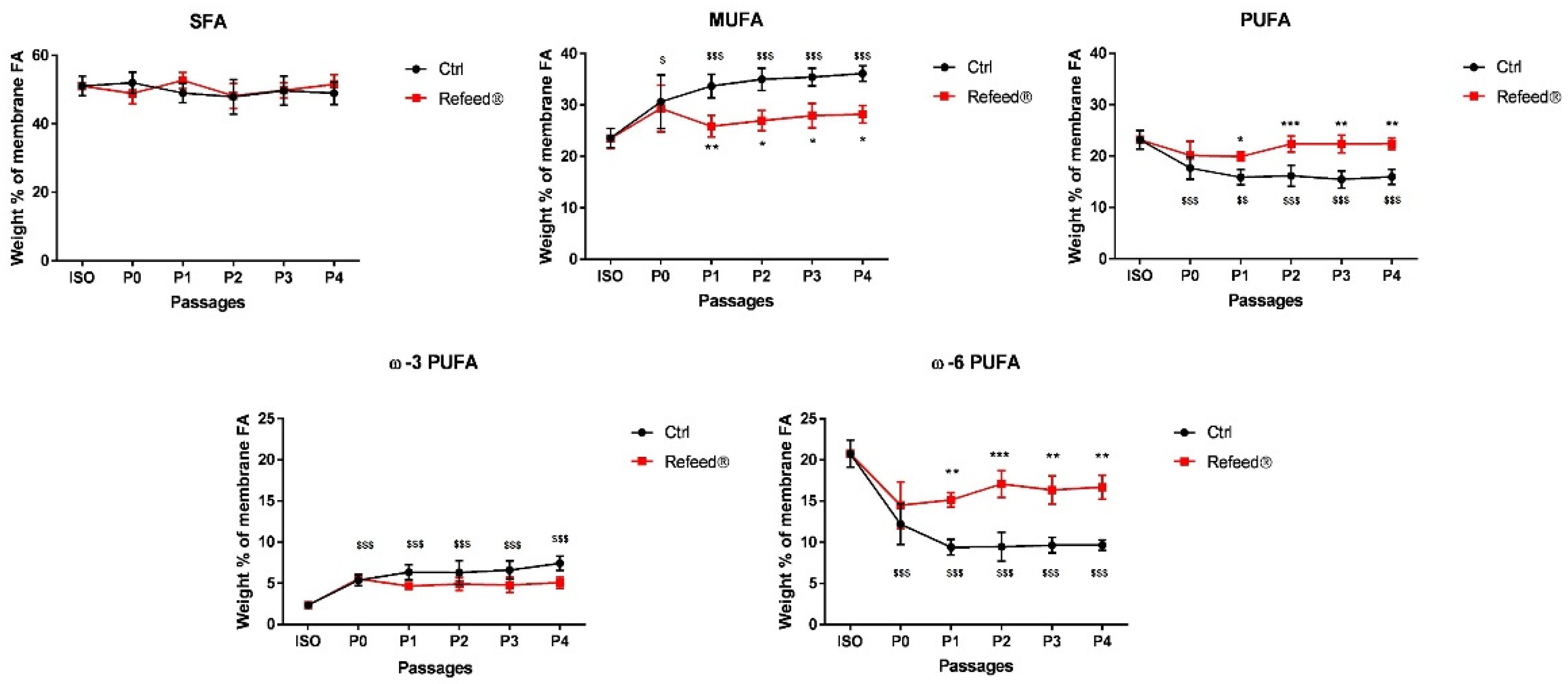
3.2. Refeed® Supplementation Restored the Membrane Fatty Acid Signature of hAECs during Cell Culture
3.3. Effect of Refeed® Supplementation on hAEC Viability and Proliferation
3.4. Expression of Pan-Cytokeratin in hAECs Cultured with or without Refeed® Supplement
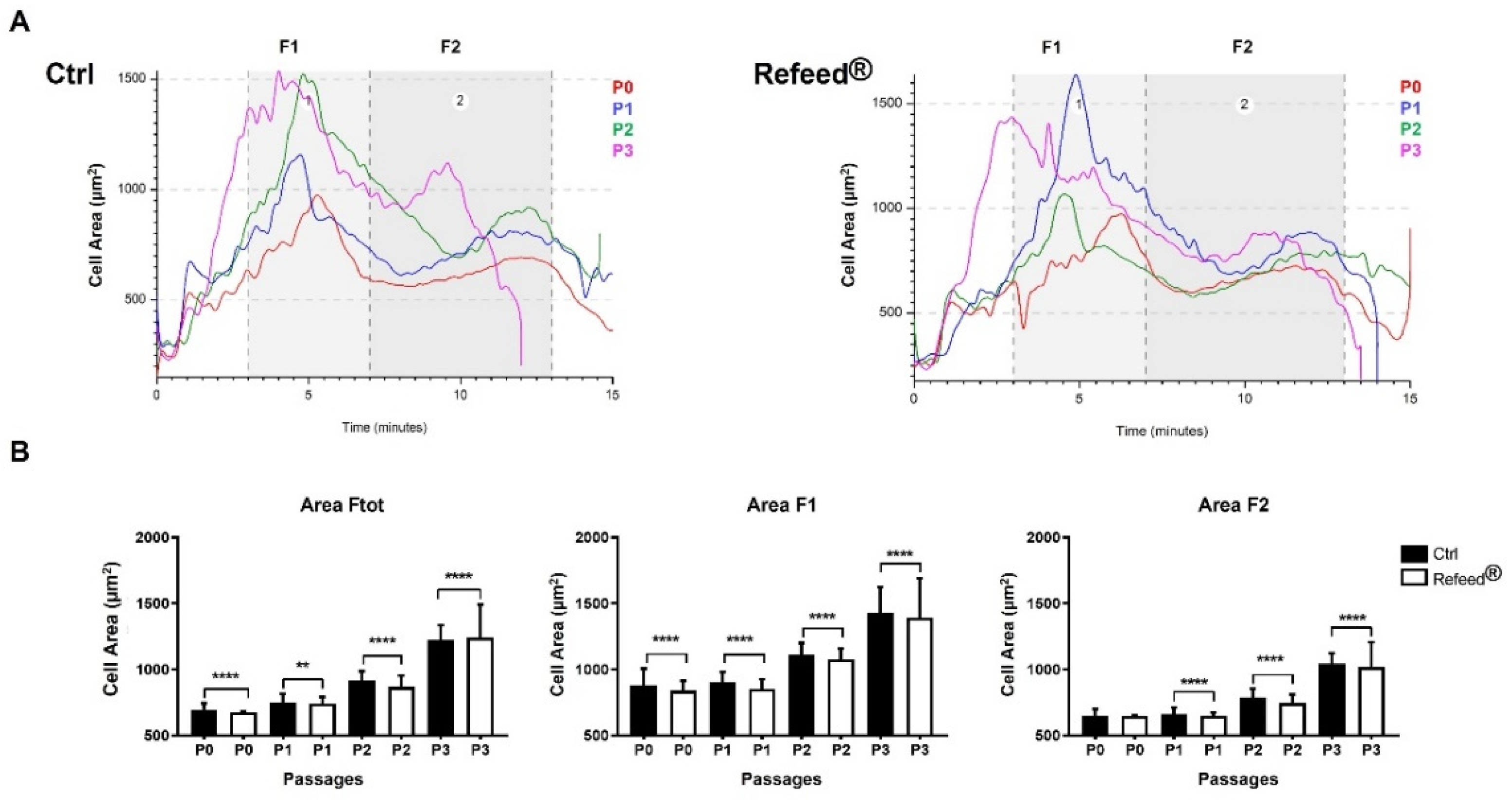
3.5. Effect of Refeed® Supplementation on hAEC Morphology
3.6. Effect of Refeed® Supplementation on hAEC Dimension
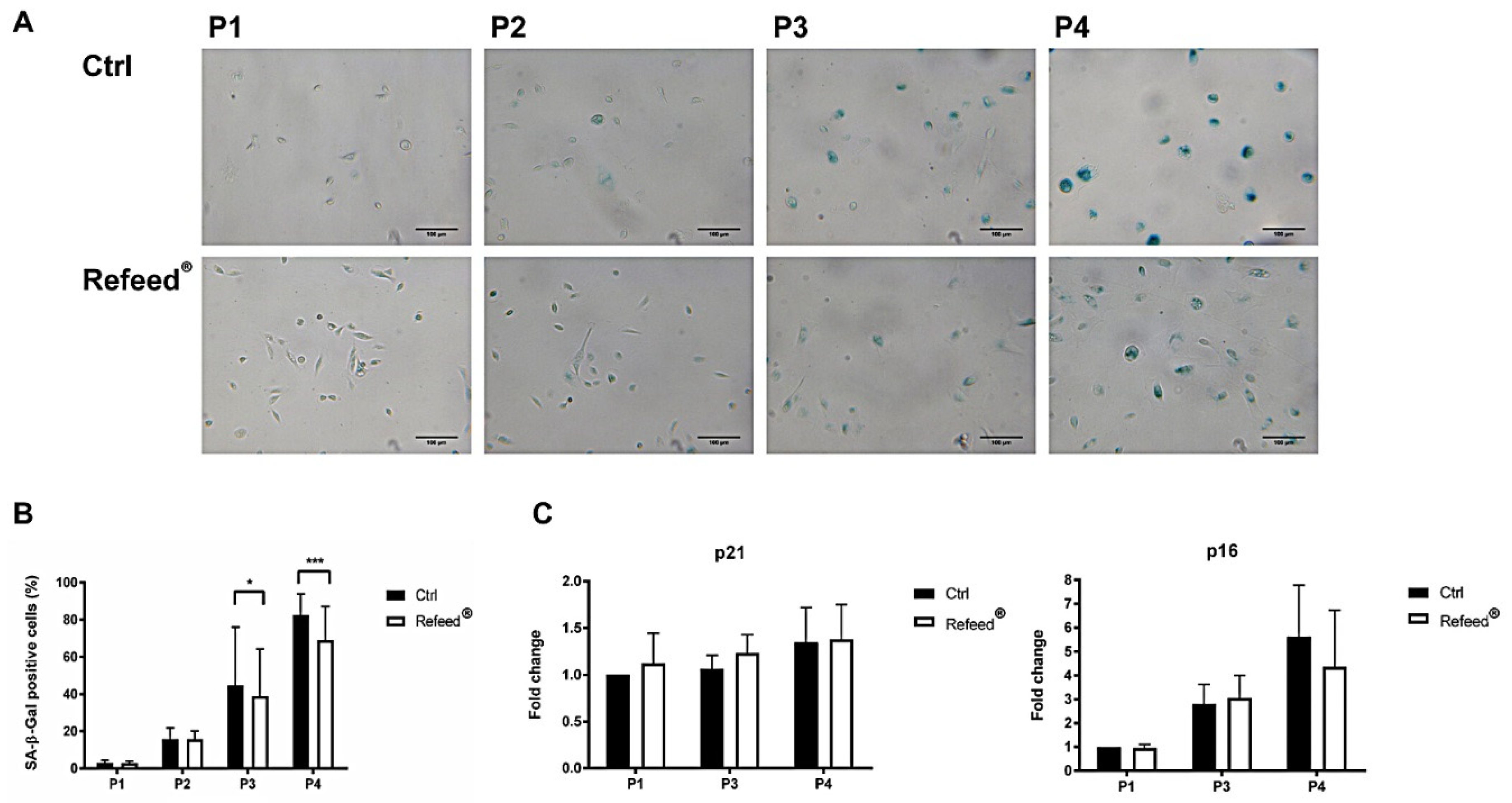
3.7. Refeed® Supplementation Reduced the Number of hAEC Senescent Cells
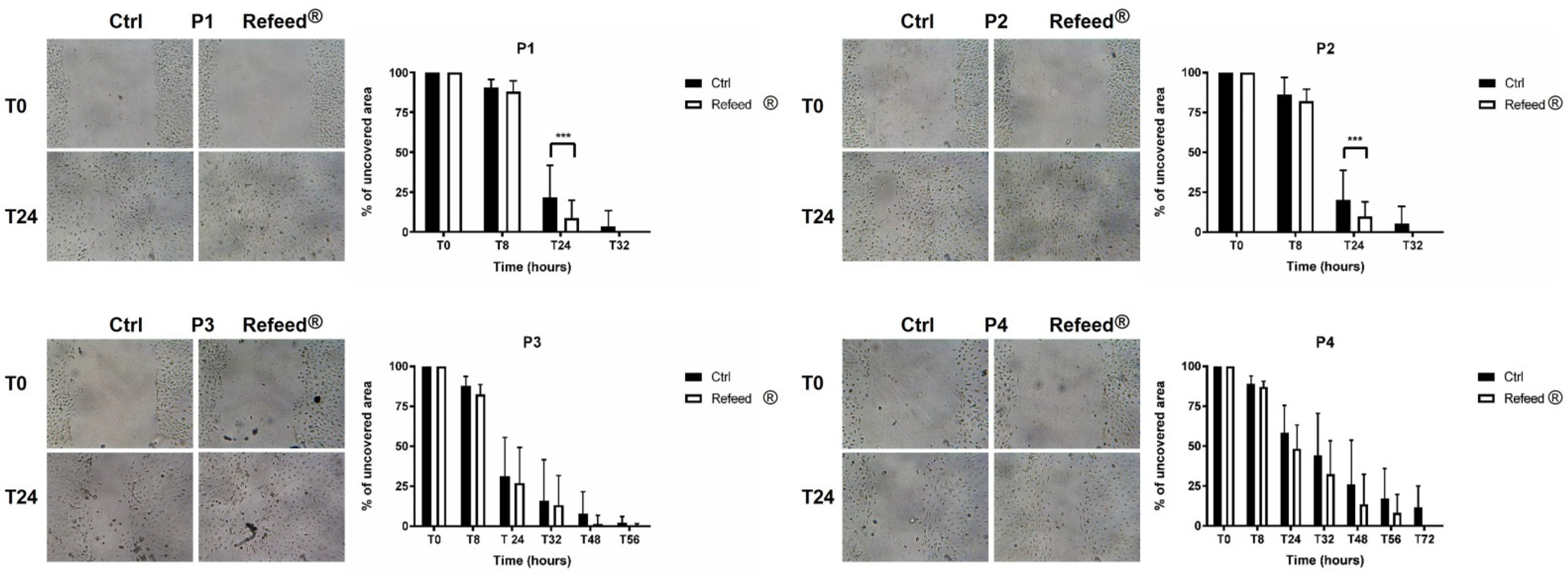
3.8. Effect of Refeed® on the Migratory Capacity of hAECs
3.9. Effect of Refeed® on the Ability of hAECs to Inhibit PBMC Proliferation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clémot, M.; Sênos Demarco, R. Lipid Mediated Regulation of Adult Stem Cell Behavior. Front. Cell Dev. Biol. 2020, 8, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singer, S.J.; Nicolson, G.L. The fluid mosaic model of the structure of cell membranes. Science 1972, 175, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Rogero, M.M.; Calder, P.C. Obesity, inflammation, toll-like receptor 4 and fatty acids. Nutrients 2018, 10, 432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasuga, K.; Suga, T. Bioanalytical insights into mediator lipidomics. J. Pharm. Biomed. Anal. 2015, 113, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Casares, D.; Escribá, P.V. Membrane lipid composition: Effect on membrane and organelle structure, function and compartmentalization and therapeutic avenues. Int. J. Mol. Sci. 2019, 20, 2167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muskiet, F.A.J.; van Goor, S.A. Long-chain polyunsaturated fatty acids in maternal and infant nutrition. Prostaglandins Leukot. Essent. Fat. Acids 2006, 75, 135–144. [Google Scholar] [CrossRef]
