Combination of HLA-DQ2/-DQ8 Haplotypes and a Single MSH5 Gene Variant in a Polish Population of Patients with Type 1 Diabetes as a First Line Screening for Celiac Disease?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Serological CD Screening
2.3. DNA Isolation
2.4. SNP Genotyping
2.5. Statistics
3. Results
3.1. HLA-DQ Haplotype Occurrence in T1D Patients
3.2. rs3130484 Typing for CD Screening in T1D Patients
3.3. Diagnostic Value of CD-Specific Haplotypes and the rs3130484 Variant of the MSH5 Gene
4. Discussion
Strengths and Limitations of the Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Green, P.H.R.; Lebwohl, B.; Greywoode, R. Celiac disease. J. Allergy Clin. Immunol. 2015, 135, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Arora, A.; Strand, T.A.; Leffler, D.A.; Catassi, C.; Green, P.H.; Kelly, C.P.; Ahuja, V.; Makharia, G.K. Global Prevalence of Celiac Disease: Systematic Review and Meta-analysis. Clin. Gastroenterol. Hepatol. 2018, 16, 823–836.e2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubio–Tapia, A.; Kyle, R.A.; Kaplan, E.L.; Johnson, D.R.; Page, W.; Erdtmann, F.; Brantner, T.L.; Kim, W.R.; Phelps, T.K.; Lahr, B.D.; et al. Increased Prevalence and Mortality in Undiagnosed Celiac Disease. Gastroenterology 2009, 137, 88–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- West, J.; Logan, R.F.A.; Smith, C.J.; Hubbard, R.B.; Card, T.R. Malignancy and mortality in people with coeliac disease: Population based cohort study. BMJ 2004, 329, 716–719. [Google Scholar] [CrossRef] [Green Version]
- Choung, R.S.; Larson, S.A.; Khaleghi, S.; Rubio-Tapia, A.; Ovsyannikova, I.G.; King, K.S.; Larson, J.J.; Lahr, B.D.; Poland, G.A.; Camilleri, M.J.; et al. Prevalence and Morbidity of Undiagnosed Celiac Disease from a Community-Based Study. Gastroenterology 2017, 152, 830–839.e5. [Google Scholar] [CrossRef] [Green Version]
- Majsiak, E.; Choina, M.; Golicki, D.; Gray, A.M.; Cukrowska, B. The impact of symptoms on quality of life before and after diagnosis of coeliac disease: The results from a Polish population survey and comparison with the results from the United Kingdom. BMC Gastroenterol. 2021, 21, 105. [Google Scholar] [CrossRef]
- Husby, S.; Koletzko, S.; Korponay-Szabó, I.R.; Mearin, M.L.; Phillips, A.; Shamir, R.; Troncone, R.; Giersiepen, K.; Branski, D.; Catassi, C.; et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J. Pediatr. Gastroenterol. Nutr. 2012, 54, 136–160. [Google Scholar] [CrossRef]
- Megiorni, F.; Pizzuti, A. HLA-DQA1 and HLA-DQB1 in Celiac disease predisposition: Practical implications of the HLA molecular typing. J. Biomed. Sci. 2012, 19, 88. [Google Scholar] [CrossRef]
- Binder, E.; Loinger, M.; Mühlbacher, A.; Edlinger, M.; Steichen, E.; Meraner, D.; Loacker, L.; Weigel, G.; Müller, T.; Fröhlich-Reiterer, E.; et al. Genotyping of coeliac-specific human leucocyte antigen in children with type 1 diabetes: Does this screening method make sense? Arch. Dis. Child. 2016, 102, 603–606. [Google Scholar] [CrossRef]
- Martín, M.B.R.; Romero, C.M.; Vilches, E.G.; Usabiaga, J.R.; Castellanos, R.B.; Frías, M.M.; Oliver, D.P.; Salces, C.C. Celiac disease screening in children and adolescents with type 1 diabetes mellitus: What test should be performed? Endocrinol. Diabetes Nutr. 2021, 68, 153–158. [Google Scholar] [CrossRef]
