Relationships between Indicators of Lower Extremity Artery Disease and miRNA Expression in Peripheral Blood Mononuclear Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. miRNA Expression Datasets
2.3. Data Analysis
3. Results
3.1. The Study Group and Expression Dataset
3.2. Relationships between miRNA Expression and Continuous Characteristics of LEAD
3.3. Relationships between miRNAs Expression and Categorical Characteristics of LEAD
3.4. The Relationships between Selected miRNAs Associated with LEAD Indicators and Risk Factors, Cardiovascular Comorbidities, Results of Laboratory Tests, and Medication
3.5. Functional Analysis of miRNAs Associated with LEAD Progression
3.6. Functional Analysis of Genes Regulated by miRNAs Associated with LEAD Indicators
4. Discussion
4.1. Comparison with Other miRNAs Previously Reported as Associated with LEAD Progression
4.2. Cellular Proliferation and Survival
4.3. Response to Hypoxia
4.4. Lipids Metabolism and Plaque Stability
4.5. Circadian Rhythm
4.6. Vascular Inflammation
4.7. Limitations of the Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aboyans, V.; Ricco, J.-B.; Bartelink, M.-L.E.L.; Björck, M.; Brodmann, M.; Cohnert, T.; Collet, J.-P.; Czerny, M.; De Carlo, M.; Debus, S.; et al. 2017 ESC Guidelines on the Diagnosis and Treatment of Peripheral Arterial Diseases, in Collaboration with the European Society for Vascular Surgery (ESVS). Eur. Heart J. 2018, 39, 763–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fowkes, F.G.R.; Rudan, D.; Rudan, I.; Aboyans, V.; Denenberg, J.O.; McDermott, M.M.; Norman, P.E.; Sampson, U.K.; Williams, L.J.; Mensah, G.A.; et al. Comparison of Global Estimates of Prevalence and Risk Factors for Peripheral Artery Disease in 2000 and 2010: A Systematic Review and Analysis. Lancet 2013, 382, 1329–1340. [Google Scholar] [CrossRef]
- Song, P.; Rudan, D.; Zhu, Y.; Fowkes, F.J.I.; Rahimi, K.; Fowkes, F.G.R.; Rudan, I. Global, Regional, and National Prevalence and Risk Factors for Peripheral Artery Disease in 2015: An Updated Systematic Review and Analysis. Lancet Glob. Health 2019, 7, e1020–e1030. [Google Scholar] [CrossRef] [Green Version]
- Vuylsteke, M.E.; Colman, R.; Thomis, S.; Guillaume, G.; Van Quickenborne, D.; Staelens, I. An Epidemiological Survey of Venous Disease Among General Practitioner Attendees in Different Geographical Regions on the Globe: The Final Results of the Vein Consult Program. Angiology 2018, 69, 779–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fowkes, F.G.R.; Aboyans, V.; Fowkes, F.J.I.; McDermott, M.M.; Sampson, U.K.A.; Criqui, M.H. Peripheral Artery Disease: Epidemiology and Global Perspectives. Nat. Rev. Cardiol. 2017, 14, 156–170. [Google Scholar] [CrossRef]
- Conte, M.S.; Bradbury, A.W.; Kolh, P.; White, J.V.; Dick, F.; Fitridge, R.; Mills, J.L.; Ricco, J.-B.; Suresh, K.R.; Murad, M.H.; et al. Global Vascular Guidelines on the Management of Chronic Limb-Threatening Ischemia. Eur. J. Vasc. Endovasc. Surg. 2019, 58, S1–S109.e33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nehler, M.R.; Duval, S.; Diao, L.; Annex, B.H.; Hiatt, W.R.; Rogers, K.; Zakharyan, A.; Hirsch, A.T. Epidemiology of Peripheral Arterial Disease and Critical Limb Ischemia in an Insured National Population. J. Vasc. Surg. 2014, 60, 686–695.e2. [Google Scholar] [CrossRef] [Green Version]
- Campia, U.; Gerhard-Herman, M.; Piazza, G.; Goldhaber, S.Z. Peripheral Artery Disease: Past, Present, and Future. Am J Med 2019, 132, 1133–1141. [Google Scholar] [CrossRef] [PubMed]
- Aboyans, V.; Criqui, M.H.; Abraham, P.; Allison, M.A.; Creager, M.A.; Diehm, C.; Fowkes, F.G.R.; Hiatt, W.R.; Jönsson, B.; Lacroix, P.; et al. Measurement and Interpretation of the Ankle-Brachial Index. Circulation 2012, 126, 2890–2909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suominen, V.; Uurto, I.; Saarinen, J.; Venermo, M.; Salenius, J. PAD as a Risk Factor for Mortality Among Patients with Elevated ABI – A Clinical Study. Eur. J. Vasc. Endovasc. Surg. 2010, 39, 316–322. [Google Scholar] [CrossRef] [Green Version]
- Alahdab, F.; Wang, A.T.; Elraiyah, T.A.; Malgor, R.D.; Rizvi, A.Z.; Lane, M.A.; Prokop, L.J.; Montori, V.M.; Conte, M.S.; Murad, M.H. A Systematic Review for the Screening for Peripheral Arterial Disease in Asymptomatic Patients. J. Vasc. Surg. 2015, 61, 42S–53S. [Google Scholar] [CrossRef] [Green Version]