- Christi, W.W.; Harwood, J.L. Oxidation of polyunsaturated fatty acids to produce lipid mediators. Essays Biochem. 2020, 64, 401–421. [Google Scholar] [CrossRef]
- Ouellette, M.È.; Bérubé, J.C. Linoleic acid supplementation of cell culture media influences the phospholipid and lipid profiles of human reconstructed adipose tissue. PLoS ONE 2019, 14, e0224228. [Google Scholar] [CrossRef] [Green Version]
- Chatgilialoglu, A.; Rossi, M. Restored in vivo-like membrane lipidomics positively influence in vitro features of cultured mesenchymal stromal/stem cells derived from human placenta. Stem Cell Res. Ther. 2017, 8, 31. [Google Scholar] [CrossRef] [Green Version]
- Smith, A.N.; Muffley, L.A. Unsaturated fatty acids induce mesenchymal stem cells to increase secretion of angiogenic mediators. J. Cell. Physiol. 2012, 227, 3225–3233. [Google Scholar] [CrossRef] [Green Version]
- Holopainen, M.; Impola, U. Human Mesenchymal Stromal Cell Secretome Promotes the Immunoregulatory Phenotype and Phagocytosis Activity in Human Macrophages. Cells 2020, 9, 2142. [Google Scholar] [CrossRef] [PubMed]
- Yanes, O.; Clark, J. Metabolic oxidation regulates embryonic stem cell differentiation. Nat. Chem. Biol. 2010, 6, 411–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Else, P.L. The highly unnatural fatty acid profile of cells in culture. Prog. Lipid Res. 2020, 77, 101017. [Google Scholar] [CrossRef] [PubMed]
- Tekkatte, C.; Gunasingh, G.P. “Humanized” stem cell culture techniques: The animal serum controversy. Stem Cells Int. 2011, 2011, 504723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patrikoski, M.; Juntunen, M. Development of fully defined xeno-free culture system for the preparation and propagation of cell therapy-compliant human adipose stem cells. Stem Cell Res. Ther. 2013, 4, 27. [Google Scholar] [CrossRef] [Green Version]