- Elias, J.; Hoorweg-Nijman, J.J.G.; Balemans, W.A. Clinical relevance and cost-effectiveness of HLA genotyping in children with Type 1 diabetes mellitus in screening for coeliac disease in the Netherlands. Diabet. Med. 2014, 32, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Wijmenga, C.; Withoff, S. From genome-wide association studies to disease mechanisms: Celiac disease as a model for autoimmune diseases. Semin. Immunopathol. 2012, 34, 567–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trynka, G.; Hunt, K.A.; Bockett, N.A.; Romanos, J.; Mistry, V.; Szperl, A.; Bakker, S.F.; Bardella, M.T.; Bhaw-Rosun, L.; Castillejo, G.; et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Genet. 2011, 43, 1193–1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almeida, R.; Ricaño-Ponce, I.; Kumar, V.; Deelen, P.; Szperl, A.; Trynka, G.; Gutierrez-Achury, J.; Kanterakis, A.; Westra, H.-J.; Franke, L.; et al. Fine mapping of the celiac disease-associated LPP locus reveals a potential functional variant. Hum. Mol. Genet. 2013, 23, 2481–2489. [Google Scholar] [CrossRef] [Green Version]
- Paziewska, A.; Cukrowska, B.; Dąbrowska, M.; Goryca, K.; Piatkowska, M.; Kluska, A.; Mikula, M.; Karczmarski, J.; Oralewska, B.; Rybak, A.; et al. Combination Testing Using a Single MSH5 Variant alongside HLA Haplotypes Improves the Sensitivity of Predicting Coeliac Disease Risk in the Polish Population. PLoS ONE 2015, 10, e0139197. [Google Scholar] [CrossRef]
- Oliynyk, R.T. Evaluating the Potential of Younger Cases and Older Controls Cohorts to Improve Discovery Power in Genome-Wide Association Studies of Late-Onset Diseases. J. Pers. Med. 2019, 9, 38. [Google Scholar] [CrossRef] [Green Version]
- Husby, S.; Koletzko, S.; Korponay-Szabó, I.; Kurppa, K.; Mearin, M.L.; Ribes-Koninckx, C.; Shamir, R.; Troncone, R.; Auricchio, R.; Castillejo, G.; et al. European Society Paediatric Gastroenterology, Hepatology and Nutrition Guidelines for Diagnosing Coeliac Disease 2020. J. Pediatr. Gastroenterol. Nutr. 2020, 70, 141–156. [Google Scholar] [CrossRef] [Green Version]
- Oberhuber, G.; Granditsch, G.; Vogelsang, H. The histopathology of coeliac disease: Time for a standardized report scheme for pathologists. Eur. J. Gastroenterol. Hepatol. 1999, 11, 1185–1194. [Google Scholar] [CrossRef]
- Poulain, C.; Johanet, C.; Delcroix, C.; Lévy-Marchal, C.; Tubiana-Rufi, N. Prevalence and clinical features of celiac disease in 950 children with type 1 diabetes in France. Diabetes Metab. 2007, 33, 453–458. [Google Scholar] [CrossRef]
- Pham-Short, A.; Donaghue, K.C.; Ambler, G.; Phelan, H.; Twigg, S.; Craig, M.E. Screening for Celiac Disease in Type 1 Diabetes: A Systematic Review. Pediatrics 2015, 136, e170–e176. [Google Scholar] [CrossRef] [Green Version]
- Oujamaa, I.; Sebbani, M.; Elmoumou, L.; Bourrahouate, A.; El Qadiry, R.; El Moussaoui, S.; Sab, I.A.; Sbihi, M.; Ennazk, L.; El Mghari-Tabib, G.; et al. The Prevalence of Celiac Disease-Specific Auto-Antibodies in Type 1 Diabetes in a Moroccan Population. Int. J. Endocrinol. 2019, 19, 7895207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elfström, P.; Sundstrom, J.; Ludvigsson, J.F. Systematic review with meta-analysis: Associations between coeliac disease and type 1 diabetes. Aliment. Pharmacol. Ther. 2014, 40, 1123–1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moravej, H.; Zamanfar, D.; Aghamahdi, F.; Hashemipour, M.; Mirrashidi, F.S.; Ghaemi, N.; Eshraghi, P.; Ilkhanipoor, H.; Amirhakimi, A.; Yazdani, N.; et al. Optimal Frequency to Screen Celiac Disease amongst Patients with Type 1 Diabetes Mellitus: A Multicenter Study. Prim. Care Diabetes 2021, 15, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, F.H.; Clarke, A.B.; Joachim, K.C.; Assor, E.; McDonald, C.; Saibil, F.; Lochnan, H.A.; Punthakee, Z.; Parikh, A.; Advani, A.; et al. Screening and Treatment Outcomes in Adults and Children with Type 1 Diabetes and Asymptomatic Celiac Disease: The CD-DIET Study. Diabetes Care 2020, 43, 1553–1556. [Google Scholar] [CrossRef]
- Mohn, A.; Cerruto, M.; Iafusco, D.; Prisco, F.; Tumini, S.; Stoppoloni, O.; Chiarelli, F. Celiac Disease in Children and Adolescents with Type I Diabetes: Importance of Hypoglycemia. J. Pediatr. Gastroenterol. Nutr. 2001, 32, 37–40. [Google Scholar] [CrossRef]