- Frank, U.; Nikol, S.; Belch, J.; Boc, V.; Brodmann, M.; Carpentier, P.H.; Chraim, A.; Canning, C.; Dimakakos, E.; Gottsäter, A.; et al. ESVM Guideline on Peripheral Arterial Disease. Vasa 2019, 48, 1–79. [Google Scholar] [CrossRef] [Green Version]
- Gerhard-Herman, M.D.; Gornik, H.L.; Barrett, C.; Barshes, N.R.; Corriere, M.A.; Drachman, D.E.; Fleisher, L.A.; Fowkes, F.G.R.; Hamburg, N.M.; Kinlay, S.; et al. 2016 AHA/ACC Guideline on the Management of Patients With Lower Extremity Peripheral Artery Disease: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation 2017, 135, e726–e779. [Google Scholar] [CrossRef] [PubMed]
- Libby, P.; Ridker, P.M.; Hansson, G.K. Progress and Challenges in Translating the Biology of Atherosclerosis. Nature 2011, 473, 317–325. [Google Scholar] [CrossRef]
- Gimbrone, M.A.; García-Cardeña, G. Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis. Circ. Res. 2016, 118, 620–636. [Google Scholar] [CrossRef] [Green Version]
- Durham, A.L.; Speer, M.Y.; Scatena, M.; Giachelli, C.M.; Shanahan, C.M. Role of Smooth Muscle Cells in Vascular Calcification: Implications in Atherosclerosis and Arterial Stiffness. Cardiovasc. Res. 2018, 114, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Moriya, J. Critical Roles of Inflammation in Atherosclerosis. J. Cardiol. 2019, 73, 22–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian MicroRNAs Predominantly Act to Decrease Target MRNA Levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Sung, Y.M.; Park, J.; Kim, S.; Kim, J.; Park, J.; Ha, H.; Bae, J.Y.; Kim, S.; Baek, D. General Rules for Functional MicroRNA Targeting. Nat Genet 2016, 48, 1517–1526. [Google Scholar] [CrossRef] [PubMed]
- Saliminejad, K.; Khorshid, H.R.K.; Fard, S.S.; Ghaffari, S.H. An Overview of MicroRNAs: Biology, Functions, Therapeutics, and Analysis Methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Thavarajah, T.; Gu, W.; Cai, J.; Xu, Q. Impact of MiRNA in Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2018, 38, e159–e170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feinberg, M.W.; Moore, K.J. MicroRNA Regulation of Atherosclerosis. Circ. Res. 2016, 118, 703–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laffont, B.; Rayner, K.J. MicroRNAs in the Pathobiology and Therapy of Atherosclerosis. Can. J. Cardiol. 2017, 33, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Churov, A.; Summerhill, V.; Grechko, A.; Orekhova, V.; Orekhov, A. MicroRNAs as Potential Biomarkers in Atherosclerosis. Int. J. Mol. Sci. 2019, 20, 5547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Yuan, P.; He, Y. Role of MicroRNAs in Peripheral Artery Disease (Review). Mol. Med. Rep. 2012, 6, 695–700. [Google Scholar] [CrossRef] [Green Version]
- Vogiatzi, G.; Oikonomou, E.; Deftereos, S.; Siasos, G.; Tousoulis, D. Peripheral Artery Disease: A Micro-RNA-Related Condition? Curr. Opin. Pharmacol. 2018, 39, 105–112. [Google Scholar] [CrossRef]
- Hamburg, N.M.; Leeper, N.J. Therapeutic Potential of Modulating MicroRNA in Peripheral Artery Disease. Curr. Vasc. Pharmacol. 2015, 13, 316–323. [Google Scholar] [CrossRef]
- Pérez-Cremades, D.; Cheng, H.S.; Feinberg, M.W. Noncoding RNAs in Critical Limb Ischemia. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 523–533. [Google Scholar] [CrossRef]
- Li, T.; Cao, H.; Zhuang, J.; Wan, J.; Guan, M.; Yu, B.; Li, X.; Zhang, W. Identification of MiR-130a, MiR-27b and MiR-210 as Serum Biomarkers for Atherosclerosis Obliterans. Clin. Chim. Acta 2011, 412, 66–70. [Google Scholar] [CrossRef]
- Stather, P.W.; Sylvius, N.; Wild, J.B.; Choke, E.; Sayers, R.D.; Bown, M.J. Differential MicroRNA Expression Profiles in Peripheral Arterial Disease. Circ. Cardiovasc. Genet. 2013, 6, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Signorelli, S.S.; Volsi, G.L.; Pitruzzella, A.; Fiore, V.; Mangiafico, M.; Vanella, L.; Parenti, R.; Rizzo, M.; Volti, G.L. Circulating MiR-130a, MiR-27b, and MiR-210 in Patients With Peripheral Artery Disease and Their Potential Relationship With Oxidative Stress: A Pilot Study. Angiology 2016, 67, 945–950. [Google Scholar] [CrossRef]
