Staphylococcus aureus-Derived Extracellular Vesicles Enhance the Efficacy of Endocrine Therapy in Breast Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Extraction from Blood Samples
2.2. Microbiome Analysis of Blood Samples
2.3. Analysis of the Bacterial Composition of the Microbiome
2.4. Extraction of EVs Derived from S. aureus
2.5. Verification of the EVs Derived from S. aureus
2.6. Cell Viability Assay after EV and Tamoxifen Treatment
2.7. Western Blotting after Combination Treatment with S. aureus EVs and Tamoxifen
2.8. Quantitative Real-Time PCR for Signaling Molecule Analysis
2.9. Statistical Analyses
3. Results
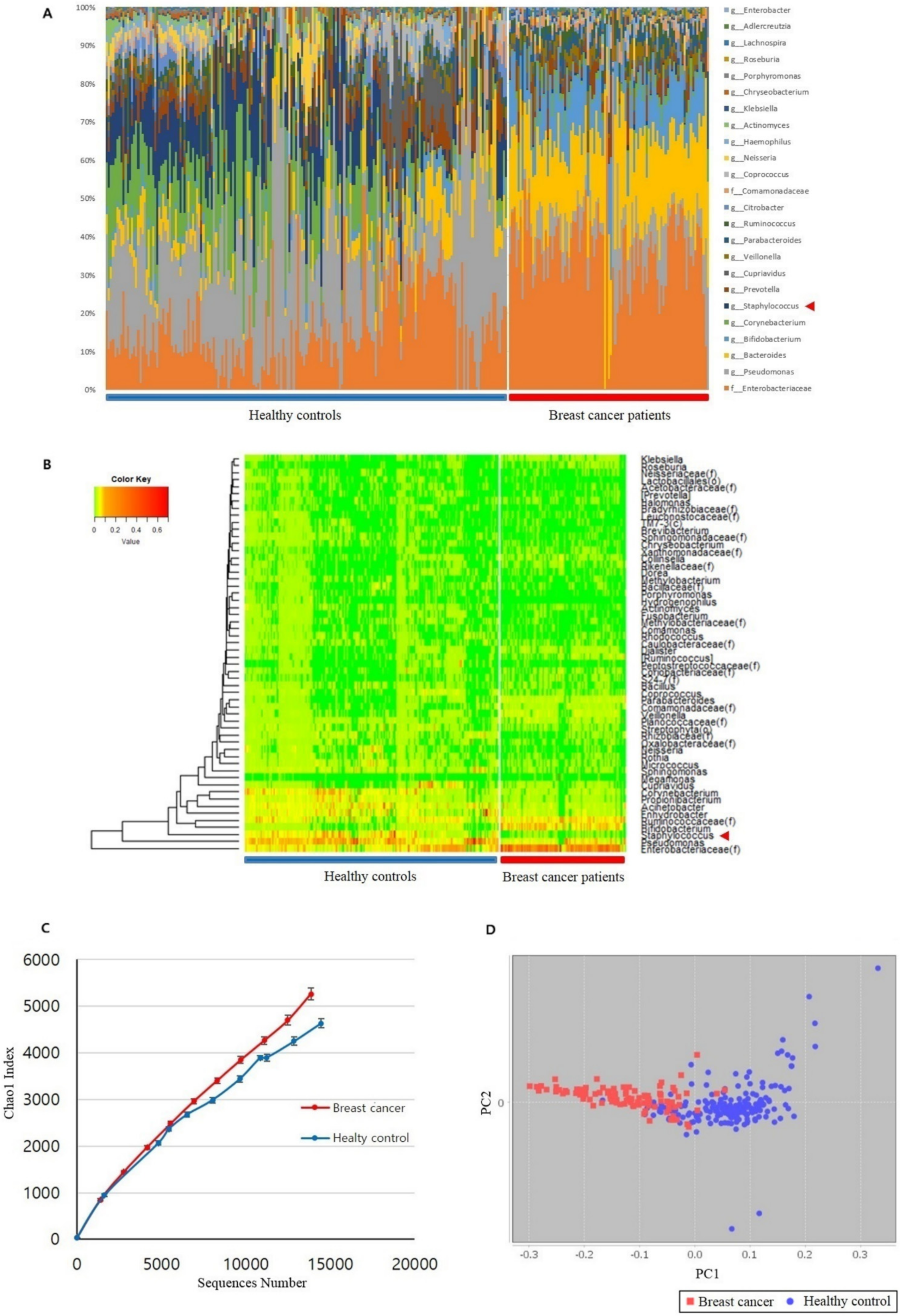
3.1. Microbiome Analysis of Breast Cancer Patients and Healthy Controls
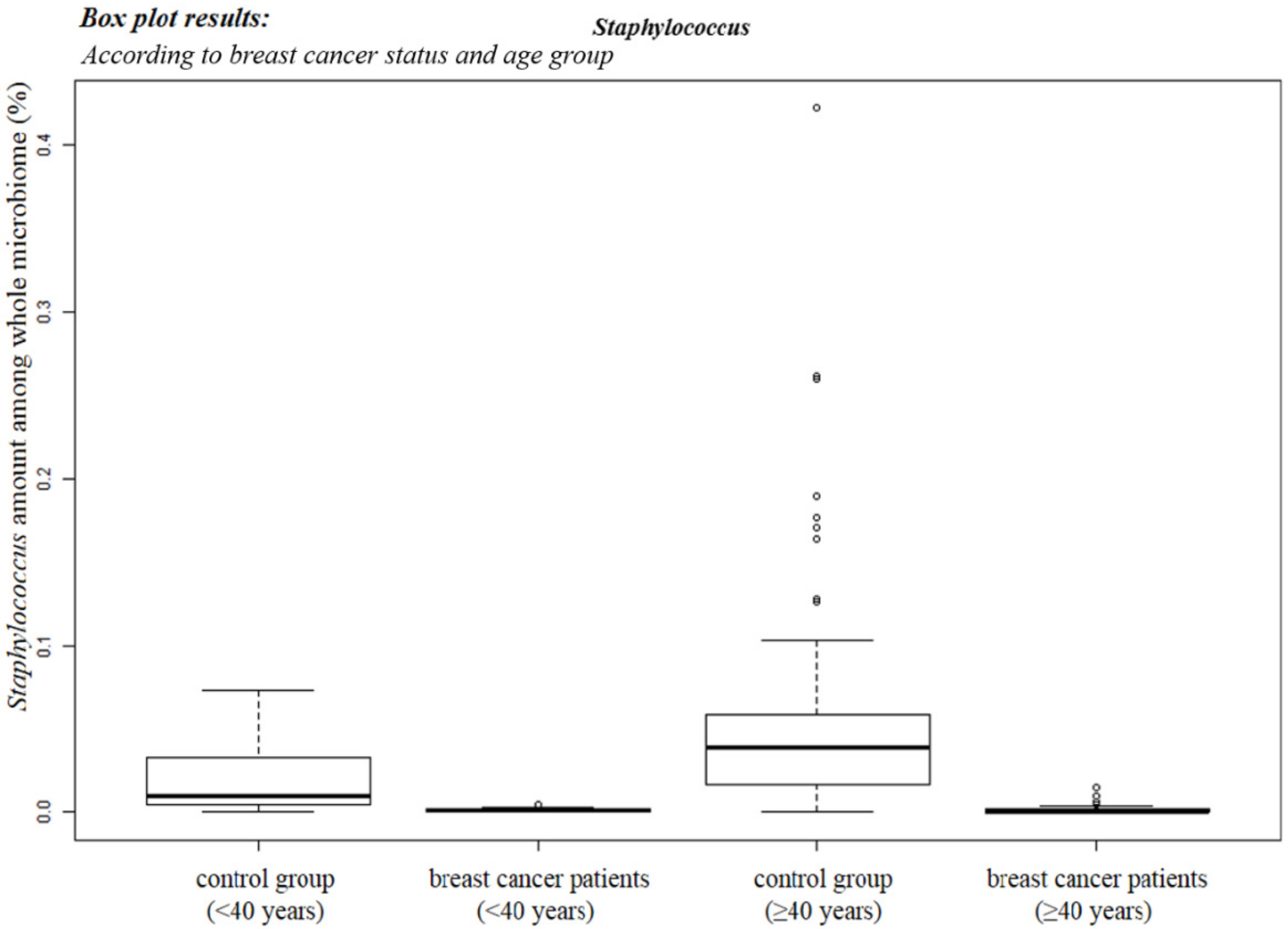
3.2. Staphylococcus sp. Abundance in the Blood Samples
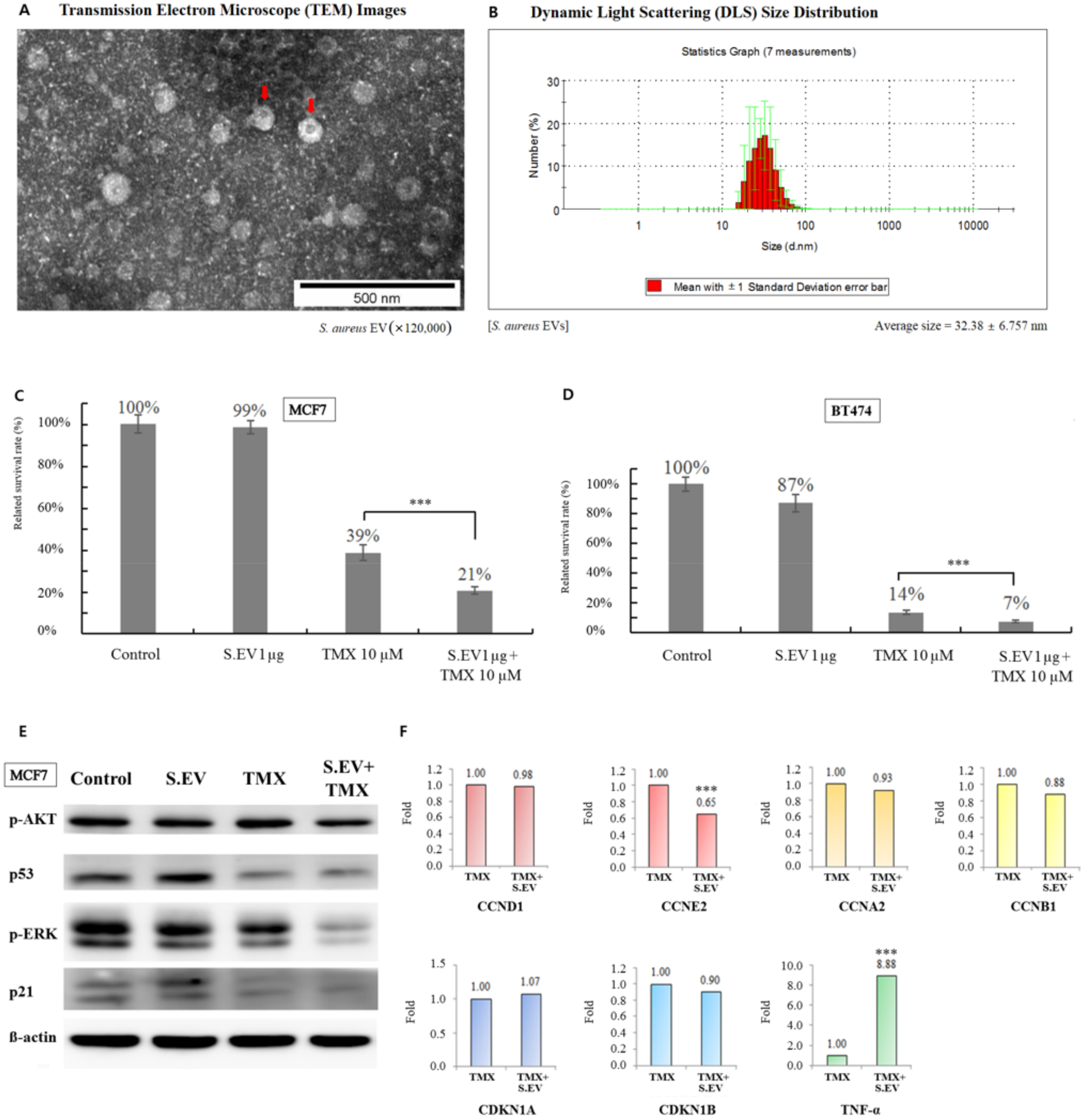
3.3. S. aureus EVs Confirmed by TEM
3.4. S. aureus EVs Enhanced Tamoxifen Efficacy for Breast Cancer Cells
3.5. Signaling Molecules Involved in Increasing Tamoxifen Efficacy
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The first microbial colonizers of the human gut: Composition, activities, and health implications of infant gut microbiota. Microbiol. Mol. Biol. Rev. 2017, 81, e00036-17. [Google Scholar] [CrossRef] [PubMed]
- Poore, G.D.; Kopylova, E.; Zhu, Q.; Carpenter, C.; Fraraccio, S.; Wandro, S.; Kosciolek, T.; Janssen, S.; Metcalf, J.; Song, S.J.; et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020, 579, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Ricci, V.; Carcione, D.; Messina, S.; Colombo, G.I.; D’Alessandra, Y. Circulating 16S RNA in biofluids: Extracellular vesicles as mirrors of human microbiome? Int. J. Mol. Sci. 2020, 21, 38959. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef] [PubMed]
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef]