- Shanbhag, S.; Stavropoulos, A. Efficacy of Humanized Mesenchymal Stem Cell Cultures for Bone Tissue Engineering: A Systematic Review with a Focus on Platelet Derivatives. Tissue Eng. Part B Rev. 2017, 23, 552–569. [Google Scholar] [CrossRef]
- Silini, A.R.; Di Pietro, R. Perinatal Derivatives: Where Do We Stand? A Roadmap of the Human Placenta and Consensus for Tissue and Cell Nomenclature. Front. Bioeng. Biotechnol. 2020, 8, 610544. [Google Scholar] [CrossRef]
- Miki, T. Stem cell characteristics and the therapeutic potential of amniotic epithelial cells. Am. J. Reprod. Immunol. 2018, 80, e13003. [Google Scholar] [CrossRef]
- Ilancheran, S.; Moodley, Y. Human Fetal Membranes: A Source of Stem Cells for Tissue Regeneration and Repair? Placenta 2009, 30, 2–10. [Google Scholar] [CrossRef]
- Insausti, C.L.; Blanquer, M. Amniotic membrane-derived stem cells: Immunomodulatory properties and potential clinical application. Stem Cells Cloning Adv. Appl. 2014, 7, 53–63. [Google Scholar] [CrossRef] [Green Version]
- Cavallini, C.; Zannini, C. Restoring In Vivo-Like Membrane Lipidomics Promotes Exosome Trophic Behavior from Human Placental Mesenchymal Stromal/Stem Cells. Cell Transplant. 2018, 27, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Miki, T.; Lehmann, T. Stem Cell Characteristics of Amniotic Epithelial Cells. Stem Cells 2005, 23, 1549–1559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, J.K.; Draper, J.S. Preimplantation Human Embryos and Embryonic Stem Cells Show Comparable Expression of Stage-Specific Embryonic Antigens. Stem Cells 2002, 20, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Fanti, M.; Gramignoli, R. Differentiation of amniotic epithelial cells into various liver cell types and potential therapeutic applications. Placenta 2017, 59, 139–145. [Google Scholar] [CrossRef]