- Cheng, B.; Li, J.; Li, X.; Wang, X.; Wang, Z.; Liu, J.; Deng, A. MiR-323b-5p Acts as a Novel Diagnostic Biomarker for Critical Limb Ischemia in Type 2 Diabetic Patients. Sci. Rep. 2018, 8, 15080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogucka-Kocka, A.; Zalewski, D.P.; Ruszel, K.P.; Stępniewski, A.; Gałkowski, D.; Bogucki, J.; Komsta, Ł.; Kołodziej, P.; Zubilewicz, T.; Feldo, M.; et al. Dysregulation of MicroRNA Regulatory Network in Lower Extremities Arterial Disease. Front. Genet. 2019, 10, 1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aquila, G.; Fortini, C.; Pannuti, A.; Delbue, S.; Pannella, M.; Morelli, M.B.; Caliceti, C.; Castriota, F.; de Mattei, M.; Ongaro, A.; et al. Distinct Gene Expression Profiles Associated with Notch Ligands Delta-like 4 and Jagged1 in Plaque Material from Peripheral Artery Disease Patients: A Pilot Study. J. Transl. Med. 2017, 15, 98. [Google Scholar] [CrossRef]
- Syed, M.H.; Zamzam, A.; Valencia, J.; Khan, H.; Jain, S.; Singh, K.K.; Abdin, R.; Qadura, M. MicroRNA Profile of Patients with Chronic Limb-Threatening Ischemia. Diagnostics 2020, 10, 230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stojkovic, S.; Jurisic, M.; Kopp, C.W.; Koppensteiner, R.; Huber, K.; Wojta, J.; Gremmel, T. Circulating MicroRNAs Identify Patients at Increased Risk of In-Stent Restenosis after Peripheral Angioplasty with Stent Implantation. Atherosclerosis 2018, 269, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Xu, X.; Luan, P.; Kou, W.; Li, M.; Yu, Q.; Zhuang, J.; Xu, Y.; Peng, W.; Jian, W. MiR-124-3p Regulates Angiogenesis in Peripheral Arterial Disease by Targeting STAT3. Mol. Med. Rep. 2020, 22, 4890–4898. [Google Scholar] [CrossRef] [PubMed]
- Saenz-Pipaon, G.; Martinez-Aguilar, E.; Orbe, J.; González Miqueo, A.; Fernandez-Alonso, L.; Paramo, J.A.; Roncal, C. The Role of Circulating Biomarkers in Peripheral Arterial Disease. Int. J. Mol. Sci. 2021, 22, 3601. [Google Scholar] [CrossRef]
- Lee, C.-Y.; Lin, S.-J.; Wu, T.-C. MiR-548j-5p Regulates Angiogenesis in Peripheral Artery Disease. Sci. Rep. 2022, 12, 838. [Google Scholar] [CrossRef]
- Shu, X.; Mao, Y.; Li, Z.; Wang, W.; Chang, Y.; Liu, S.; Li, X.-Q. MicroRNA-93 Regulates Angiogenesis in Peripheral Arterial Disease by Targeting CDKN1A. Mol. Med. Rep. 2019, 19, 5195–5202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zampetaki, A.; Kiechl, S.; Drozdov, I.; Willeit, P.; Mayr, U.; Prokopi, M.; Mayr, A.; Weger, S.; Oberhollenzer, F.; Bonora, E.; et al. Plasma MicroRNA Profiling Reveals Loss of Endothelial MiR-126 and Other MicroRNAs in Type 2 Diabetes. Circ. Res. 2010, 107, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, T.A.; Duong, P.; Bouchareychas, L.; Chen, M.; Chung, A.; Schaller, M.S.; Oskowitz, A.; Raffai, R.L.; Conte, M.S. Circulating Exosomes from Patients with Peripheral Artery Disease Influence Vascular Cell Migration and Contain Distinct MicroRNA Cargo. JVS Vasc. Sci. 2020, 1, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. PROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. MiRNet 2.0: Network-Based Visual Analytics for MiRNA Functional Analysis and Systems Biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Stather, P.W.; Sylvius, N.; Sidloff, D.A.; Dattani, N.; Verissimo, A.; Wild, J.B.; Butt, H.Z.; Choke, E.; Sayers, R.D.; Bown, M.J. Identification of MicroRNAs Associated with Abdominal Aortic Aneurysms and Peripheral Arterial Disease. Br. J. Surg. 2015, 102, 755–766. [Google Scholar] [CrossRef]
- Sen, A.; Ren, S.; Lerchenmüller, C.; Sun, J.; Weiss, N.; Most, P.; Peppel, K. MicroRNA-138 Regulates Hypoxia-Induced Endothelial Cell Dysfunction By Targeting S100A1. PLOS ONE 2013, 8, e78684. [Google Scholar] [CrossRef]
- Sha, H.-H.; Wang, D.-D.; Chen, D.; Liu, S.-W.; Wang, Z.; Yan, D.-L.; Dong, S.-C.; Feng, J.-F. MiR-138: A Promising Therapeutic Target for Cancer. Tumour Biol. 2017, 39, 1010428317697575. [Google Scholar] [CrossRef] [Green Version]
- Yeh, M.; Oh, C.S.; Yoo, J.Y.; Kaur, B.; Lee, T.J. Pivotal Role of MicroRNA-138 in Human Cancers. Am. J. Cancer Res. 2019, 9, 1118–1126. [Google Scholar]