- Słomka, A.; Urban, S.K.; Lukacs-Kornek, V.; Żekanowska, E.; Kornek, M. Large extracellular vesicles: Have we found the holy grail of inflammation? Front. Immunol. 2018, 9, 2723. [Google Scholar] [CrossRef]
- Zitvogel, L.; Daillère, R.; Roberti, M.P.; Routy, B.; Kroemer, G. Anticancer effects of the microbiome and its products. Nat. Rev. Microbiol. 2017, 15, 465–478. [Google Scholar] [CrossRef]
- Chen, K.L.; Madak-Erdogan, Z. Estrogen and microbiota crosstalk: Should we pay attention? Trends Endocrinol. Metab. 2016, 27, 752–755. [Google Scholar] [CrossRef]
- Kwa, M.; Plottel, C.S.; Blaser, M.J.; Adams, S. The intestinal microbiome and estrogen receptor-positive female breast cancer. J. Natl. Cancer Inst. 2016, 108, djw029. [Google Scholar] [CrossRef]
- Bodai, B.I.; Nakata, T.E. Breast cancer: Lifestyle, the human gut microbiota/microbiome, and survivorship. Perm. J. 2020, 24, 19129. [Google Scholar] [CrossRef] [PubMed]
- Schreurs, M.P.H.; De Vos van Steenwijk, P.J.; Romano, A.; Dieleman, S.; Werner, H.M.J. How the gut microbiome links to menopause and obesity, with possible implications for endometrial cancer development. J. Clin. Med. 2021, 10, 2916. [Google Scholar] [CrossRef] [PubMed]
- Vaz-Luis, I.; Francis, P.A.; Di Meglio, A.; Stearns, V. Challenges in adjuvant therapy for premenopausal women diagnosed with luminal breast cancers. Am. Soc. Clin. Oncol. Educ. Book 2021, 41, e47–e61. [Google Scholar] [CrossRef] [PubMed]
- Laborda-Illanes, A.; Sanchez-Alcoholado, L.; Dominguez-Recio, M.E.; Jimenez-Rodriguez, B.; Lavado, R.; Comino-Méndez, I.; Alba, E.; Queipo-Ortuño, M.I. Breast and gut microbiota action mechanisms in breast cancer pathogenesis and treatment. Cancers 2020, 12, 2465. [Google Scholar] [CrossRef]
- Philley, J.V.; Kannan, A.; Olusola, P.; McGaha, P.; Singh, K.P.; Samten, B.; Griffith, D.E.; Dasgupta, S. Microbiome diversity in sputum of nontuberculous mycobacteria infected women with a history of breast cancer. Cell Physiol. Biochem. 2019, 52, 263–279. [Google Scholar] [CrossRef]