- Okere, B.; Alviano, F. In vitro differentiation of human amniotic epithelial cells into insulinproducing 3D spheroids. Int. J. Immunopathol. Pharmacol. 2015, 28, 390–402. [Google Scholar] [CrossRef] [Green Version]
- Marongiu, F.; Gramignoli, R. Hepatic differentiation of amniotic epithelial cells. Hepatology 2011, 53, 1719–1729. [Google Scholar] [CrossRef] [Green Version]
- Moodley, Y.; Ilancheran, S. Human amnion epithelial cell transplantation abrogates lung fibrosis and augments repair. Am. J. Respir. Crit. Care Med. 2010, 182, 643–651. [Google Scholar] [CrossRef]
- Evans, M.A.; Lim, R. Acute or delayed systemic administration of human amnion epithelial cells improves outcomes in experimental stroke. Stroke 2018, 49, 700–709. [Google Scholar] [CrossRef]
- Motedayyen, H.; Zarnani, A.H. Immunomodulatory effects of human amniotic epithelial cells on naive CD4+ T cells from women with unexplained recurrent spontaneous abortion. Placenta 2018, 71, 31–40. [Google Scholar] [CrossRef]
- Liu, Y.H.; Vaghjiani, V. Amniotic epithelial cells from the human placenta potently suppress a mouse model of multiple sclerosis. PLoS ONE 2012, 7, e35758. [Google Scholar] [CrossRef] [Green Version]
- Motedayyen, H.; Fathi, F. The effect of lipopolysaccharide on anti-inflammatory and pro-inflammatory cytokines production of human amniotic epithelial cells. Reprod. Biol. 2018, 18, 404–409. [Google Scholar] [CrossRef] [PubMed]
- Pratama, G.; Vaghjiani, V. Changes in culture expanded human amniotic epithelial cells: Implications for potential therapeutic applications. PLoS ONE 2011, 6, e26136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilic, G.; Zeisberger, S.M. Comparative characterization of cultured human term amnion epithelial and mesenchymal stromal cells for application in cell therapy. Cell Transplant. 