- Yang, P.; Ni, F.; Deng, R.; Qiang, G.; Zhao, H.; Yang, M.; Wang, X.; Xu, Y.; Chen, L.; Chen, D.; et al. MiR-362-5p Promotes the Malignancy of Chronic Myelocytic Leukaemia via down-Regulation of GADD45α. Mol. Cancer 2015, 14, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.; Yin, C.; Qi, J.; Duan, D.; Jiang, X.; Yu, J.; Luo, Z. MiR-362-5p Promotes Cell Proliferation and Cell Cycle Progression by Targeting GAS7 in Acute Myeloid Leukemia. Hum. Cell 2020, 33, 405–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Gao, Y.; Li, K.; Lin, A.; Jiang, Z. Genomic Analysis of Biomarkers Related to the Prognosis of Acute Myeloid Leukemia. Oncol. Lett. 2020, 20, 1824–1834. [Google Scholar] [CrossRef] [PubMed]
- Gassner, F.J.; Zaborsky, N.; Feldbacher, D.; Greil, R.; Geisberger, R. RNA Editing Alters MiRNA Function in Chronic Lymphocytic Leukemia. Cancers 2020, 12, 1159. [Google Scholar] [CrossRef]
- Plummer, P.N.; Freeman, R.; Taft, R.J.; Vider, J.; Sax, M.; Umer, B.A.; Gao, D.; Johns, C.; Mattick, J.S.; Wilton, S.D.; et al. MicroRNAs Regulate Tumor Angiogenesis Modulated by Endothelial Progenitor Cells. Cancer Res. 2013, 73, 341–352. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Xie, K.-L.; Zheng, F.; Liu, S.-X. Aerobic Exercise Prevents Insulin Resistance Through the Regulation of MiR-492/Resistin Axis in Aortic Endothelium. J. Cardiovasc. Trans. Res. 2018, 11, 450–458. [Google Scholar] [CrossRef]
- Zhang, T.; Tian, F.; Wang, J.; Jing, J.; Zhou, S.-S.; Chen, Y.-D. Atherosclerosis-Associated Endothelial Cell Apoptosis by MiR-429-Mediated Down Regulation of Bcl-2. CPB 2015, 37, 1421–1430. [Google Scholar] [CrossRef] [PubMed]
- Patella, F.; Leucci, E.; Evangelista, M.; Parker, B.; Wen, J.; Mercatanti, A.; Rizzo, M.; Chiavacci, E.; Lund, A.H.; Rainaldi, G. MiR-492 Impairs the Angiogenic Potential of Endothelial Cells. J. Cell. Mol. Med. 2013, 17, 1006–1015. [Google Scholar] [CrossRef]
- Hu, Y.-W.; Hu, Y.-R.; Zhao, J.-Y.; Li, S.-F.; Ma, X.; Wu, S.-G.; Lu, J.-B.; Qiu, Y.-R.; Sha, Y.-H.; Wang, Y.-C.; et al. An Agomir of MiR-144-3p Accelerates Plaque Formation through Impairing Reverse Cholesterol Transport and Promoting Pro-Inflammatory Cytokine Production. PLOS ONE 2014, 9, e94997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, J.; Cheng, A.; Clifford, B.L.; Wu, X.; Hedin, U.; Maegdefessel, L.; Pamir, N.; Sallam, T.; Tarling, E.J.; de Aguiar Vallim, T.Q. MicroRNA-144 Silencing Protects Against Atherosclerosis in Male, but Not Female Mice. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 412–425. [Google Scholar] [CrossRef]
- Chen, B.; Luo, L.; Wei, X.; Gong, D.; Jin, L. Altered Plasma MiR-144 as a Novel Biomarker for Coronary Artery Disease. Ann. Clin. Lab. Sci. 2018, 48, 440–445. [Google Scholar] [PubMed]
- Fu, W.; Liu, Z.; Zhang, J.; Shi, Y.; Zhao, R.; Zhao, H. Effect of MiR-144-5p on the Proliferation, Migration, Invasion and Apoptosis of Human Umbilical Vein Endothelial Cells by Targeting RICTOR and Its Related Mechanisms. Exp. Ther. Med. 2020, 19, 1817–1823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, W.; Liu, Z.; Zhang, J.; Shi, Y.; Zhao, R.; Zhao, H. Effect of MicroRNA-144-5p on the Proliferation, Invasion and Migration of Human Umbilical Vein Endothelial Cells by Targeting SMAD1. Exp. Ther. Med. 2020, 19, 165–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanaoka, R.; Iinuma, H.; Dejima, H.; Sakai, T.; Uehara, H.; Matsutani, N.; Kawamura, M. Usefulness of Plasma Exosomal MicroRNA-451a as a Noninvasive Biomarker for Early Prediction of Recurrence and Prognosis of Non-Small Cell Lung Cancer. OCL 2018, 94, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Li, J.; Zhang, G.; Wang, Q.; Wu, C.; Zhang, Q.; Wang, H.; Sun, P.; Xiang, R.; Yang, S. Exosomal MiR-451a Functions as a Tumor Suppressor in Hepatocellular Carcinoma by Targeting LPIN1 | Cell Physiol Biochem. Cell. Physiol. Biochem. 2019, 53, 19–35. [Google Scholar]
- Wei, Z.; Lyu, B.; Hou, D.; Liu, X. Mir-5100 Mediates Proliferation, Migration and Invasion of Oral Squamous Cell Carcinoma Cells Via Targeting SCAI. J. Investig. Surg. 2021, 34, 834–841. [Google Scholar] [CrossRef]