- Ervin, S.M.; Li, H.; Lim, L.; Roberts, L.R.; Liang, X.; Mani, S.; Redinbo, M.R. Gut microbial β-glucuronidases reactivate estrogens as components of the estrobolome that reactivate estrogens. J. Biol. Chem. 2019, 294, 18586–18599. [Google Scholar] [CrossRef]
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef]
- Schneider, D.; Liaw, L.; Daniel, C.; Athanasopoulos, A.N.; Herrmann, M.; Preissner, K.T.; Nawroth, P.P.; Chavakis, T. Inhibition of breast cancer cell adhesion and bone metastasis by the extracellular adherence protein of Staphylococcus aureus. Biochem. Biophys. Res. Commun. 2007, 357, 282–288. [Google Scholar] [CrossRef]
- Wang, X.; Thompson, C.D.; Weidenmaier, C.; Lee, J.C. Release of Staphylococcus aureus extracellular vesicles and their application as a vaccine platform. Nat. Commun. 2018, 9, 1379. [Google Scholar] [CrossRef]
- Jung, A.L.; Schmeck, B.; Wiegand, M.; Bedenbender, K.; Benedikter, B.J. The clinical role of host and bacterial-derived extracellular vesicles in pneumonia. Adv. Drug Deliv. Rev. 2021, 176, 113811. [Google Scholar] [CrossRef]
- Zeng, H.; Umar, S.; Rust, B.; Lazarova, D.; Bordonaro, M. Secondary bile acids and short chain fatty acids in the colon: A focus on colonic microbiome, cell proliferation, inflammation, and cancer. Int. J. Mol. Sci. 2019, 20, 51214. [Google Scholar] [CrossRef] [PubMed]
- Xavier, J.B.; Young, V.B.; Skufca, J.; Ginty, F.; Testerman, T.; Pearson, A.T.; Macklin, P.; Mitchell, A.; Shmulevich, I.; Xie, L.; et al. The cancer microbiome: Distinguishing direct and indirect effects requires a systemic view. Trends Cancer 2020, 6, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Kim, O.Y.; Park, H.T.; Dinh, N.T.H.; Choi, S.J.; Lee, J.; Kim, J.H.; Lee, S.W.; Gho, Y.S. Bacterial outer membrane vesicles suppress tumor by interferon-γ-mediated antitumor response. Nat. Commun. 2017, 8, 626. [Google Scholar] [CrossRef] [PubMed]