2008, 17, 955–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terada, S.; Matsuura, K. Inducing proliferation of human amniotic epithelial (HAE) cells for cell therapy. Cell Transplant. 2000, 9, 701–704. [Google Scholar] [CrossRef] [PubMed]
- Fatimah, S.S.; Tan, G.C.; Chua, K.H.; Tan, A.E.; Hayati, A.R. Effects of epidermal growth factor on the proliferation and cell cycle regulation of cultured human amnion epithelial cells. J Biosci Bioeng. 2012, 114, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Strober, W. Trypan Blue Exclusion Test of Cell Viability. Curr Protoc Immunol. 2015, 111, A3.B.1–A3.B.3. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.I.; Kim, H.J.; Lee, H.J.; Lee, K.; Hong, D.; Lim, H.; Cho, K.; Jung, N.; Yi, Y.W. Application of a non-hazardous vital dye for cell counting with automated cell counters. Anal Biochem. 2016, 492, 8–12. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Roda, B.; Reschiglian, P. Gravitational field-flow fractionation of human hemopoietic stem cells. J. Chromatogr. A 2009, 1216, 9081–9087. [Google Scholar] [CrossRef]
- Facchin, F.; Alviano, F.; Canaider, S.; Bianconi, E.; Rossi, M.; Bonsi, L.; Casadei, R.; Biava, P.M.; Ventura, C. Early Developmental Zebrafish Embryo Extract to Modulate Senescence in Multisource Human Mesenchymal Stem Cells. Int. J. Mol. Sci. 2019, 20, 2646. [Google Scholar] [CrossRef] [Green Version]
- Bolotta, A.; Abruzzo, P.M.; Baldassarro, V.A.; Ghezzo, A.; Scotlandi, K.; Marini, M.; Zucchini, C. New Insights into the Hepcidin-Ferroportin Axis and Iron Homeostasis in iPSC-Derived Cardiomyocytes from Friedreich’s Ataxia Patient. Oxid. Med. Cell Longev. 2019, 2019, 7623023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zia, S.; Martini, G. A New Predictive Technology for Perinatal Stem Cell Isolation Suited for Cell Therapy Approaches. Micromachines 2021, 12, 782. [Google Scholar] [CrossRef] [PubMed]