- Huang, H.; Jiang, Y.; Wang, Y.; Chen, T.; Yang, L.; He, H.; Lin, Z.; Liu, T.; Yang, T.; Kamp, D.W.; et al. MiR-5100 Promotes Tumor Growth in Lung Cancer by Targeting Rab6. Cancer Lett. 2015, 362, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jiang, F.; Ma, F.; Zhang, B. MiR-873-5p Suppresses Cell Proliferation and Epithelial–Mesenchymal Transition via Directly Targeting Jumonji Domain-Containing Protein 8 through the NF-ΚB Pathway in Colorectal Cancer. J. Cell Commun. Signal. 2019, 13, 549–560. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Liu, W.; Wang, C.; Ai, Z. MiR-873-5p Inhibits Cell Migration and Invasion of Papillary Thyroid Cancer via Regulation of CXCL16. OTT 2020, 13, 1037–1046. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Zhang, X.; Qi, M.; Zhang, Y.; Ding, F. MiR-873-5p Inhibits the Progression of Colon Cancer via Repression of Tumor Suppressor Candidate 3/AKT Signaling. J. Gastroenterol. Hepatol. 2019, 34, 2126–2134. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.-H.; Sun, J.; Zhang, J.; Hua, F.-Z.; Liu, Q.; Liang, Y.-P. Long Non-Coding RNA ROR Sponges MiR-138 to Aggravate Hypoxia/Reoxygenation-Induced Cardiomyocyte Apoptosis via Upregulating Mst1. Exp. Mol. Pathol. 2020, 114, 104430. [Google Scholar] [CrossRef] [PubMed]
- Paradis, S.; Charles, A.-L.; Meyer, A.; Lejay, A.; Scholey, J.W.; Chakfé, N.; Zoll, J.; Geny, B. Chronology of Mitochondrial and Cellular Events during Skeletal Muscle Ischemia-Reperfusion. Am. J. Physiol.-Cell Physiol. 2016, 310, C968–C982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiatt, W.R.; Armstrong, E.J.; Larson, C.J.; Brass, E.P. Pathogenesis of the Limb Manifestations and Exercise Limitations in Peripheral Artery Disease. Circ. Res. 2015, 116, 1527–1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDermott, M.M. Lower Extremity Manifestations of Peripheral Artery Disease. Circ. Res. 2015, 116, 1540–1550. [Google Scholar] [CrossRef]
- Kumar, S.; Williams, D.; Sur, S.; Wang, J.-Y.; Jo, H. Role of Flow-Sensitive MicroRNAs and Long Noncoding RNAs in Vascular Dysfunction and Atherosclerosis. Vasc. Pharmacol. 2019, 114, 76–92. [Google Scholar] [CrossRef]
- Kuo, J.-T.; Tsai, H.-E.; Lin, C.-T.; Lee, C.-I.; Lee, P.-L.; Ruan, Y.-R.; Chiu, J.-J.; Lee, D.-Y. Low Levels of MicroRNA-10a in Cardiovascular Endothelium and Blood Serum Are Related to Human Atherosclerotic Disease. Cardiol. Res. Pract. 2021, 2021, e1452917. [Google Scholar] [CrossRef]
- Xu, D.; Li, W.; Zhang, T.; Wang, G. MiR-10a Overexpression Aggravates Renal Ischemia–Reperfusion Injury Associated with Decreased PIK3CA Expression. BMC Nephrol. 2020, 21, 248. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Toscano, J.F.; Song, S.S.; Schlick, K.H.; Dumitrascu, O.M.; Pan, J.; Lyden, P.D.; Saver, J.L.; Gonzalez, N.R. Differential Expression of Circulating Exosomal MicroRNAs in Refractory Intracranial Atherosclerosis Associated with Antiangiogenesis. Sci. Rep. 2019, 9, 19429. [Google Scholar] [CrossRef]
- Aryal, B.; Singh, A.K.; Rotllan, N.; Price, N.; Fernández-Hernando, C. MicroRNAs and Lipid Metabolism. Curr. Opin. Lipidol. 2017, 28, 273–280. [Google Scholar] [CrossRef]
- Doghish, A.S.; Elsisi, A.M.; Amin, A.I.; Abulsoud, A.I. Circulating MiR-148a-5p and MiR-21-5p as Novel Diagnostic Biomarkers in Adult Egyptian Male Patients With Metabolic Syndrome. Can. J. Diabetes 2021, 45, 614–618. [Google Scholar] [CrossRef]
- Shi, C.; Zhang, M.; Tong, M.; Yang, L.; Pang, L.; Chen, L.; Xu, G.; Chi, X.; Hong, Q.; Ni, Y.; et al. MiR-148a Is Associated with Obesity and Modulates Adipocyte Differentiation of Mesenchymal Stem Cells through Wnt Signaling. Sci. Rep. 2015, 5, 9930. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Li, F.; Wang, S.; Wang, C.; Xie, Y.; Zhou, J.; Li, X.; Wang, B. MiR-196b-5p Controls Adipocyte Differentiation and Lipogenesis through Regulating MTORC1 and TGF-β Signaling. FASEB J. 2020, 34, 9207–9222. [Google Scholar] [CrossRef]
- Wu, Y.; Jiang, L.; Zhang, H.; Cheng, S.; Wen, W.; Xu, L.; Zhang, F.; Yang, Y.; Wang, L.; Chen, J. Integrated Analysis of MicroRNA and MRNA Expression Profiles in Homozygous Familial Hypercholesterolemia Patients and Validation of Atherosclerosis Associated Critical Regulatory Network. Genomics 2021, 113, 2572–2582. [Google Scholar] [CrossRef] [PubMed]
- Bai, R.; Yang, Q.; Xi, R.; Li, L.; Shi, D.; Chen, K. MiR-941 as a Promising Biomarker for Acute Coronary Syndrome. BMC Cardiovasc. Disord. 2017, 17, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Xie, J.; Sun, H. Three MiRNAs Cooperate with Host Genes Involved in Human Cardiovascular Disease. Hum. Genom. 2019, 13, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Cai, H.; Zhu, M.; Qian, Y.; Lin, S.; Li, X. Circulating MicroRNAs as Biomarkers for Severe Coronary Artery Disease. Medicine 2020, 99, e19971. [Google Scholar] [CrossRef]