- Tulkens, J.; De Wever, O.; Hendrix, A. Analyzing bacterial extracellular vesicles in human body fluids by orthogonal biophysical separation and biochemical characterization. Nat. Protoc. 2020, 15, 40–67. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Defourny, K.A.Y.; Smid, E.J.; Abee, T. Gram-positive bacterial extracellular vesicles and their impact on health and disease. Front. Microbiol. 2018, 9, 1502. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Hong, Y.; Cho, E.; Kim, G.B.; Kim, I.S. Extracellular vesicles as a platform for membrane-associated therapeutic protein delivery. J. Extracell. Vesicles 2018, 7, 1440131. [Google Scholar] [CrossRef]
- Bierman, H.R.; Crile, D.M.; Dod, K.S.; Kelly, K.H.; Petrakis, N.L.; White, L.P.; Shimkin, M.B. Remissions in leukemia of childhood following acute infectious disease: Staphylococcus and streptococcus, varicella, and feline panleukopenia. Cancer 1953, 6, 591–605. [Google Scholar] [CrossRef]
- Hassan, Z.; Mustafa, S.; Rahim, R.A.; Isa, N.M. Anti-breast cancer effects of live, heat-killed and cytoplasmic fractions of Enterococcus faecalis and Staphylococcus hominis isolated from human breast milk. In Vitro Cell. Dev. Biol. Anim. 2016, 52, 337–348. [Google Scholar] [CrossRef]
- Lee, W.H.; Choi, H.I.; Hong, S.W.; Kim, K.S.; Gho, Y.S.; Jeon, S.G. Vaccination with Klebsiella pneumoniae-derived extracellular vesicles protects against bacteria-induced lethality via both humoral and cellular immunity. Exp. Mol. Med. 2015, 47, e183. [Google Scholar] [CrossRef]
- An, J.; Kim, J.B.; Yang, E.Y.; Kim, H.O.; Lee, W.H.; Yang, J.; Kwon, H.; Paik, N.S.; Lim, W.; Kim, Y.K.; et al. Bacterial extracellular vesicles affect endocrine therapy in MCF7 cells. Medicine 2021, 100, e25835. [Google Scholar] [CrossRef]