- Gaggi, G.; Di Credico, A. Epigenetic Features of Human Perinatal Stem Cells Redefine Their Stemness Potential. Cells 2020, 9, 1304. [Google Scholar] [CrossRef]
- Marinho, P.A.; Chailangkarn, T. Systematic optimization of human pluripotent stem cells media using Design of Experiments. Sci. Rep. 2015, 5, 9834. [Google Scholar] [CrossRef] [Green Version]
- Grammatikos, S.I.; Subbaiah, P.V. Diverse effects of essential (n-6 and n-3) fatty acids on cultured cells. Cytotechnology 1994, 15, 31–50. [Google Scholar] [CrossRef]
- De Antueno, R.J.; Knickle, L.C. Activity of human Δ5 and Δ6 desaturases on multiple n-3 and n-6 polyunsaturated fatty acids. FEBS Lett. 2001, 509, 77–80. [Google Scholar] [CrossRef]
- Wada, M.; DeLong, C.J. Enzymes and receptors of prostaglandin pathways with arachidonic acid-derived versus eicosapentaenoic acid-derived substrates and products. J. Biol. Chem. 2007, 282, 22254–22266. [Google Scholar] [CrossRef] [Green Version]
- Poggi, P.; Mirabella, R. Membrane fatty acid heterogeneity of leukocyte classes is altered during in vitro cultivation but can be restored with ad-hoc lipid supplementation. Lipids Health Dis. 2015, 14, 165. [Google Scholar] [CrossRef] [Green Version]
- Avila-González, D.; García-López, G. In vitro culture of epithelial cells from different anatomical regions of the human amniotic membrane. J. Vis. Exp. 2019, 153, e60551. [Google Scholar] [CrossRef]
- González-Gualda, E.; Baker, A.G. A guide to assessing cellular senescence in vitro and in vivo. FEBS J. 2021, 288, 56–80. [Google Scholar] [CrossRef] [PubMed]