- Li, P.; Teng, F.; Gao, F.; Zhang, M.; Wu, J.; Zhang, C. Identification of Circulating MicroRNAs as Potential Biomarkers for Detecting Acute Ischemic Stroke. Cell Mol. Neurobiol. 2015, 35, 433–447. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Liu, Y.; Gao, L.; Wei, X.; Xu, Y.; Cai, R.; Su, Q. MiR-32-3p Regulates Myocardial Injury Induced by Microembolism and Microvascular Obstruction by Targeting RNF13 to Regulate the Stability of Atherosclerotic Plaques. J. Cardiovasc. Trans. Res. 2021. [Google Scholar] [CrossRef]
- Di Gregoli, K.; Mohamad Anuar, N.N.; Bianco, R.; White, S.J.; Newby, A.C.; George, S.J.; Johnson, J.L. MicroRNA-181b Controls Atherosclerosis and Aneurysms Through Regulation of TIMP-3 and Elastin. Circ. Res. 2017, 120, 49–65. [Google Scholar] [CrossRef] [Green Version]
- Cheng, H.-P.; Gong, D.; Zhao, Z.-W.; He, P.-P.; Yu, X.-H.; Ye, Q.; Huang, C.; Zhang, X.; Chen, L.-Y.; Xie, W.; et al. MicroRNA-182 Promotes Lipoprotein Lipase Expression and Atherogenesisby Targeting Histone Deacetylase 9 in Apolipoprotein E-Knockout Mice. Circ. J. 2018, 82, 28–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, C.; Gao, S.; Li, D.; Shi, X.; Hu, Z.; Wang, C.; Xiao, J.; Sheng, Z.; Ding, Z.; Zhang, D.; et al. MiR-182-5p Inhibits the Proliferation of Vascular Smooth Muscle Cells Induced by Ox-LDL Through Targeting PAPPA. Int. Heart J. 2020, 61, 822–830. [Google Scholar] [CrossRef]
- Qin, S.-B.; Peng, D.-Y.; Lu, J.-M.; Ke, Z.-P. MiR-182-5p Inhibited Oxidative Stress and Apoptosis Triggered by Oxidized Low-Density Lipoprotein via Targeting Toll-like Receptor 4. J. Cell. Physiol. 2018, 233, 6630–6637. [Google Scholar] [CrossRef] [PubMed]
- McAlpine, C.S.; Swirski, F.K. Circadian Influence on Metabolism and Inflammation in Atherosclerosis. Circ. Res. 2016, 119, 131–141. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Yu, B.; Wang, X.; Luo, C.; Zhou, T.; Zheng, X.; Ding, J. Circadian Rhythm and Atherosclerosis (Review). Exp. Ther. Med. 2020, 20, 1-1. [Google Scholar] [CrossRef]
- Man, A.W.C.; Li, H.; Xia, N. Circadian Rhythm: Potential Therapeutic Target for Atherosclerosis and Thrombosis. Int. J. Mol. Sci. 2021, 22, 676. [Google Scholar] [CrossRef]
- Oyama, Y.; Bartman, C.M.; Gile, J.; Eckle, T. Circadian MicroRNAs in Cardioprotection. Curr. Pharm. Des. 2017, 23, 3723–3730. [Google Scholar] [CrossRef] [Green Version]
- Chinnapaiyan, S.; Dutta, R.K.; Devadoss, D.; Chand, H.S.; Rahman, I.; Unwalla, H.J. Role of Non-Coding RNAs in Lung Circadian Clock Related Diseases. Int. J. Mol. Sci. 2020, 21, 3013. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.-Y.M.; Obrietan, K. Revealing a Role of MicroRNAs in the Regulation of the Biological Clock. Cell Cycle 2007, 6, 3034–3038. [Google Scholar] [CrossRef]
- Shi, F.; Chen, X.; Fu, A.; Hansen, J.; Stevens, R.; Tjonneland, A.; Vogel, U.B.; Zheng, T.; Zhu, Y. Aberrant DNA Methylation of MiR-219 Promoter in Long-Term Night Shiftworkers. Environ. Mol. Mutagenesis 2013, 54, 406–413. [Google Scholar] [CrossRef]
- Li, D.; Huang, W.; Yang, F.; Li, B.; Cai, S. Study of the Modulatory Mechanism of the MiR-182-Clock Axis in Circadian Rhythm Disturbance after Hypoxic–Ischemic Brain Damage. Eur. J. Inflamm. 2020, 18, 2058739220929159. [Google Scholar] [CrossRef]
- Sun, P.; Li, L.; Liu, Y.-Z.; Li, G.-Z.; Xu, Q.-H.; Wang, M.; Gong, Y. MiR-181b Regulates Atherosclerotic Inflammation and Vascular Endothelial Function through Notch1 Signaling Pathway. Eur. Rev. Med. Pharm. Sci. 2019, 23, 3051–3057. [Google Scholar] [CrossRef]
- Xu, X.; Yang, Y.; Wang, G.; Yin, Y.; Han, S.; Zheng, D.; Zhou, S.; Zhao, Y.; Chen, Y.; Jin, Y. Low Shear Stress Regulates Vascular Endothelial Cell Pyroptosis through MiR-181b-5p/STAT-3 Axis. J. Cell. Physiol. 2021, 236, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Wu, F.; Shan, Y. Identification of Key Genes and MiRNAs Associated with Carotid Atherosclerosis Based on MRNA-Seq Data. Medicine 2018, 97, e9832. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, S.; Hartmann, P.; Karshovska, E.; Rinderknecht, F.-A.; Subramanian, P.; Gremse, F.; Grommes, J.; Jacobs, M.; Kiessling, F.; Weber, C.; et al. Endothelial Hypoxia-Inducible Factor-1α Promotes Atherosclerosis and Monocyte Recruitment by Upregulating MicroRNA-19a. Hypertension 2015, 66, 1220–1226. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Li, X.; Liu, S.; Gu, L.; Zhou, X. MircroRNA-19a Promotes Vascular Inflammation and Foam Cell Formation by Targeting HBP-1 in Atherogenesis. Sci. Rep. 2017, 7, 12089. [Google Scholar] [CrossRef]