- Czeczuga-Semeniuk, E.; Lemancewicz, D.; Wołczyński, S. Estradiol and tamoxifen differently affects the inhibitory effects of vitamin A and their metabolites on the proliferation and expression of alpha2beta1 integrins in MCF-7 breast cancer cells. Adv. Med. Sci. 2009, 54, 91–98. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lu, Y.C.; Jiann, B.P.; Chang, H.T.; Huang, J.K.; Chen, W.C.; Su, W.; Jan, C.R. Effect of the anti-breast cancer drug tamoxifen on Ca(2+) movement in human osteosarcoma cells. Pharmacol. Toxicol. 2002, 91, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, B.V.; Kuehn, M.J. Staphylococcus aureus secretes immunomodulatory RNA and DNA via membrane vesicles. Sci. Rep. 2020, 10, 18293. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Sun, T.; Xu, J. Tumor-related microbiome in the breast microenvironment and breast cancer. J. Cancer 2021, 12, 4841–4848. [Google Scholar] [CrossRef]
- Chan, A.A.; Bashir, M.; Rivas, M.N.; Duvall, K.; Sieling, P.A.; Pieber, T.R.; Vaishampayan, P.A.; Love, S.M.; Lee, D.J. Characterization of the microbiome of nipple aspirate fluid of breast cancer survivors. Sci. Rep. 2016, 6, 28061. [Google Scholar] [CrossRef]
- Bleach, R.; Sherlock, M.; O’Reilly, M.W.; McIlroy, M. Growth hormone/insulin growth factor axis in sex steroid associated disorders and related cancers. Front. Cell Dev. Biol. 2021, 9, 630503. [Google Scholar] [CrossRef]
- Kahlert, S.; Nuedling, S.; Van Eickels, M.; Vetter, H.; Meyer, R.; Grohe, C. Estrogen receptor alpha rapidly activates the IGF-1 receptor pathway. J. Biol. Chem. 2000, 275, 18447–18453. [Google Scholar] [CrossRef]
- Shi, D.; Zhao, P.; Cui, L.; Li, H.; Sun, L.; Niu, J.; Chen, M. Inhibition of PI3K/AKT molecular pathway mediated by membrane estrogen receptor GPER accounts for cryptotanshinone induced antiproliferative effect on breast cancer SKBR-3 cells. BMC Pharmacol. Toxicol. 2020, 21, 32. [Google Scholar] [CrossRef]