- Campisi, J.; D’Adda Di Fagagna, F. Cellular senescence: When bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007, 8, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Jat, P. Mechanisms of Cellular Senescence: Cell Cycle Arrest and Senescence Associated Secretory Phenotype. Front. Cell Dev. Biol. 2021, 9, 645593. [Google Scholar] [CrossRef] [PubMed]
- Abruzzo, P.M.; Canaider, S. Herb-Derived Products: Natural Tools to Delay and Counteract Stem Cell Senescence. Stem Cells Int. 2020, 2020, 8827038. [Google Scholar] [CrossRef]
- Di Micco, R.; Krizhanovsky, V.; Baker, D.; d’Adda di Fagagna, F. Cellular senescence in ageing: From mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 2021, 22, 75–95. [Google Scholar] [CrossRef]
- Menon, R.; Boldogh, I. Histological evidence of oxidative stress and premature senescence in preterm premature rupture of the human fetal membranes recapitulated in vitro. Am. J. Pathol. 2014, 184, 1740–1751. [Google Scholar] [CrossRef]
- Fontaine, E.M.; Moussa, M. Effect of polyunsaturated fatty acids deficiency on oxidative phosphorylation in rat liver mitochondria. Biochim. Biophys. Acta Bioenerg. 1996, 1276, 181–187. [Google Scholar] [CrossRef] [Green Version]
- Nelson, G.; Kucheryavenko, O. The senescent bystander effect is caused by ROS-activated NF-κB signalling. Mech. Ageing Dev. 2018, 170, 30–36. [Google Scholar] [CrossRef]
- Davalli, P.; Mitic, T. ROS, Cell Senescence, and Novel Molecular Mechanisms in Aging and Age-Related Diseases. Oxid. Med. Cell. Longev. 2016, 2016, 3565127. [Google Scholar] [CrossRef] [Green Version]
- Rajendran, P.; Alzahrani, A.M. Autophagy and senescence: A new insight in selected human diseases. J. Cell Physiol. 2019, 234, 21485–21492. [Google Scholar] [CrossRef]