- Ren, Z.-Q.; Liu, N.; Zhao, K. Micro RNA-19a Suppresses IL-10 in Peripheral B Cells from Patients with Atherosclerosis. Cytokine 2016, 86, 86–91. [Google Scholar] [CrossRef]
- Tang, Y.; Zhang, Y.; Chen, Y.; Xiang, Y.; Shen, C.; Li, Y. The Role of MiR-19b in the Inhibition of Endothelial Cell Apoptosis and Its Relationship with Coronary Artery Disease. Sci. Rep. 2015, 5, 15132. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, S.; Zhang, F.; Wu, M.; Liang, H.; Song, J.; Lee, C.; Chen, H. Endothelial Microparticles-Mediated Transfer of MicroRNA-19b Promotes Atherosclerosis via Activating Perivascular Adipose Tissue Inflammation in ApoE−/−Mice. Biochem. Biophys. Res. Commun. 2018, 495, 1922–1929. [Google Scholar] [CrossRef]



| Characteristic | Value |
|---|---|
| Clinical Parameters of Disease | |
| Ankle brachial index (ABI) | 0.68 ± 0.05, 0.59–0.8 1 |
| Claudication distance (m) | 153.63 ± 33.01, 90–200 1 |
| Length of occlusion (cm) | 11.25 ± 5.11, 3–25 1 |
| Rutherford category 2 Rutherford category 3 | 34 (85%) 6 (15%) |
| Plaque localization: iliac artery femoral artery iliac and femoral artery popliteal artery femoral and popliteal artery | 7 (17.5%) 25 (62.5%) 1 (2.5) 5 (12.5%) 2 (5%) |
| Risk Factors and Cardiovascular Comorbidities | |
| Age | 57.6 ± 9.82, 43–71 1 |
| Sex: male female | 35 (87.5%) 5 (12.5%) |
| Body mass index (BMI) | 27.2 ± 2.62, 21.9–31.6 1 |
| Former or current smoker | 18 (45%)/22 (55%) |
| Diabetes type 2 | 5 (12.5%) |
| Hypertension | 36 (90%) |
| Coronary artery disease | 11 (27.5%) |
| Myocardial infarction | 8 (20%) |
| Stroke/transient ischemic attack | 2 (5%) |
| Results of Laboratory Tests | |
| Red blood cells (M/µL) | 4.74 ± 0.30, 4.11–5.18 1 |
| White blood cells (K/µL) | 5.49 ± 0.69, 4.45–6.89 1 |
| Lymphocytes (K/µL) | 3.04 ± 0.54, 2.01–3.99 1 |
| Monocytes (K/µL) | 0.47 ± 0.15, 0.22–0.87 1 |
| Neutrophils (K/µL) | 4.21 ± 0.47, 3.51–5.21 1 |
| Eosinophils (K/µL) | 0.21 ± 0.09, 0.10–0.56 1 |
| Basophils (K/µL) | 0.10 ± 0.03, 0.07–0.19 1 |
| Platelets (K/µL) | 309.3 ± 75.7, 179–561 1 |
| Hemoglobin (g/dL) | 14.12 ± 0.52, 12.99–14.99 1 |
| Hematocrit (%) | 41.33 ± 1.42, 38.4–43.8 1 |
| Creatinine (mmol/L) | 78.70 ± 12.64, 56–99 1 |
| Urea (mmol/L) | 4.69 ± 0.83, 2.99–6.02 1 |
| Medication | |
| Statins | 34 (85%) |
| Acetylsalicylic acid | 40 (100%) |
| Clopidogrel | 8 (20%) |
| Beta-adrenergic blockers | 27 (67.5%) |
| Angiotensin-converting enzyme inhibitor | 20 (50%) |
| Ca2+ channel blockers | 11 (27.5%) |
| Fibrates | 5 (12.5%) |
| Metformin | 2 (5%) |
| Gliclazide | 4 (10%) |
| Indicator | miRNA Transcript | miRNA ID 1 | Correlation Analysis | Regression Analysis | ||
|---|---|---|---|---|---|---|
| R | p | Intercept | β | |||
| ABI | hsa-mir-148a_hsa-miR-148a-5p | hsa-miR-148a-5p | −0.44 | 4.26 × 10−3 | 0.831 | −0.00174 |
| hsa-mir-362_hsa-miR-362-5p | hsa-miR-362-5p | −0.40 | 9.55 × 10−3 | 0.735 | −0.000263 | |
| Claudication distance | hsa-mir-3182_hsa-miR-3182 | hsa-miR-3182 | 0.49 | 1.34 × 10−3 | 113.9 | 0.290 |
| hsa-mir-10a_hsa-miR-10a-5p | hsa-miR-10a-5p | −0.49 | 1.47 × 10−3 | 167.0 | −0.044 | |
| hsa-mir-32_hsa-miR-32-3p | hsa-miR-32-3p | 0.43 | 6.16 × 10−3 | 75.2 | 0.568 | |
| hsa-mir-196b_hsa-miR-196b-5p | hsa-miR-196b-5p | −0.42 | 6.70 × 10−3 | 182.0 | −0.581 | |
| hsa-mir-941-4_hsa-miR-941 | hsa-miR-941 | −0.42 | 6.84 × 10−3 | 127.1 | −1.222 | |
| hsa-mir-3157_hsa-miR-3157-5p | hsa-miR-3157-5p | −0.40 | 1.05 × 10−2 | 112.9 | −2.636 | |
| Length of occlusion | hsa-mir-182_hsa-miR-182-5p | hsa-miR-182-5p | 0.48 | 1.82 × 10−3 | −2.75 | 0.106 |
| hsa-mir-19a_hsa-miR-19a-5p | hsa-miR-19a-5p | −0.47 | 2.01 × 10−3 | 19.11 | −0.132 | |
| hsa-mir-3620_hsa-miR-3620-3p | hsa-miR-3620-3p | 0.44 | 4.43 × 10−3 | 3.77 | 0.259 | |
| hsa-mir-181b-2_hsa-miR-181b-5p | hsa-miR-181b-5p | −0.44 | 4.51 × 10−3 | 17.57 | −0.00128 | |