- Lam, L.; Hu, X.; Aktary, Z.; Andrews, D.W.; Pasdar, M. Tamoxifen and ICI 182,780 increase Bcl-2 levels and inhibit growth of breast carcinoma cells by modulating PI3K/AKT, ERK and IGF-1R pathways independent of ERalpha. Breast Cancer Res. Treat. 2009, 118, 605–621. [Google Scholar] [CrossRef]
- Radenkovic, S.; Konjevic, G.; Nikitovic, M.; Stojanovic Rundic, S.; Plesinac Karapandzic, V.; Milovic Kovacevic, M.; Jurisic, V. Evaluation of Cyclin D1 expression by western blotting methods and immunohistochemistry in breast cancer patients. J. BUON 2021, 26, 475–482. [Google Scholar]
- Caldon, C.E.; Sergio, C.M.; Kang, J.; Muthukaruppan, A.; Boersma, M.N.; Stone, A.; Barraclough, J.; Lee, C.S.; Black, M.A.; Miller, L.D.; et al. Cyclin E2 overexpression is associated with endocrine resistance but not insensitivity to CDK2 inhibition in human breast cancer cells. Mol. Cancer Ther. 2012, 11, 1488–1499. [Google Scholar] [CrossRef] [PubMed]


| Genus | β-glucuronidase | β-galactosidase |
|---|---|---|
| Actinomyces | − | + |
| Alistipes | − | + |
| Anaerostipes | − | + |
| Bacteroides | − | + |
| Bifidobacterium | − | + |
| Blautia | − | + |
| Catenibacterium | − | + |
| Citrobacter | + | + |
| Clostridium | + | + |
| Collinsella | + | − |
| Coprococcus | − | + |
| Dorea | − | + |
| Enterococcus | − | + |
| Eubacterium | − | + |
| Faecalibacterium | + | + |
| Fusobacterium | − | + |
| Holdemania | − | + |
| Klebsiella | − | + |
| Lactobacillus | + | + |
| Megamonas | − | + |
| Parabacteroides | − | + |
| Paraprevotella | − | + |
| Porphyromonas | − | + |
| Prevotella | − | + |
| Roseburia | + | + |
| Ruminococcus | − | + |
| Staphylococcus | − | + |
| Subdoligranulum | − | + |
| Weissella | − | + |
| Gene | Type | Sequence (5′–3′) | MER | TM (°C) | GC (%) | Size (bp) |
|---|---|---|---|---|---|---|
| CCND1 | Fw | CTCTGTGCCACAGATGTGAAG | 21 | 57.9 | 52 | 170 |