- Rastaldo, R.; Vitale, E. Dual Role of Autophagy in Regulation of Mesenchymal Stem Cell Senescence. Front Cell Dev. Biol. 2020, 8, 276. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, E.J.; Kuballa, P. ω-6 Polyunsaturated fatty acids extend life span through the activation of autophagy. Genes Dev. 2013, 27, 429–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Lei, H. Adipose-Derived Mesenchymal Stem Cells from the Elderly Exhibit Decreased Migration and Differentiation Abilities with Senescent Properties. Cell Transplant. 2017, 26, 1505–1519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres Crigna, A.; Uhlig, S. Human Adipose Tissue-Derived Stromal Cells Suppress Human, but Not Murine Lymphocyte Proliferation, via Indoleamine 2,3-Dioxygenase Activity. Cells 2020, 9, 2419. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pizzuti, V.; Abruzzo, P.M.; Chatgilialoglu, A.; Zia, S.; Marrazzo, P.; Petrocelli, G.; Zannini, C.; Marchionni, C.; Poggi, P.; Simonazzi, G.; et al. A Tailored Lipid Supplement Restored Membrane Fatty Acid Composition and Ameliorates In Vitro Biological Features of Human Amniotic Epithelial Cells. J. Clin. Med. 2022, 11, 1236. https://doi.org/10.3390/jcm11051236
Pizzuti V, Abruzzo PM, Chatgilialoglu A, Zia S, Marrazzo P, Petrocelli G, Zannini C, Marchionni C, Poggi P, Simonazzi G, et al. A Tailored Lipid Supplement Restored Membrane Fatty Acid Composition and Ameliorates In Vitro Biological Features of Human Amniotic Epithelial Cells. Journal of Clinical Medicine. 2022; 11(5):1236. https://doi.org/10.3390/jcm11051236
Chicago/Turabian StylePizzuti, Valeria, Provvidenza Maria Abruzzo, Alexandros Chatgilialoglu, Silvia Zia, Pasquale Marrazzo, Giovannamaria Petrocelli, Chiara Zannini, Cosetta Marchionni, Paola Poggi, Giuliana Simonazzi, and et al. 2022. "A Tailored Lipid Supplement Restored Membrane Fatty Acid Composition and Ameliorates In Vitro Biological Features of Human Amniotic Epithelial Cells" Journal of Clinical Medicine 11, no. 5: 1236. https://doi.org/10.3390/jcm11051236