| hsa-mir-548au_hsa-miR-548au-5p | hsa-miR-548au-5p | −0.43 | 5.26 × 10−3 | 12.90 | −0.499 | |
| hsa-mir-6513_hsa-miR-6513-3p | hsa-miR-6513-3p | 0.42 | 6.28 × 10−3 | 9.47 | 0.264 | |
| hsa-mir-19b-1_hsa-miR-19b-1-5p | hsa-miR-19b-1-5p | −0.42 | 6.39 × 10−3 | 18.26 | −0.045 | |
| hsa-mir-138-1_hsa-miR-138-5p | hsa-miR-138-5p | −0.42 | 7.50 × 10−3 | 20.57 | −0.184 | |
| hsa-mir-429_hsa-miR-429 | hsa-miR-429 | 0.41 | 8.88 × 10−3 | 11.98 | 0.135 | |
| hsa-mir-219a-2_hsa-miR-219a-5p | hsa-miR-219a-5p | −0.40 | 1.03 × 10−2 | 20.19 | −0.122 | |
| hsa-mir-219b_hsa-miR-219b-3p | hsa-miR-219b-3p | −0.40 | 1.03 × 10−2 | 20.19 | −0.122 | |
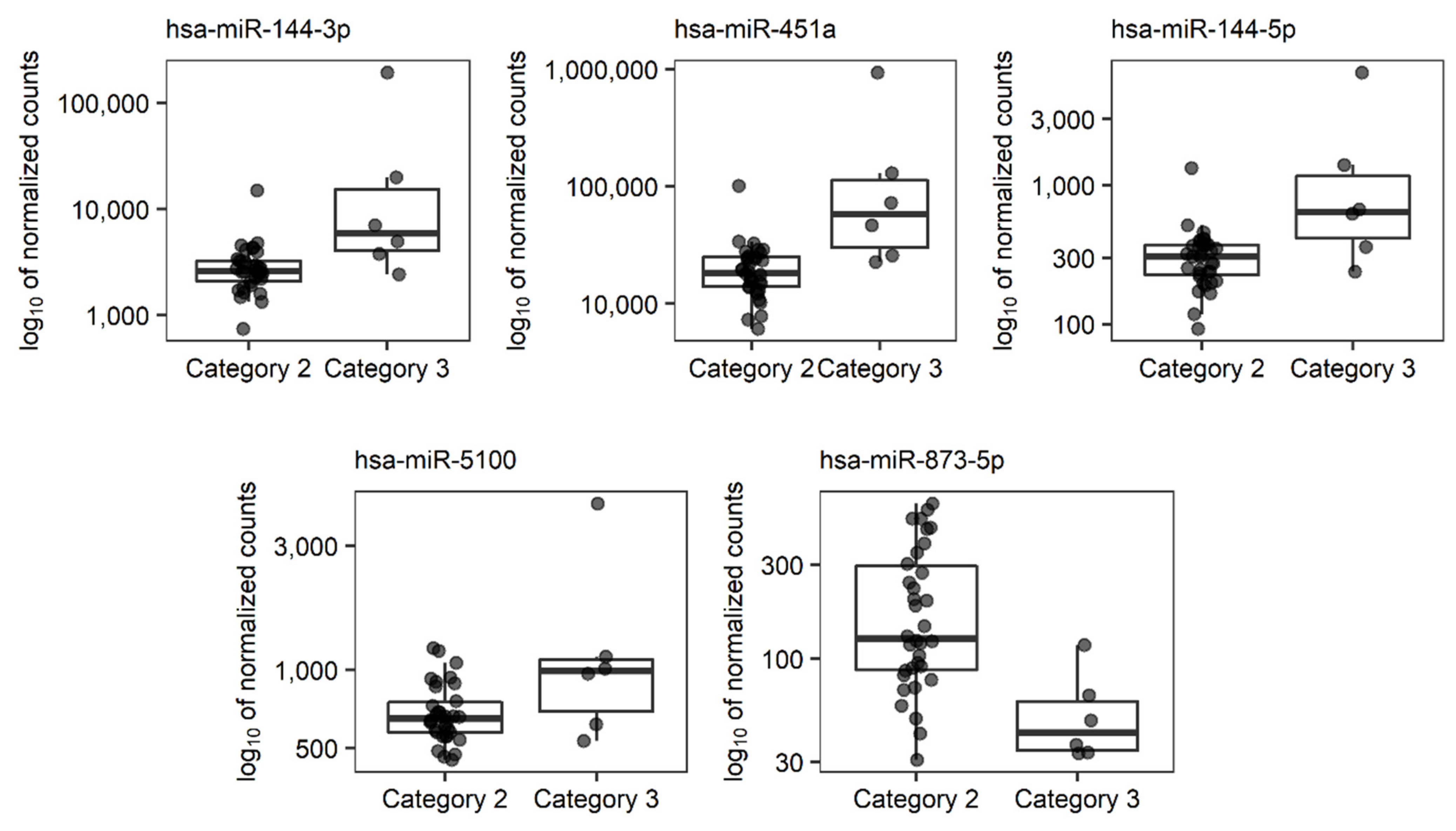
| miRNA Transcript | miRNA ID 1 | p | Fold Change | ROC-AUC |
|---|---|---|---|---|
| hsa-mir-144_hsa-miR-144-3p | hsa-miR-144-3p | 1.256 × 10−10 | 12.435 | 0.843 |
| hsa-mir-451a_hsa-miR-451a | hsa-miR-451a | 5.855 × 10−10 | 9.546 | 0.892 |
| hsa-mir-144_hsa-miR-144-5p | hsa-miR-144-5p | 2.784 × 10−6 | 5.016 | 0.824 |
| hsa-mir-5100_hsa-miR-5100 | hsa-miR-5100 | 1.076 × 10−2 | 2.062 | 0.711 |
| hsa-mir-873_hsa-miR-873-5p | hsa-miR-873-5p | 1.388 × 10−2 | 0.261 | 0.892 |
| Characteristic | miRNA Transcript | miRNA ID 1 | R | p |
|---|---|---|---|---|
| Basophils | hsa-mir-138-1_hsa-miR-138-5p | hsa-miR-138-5p | 0.45 | 3.935 × 10−3 |
| hsa-mir-19b-1_hsa-miR-19b-1-5p | hsa-miR-19b-1-5p | 0.40 | 9.865 × 10−3 | |
| hsa-mir-181b-2_hsa-miR-181b-5p | hsa-miR-181b-5p | 0.40 | 1.002 × 10−2 | |
| Eosinophils | hsa-mir-144_hsa-miR-144-5p | hsa-miR-144-5p | −0.45 | 3.850 × 10−3 |
| hsa-mir-3157_hsa-miR-3157-5p | hsa-miR-3157-5p | 0.41 | 9.200 × 10−3 | |
| hsa-mir-3620_hsa-miR-3620-3p | hsa-miR-3620-3p | −0.40 | 9.839 × 10−3 | |
| Monocytes | hsa-mir-19b-1_hsa-miR-19b-1-5p | hsa-miR-19b-1-5p | 0.66 | 3.432 × 10−6 |
| hsa-mir-181b-2_hsa-miR-181b-5p | hsa-miR-181b-5p | 0.43 | 5.291 × 10−3 | |
| hsa-mir-548au_hsa-miR-548au-5p | hsa-miR-548au-5p | 0.43 | 5.358 × 10−3 | |
| Red blood cells | hsa-mir-5100_hsa-miR-5100 | hsa-miR-5100 | −0.41 | 9.129 × 10−3 |
| Hemoglobin | hsa-mir-5100_hsa-miR-5100 | hsa-miR-5100 | −0.42 | 7.436 × 10−3 |
| Characteristic | miRNA Transcript | miRNA ID 1 | p | Fold Change |
|---|---|---|---|---|
| Hypertension | hsa-mir-5100_hsa-miR-5100 | hsa-miR-5100 | 4.823 × 10−4 | 0.444 |
| Myocardial infarction | hsa-mir-219a-2_hsa-miR-219a-5p | hsa-miR-219a-5p | 4.396 × 10−2 | 0.493 |
| hsa-mir-219b_hsa-miR-219b-3p | hsa-miR-219b-3p | 4.396 × 10−2 | 0.493 | |
| Medication with fibrates | hsa-mir-144_hsa-miR-144-3p | hsa-miR-144-3p | 4.872 × 10−9 | 11.014 |
| hsa-mir-451a_hsa-miR-451a | hsa-miR-451a | 7.336 × 10−7 | 7.602 | |
| hsa-mir-144_hsa-miR-144-5p | hsa-miR-144-5p | 6.029 × 10−6 | 4.415 | |
| hsa-mir-362_hsa-miR-362-5p | hsa-miR-362-5p | 1.280 × 10−2 | 0.327 | |
| Medication with metformin | hsa-mir-362_hsa-miR-362-5p | hsa-miR-362-5p | 5.750 × 10−3 | 0.122 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zalewski, D.P.; Ruszel, K.P.; Stępniewski, A.; Gałkowski, D.; Feldo, M.; Kocki, J.; Bogucka-Kocka, A. Relationships between Indicators of Lower Extremity Artery Disease and miRNA Expression in Peripheral Blood Mononuclear Cells. J. Clin. Med. 2022, 11, 1619. https://doi.org/10.3390/jcm11061619
Zalewski DP, Ruszel KP, Stępniewski A, Gałkowski D, Feldo M, Kocki J, Bogucka-Kocka A. Relationships between Indicators of Lower Extremity Artery Disease and miRNA Expression in Peripheral Blood Mononuclear Cells. Journal of Clinical Medicine. 2022; 11(6):1619. https://doi.org/10.3390/jcm11061619
Chicago/Turabian StyleZalewski, Daniel P., Karol P. Ruszel, Andrzej Stępniewski, Dariusz Gałkowski, Marcin Feldo, Janusz Kocki, and Anna Bogucka-Kocka. 2022. "Relationships between Indicators of Lower Extremity Artery Disease and miRNA Expression in Peripheral Blood Mononuclear Cells" Journal of Clinical Medicine 11, no. 6: 1619. https://doi.org/10.3390/jcm11061619
APA StyleZalewski, D. P., Ruszel, K. P., Stępniewski, A., Gałkowski, D., Feldo, M., Kocki, J., & Bogucka-Kocka, A. (2022). Relationships between Indicators of Lower Extremity Artery Disease and miRNA Expression in Peripheral Blood Mononuclear Cells. Journal of Clinical Medicine, 11(6), 1619. https://doi.org/10.3390/jcm11061619