| Rv | GAGGCAGTCCGGGTCACAC | 19 | 57.2 | 68 | ||
| CCNE2 | Fw | CTGGCTTTTAGAGGTATGTGAAG | 23 | 56.0 | 43 | 162 |
| Rv | AGCATAGATTTCCTCAAGTTTGG | 23 | 55.3 | 39 | ||
| CCNA2 | Fw | GGACAAAGCTGGCCTGAATC | 20 | 55.9 | 55 | 166 |
| Rv | GGAGAGAAACACCATGATACTTTG | 24 | 55.2 | 42 | ||
| CCNB1 | Fw | ATAATGGTGAATGGACACCAACTC | 24 | 55.1 | 42 | 143 |
| Rv | ATACTTGTTCTTGACAGTCATGTG | 24 | 54.8 | 38 | ||
| CDKN1A | Fw | ACCATGTGGACCTGTCACTG | 20 | 54.8 | 55 | 135 |
| Rv | TGGAGTGGTAGAAATCTGTCATG | 23 | 55.5 | 43 | ||
| CDKN1B | Fw | GACCTGCAACCGACGATTC | 19 | 55.2 | 58 | 156 |
| Rv | TATTCTTAATTCGAGCTGTTTACG | 24 | 55.3 | 33 | ||
| TNF-α | Fw | AGGCAGTCAGATCATCTTCTC | 21 | 55.6 | 48 | 162 |
| Rv | CTGATGGCACCACCAGCTG | 19 | 57.9 | 63 | ||
| GAPDH | Fw | GAGTCAACGGATTTGGTCG | 19 | 57.5 | 53 | 133 |
| Rv | TGGAATCATATTGGAACATGTAAAC | 25 | 57.8 | 32 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, J.; Kwon, H.; Lim, W.; Moon, B.-I. Staphylococcus aureus-Derived Extracellular Vesicles Enhance the Efficacy of Endocrine Therapy in Breast Cancer Cells. J. Clin. Med. 2022, 11, 2030. https://doi.org/10.3390/jcm11072030
An J, Kwon H, Lim W, Moon B-I. Staphylococcus aureus-Derived Extracellular Vesicles Enhance the Efficacy of Endocrine Therapy in Breast Cancer Cells. Journal of Clinical Medicine. 2022; 11(7):2030. https://doi.org/10.3390/jcm11072030
Chicago/Turabian StyleAn, Jeongshin, Hyungju Kwon, Woosung Lim, and Byung-In Moon. 2022. "Staphylococcus aureus-Derived Extracellular Vesicles Enhance the Efficacy of Endocrine Therapy in Breast Cancer Cells" Journal of Clinical Medicine 11, no. 7: 2030. https://doi.org/10.3390/jcm11072030
APA StyleAn, J., Kwon, H., Lim, W., & Moon, B.-I. (2022). Staphylococcus aureus-Derived Extracellular Vesicles Enhance the Efficacy of Endocrine Therapy in Breast Cancer Cells. Journal of Clinical Medicine, 11(7), 2030. https://doi.org/10.3390/jcm11072030