- Hansen, D.; Brock-Jacobsen, B.; Lund, E.; Bjørn, C.; Hansen, L.P.; Nielsen, C.; Fenger, C.; Lillevang, S.T.; Husby, S. Clinical Benefit of a Gluten-Free Diet in Type 1 Diabetic Children with Screening-Detected Celiac Disease: A population-based screening study with 2 years’ follow-up. Diabetes Care 2006, 29, 2452–2456. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Puttha, R.; Ghezaiel, S.; Skae, M.; Cooper, C.; Amin, R.; on behalf of the North West England Paediatric Diabetes Network. The effect of biopsy-positive silent coeliac disease and treatment with a gluten-free diet on growth and glycaemic control in children with Type 1 diabetes. Diabet. Med. 2009, 26, 1250–1254. [Google Scholar] [CrossRef]
- Mahmud, F.H.; Elbarbary, N.S.; Fröhlich-Reiterer, E.; Holl, R.W.; Kordonouri, O.; Knip, M.; Simmons, K.; Craig, M.E. ISPAD Clinical Practice Consensus Guidelines 2018: Other complications and associated conditions in children and adolescents with type 1 diabetes. Pediatr. Diabetes 2018, 19, 275–286. [Google Scholar] [CrossRef]
- American Diabetes Association 12. Children and Adolescents: Standards of Medical Care in Diabetes—2018. Diabetes Care 2017, 41, S126–S136. [Google Scholar] [CrossRef] [Green Version]
- Tye-Din, J.; Cameron, D.J.S.; Daveson, A.J.; Day, A.S.; Dellsperger, P.; Hogan, C.; Newnham, E.D.; Shepherd, S.J.; Steele, R.H.; Wienholt, L.; et al. Appropriate clinical use of human leukocyte antigen typing for coeliac disease: An Australasian perspective. Intern. Med. J. 2015, 45, 441–450. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, R.T.; Sun, A.; Mayo, A.; Forgan, M.; Comrie, A.; Gillett, P.M. Coeliac screening in a Scottish cohort of children with type 1 diabetes mellitus: Is DQ typing the way forward? Arch. Dis. Child. 2015, 101, 230–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Contreas, G.; Valletta, E.; Ulmi, D.; Cantoni, S.; Pinelli, L. Screening of coeliac disease in north Italian children with type 1 diabetes: Limited usefulness of HLA-DQ typing. Acta Paediatr. 2004, 93, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Deja, G.; Sikora, D.; Pyziak-Skupien, A.; Klenczar, K.; Deja, R.; Jarosz-Chobot, P. The Usefulness of Genotyping of Celiac Disease-Specific HLA among Children with Type 1 Diabetes in Various Clinical Situations. J. Diabetes Res. 2020, 2020, 7869350. [Google Scholar] [CrossRef] [PubMed]
- Romanos, J.; Rosén, A.; Kumar, V.; Trynka, G.; Franke, L.; Szperl, A.; Gutierrez-Achury, J.; van Diemen, C.; Kanninga, R.; Jankipersadsing, S.; et al. Improving coeliac disease risk prediction by testing non-HLA variants additional to HLA variants. Gut 2013, 63, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Sekine, H.; Ferreira, R.C.; Pan-Hammarström, Q.; Graham, R.R.; Ziemba, B.; de Vries, S.S.; Liu, J.; Hippen, K.; Koeuth, T.; Ortmann, W.; et al. Role for Msh5 in the regulation of Ig class switch recombination. Proc. Natl. Acad. Sci. USA 2007, 104, 7193–7198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Her, C. MutS Homologues hMSH4 and hMSH5: Genetic Variations, Functions, and Implications in Human Diseases. Curr. Genom. 2013, 14, 81–90. [Google Scholar] [CrossRef] [Green Version]
- Valdes, A.M.; Thomson, G. Type 1 Diabetes Genetics Consortium Several loci in the HLA class III region are associated with T1D risk after adjusting for DRB1-DQB1. Diabetes Obes. Metab. 2009, 11, 46–52. [Google Scholar] [CrossRef] [Green Version]
- Monsuur, A.J.; de Bakker, P.I.W.; Zhernakova, A.; Pinto, D.; Verduijn, W.; Romanos, J.; Auricchio, R.; Lopez, A.; van Heel, D.A.; Crusius, J.B.A.; et al. Effective Detection of Human Leukocyte Antigen Risk Alleles in Celiac Disease Using Tag Single Nucleotide Polymorphisms. PLoS ONE 2008, 3, e2270. [Google Scholar] [CrossRef] [Green Version]
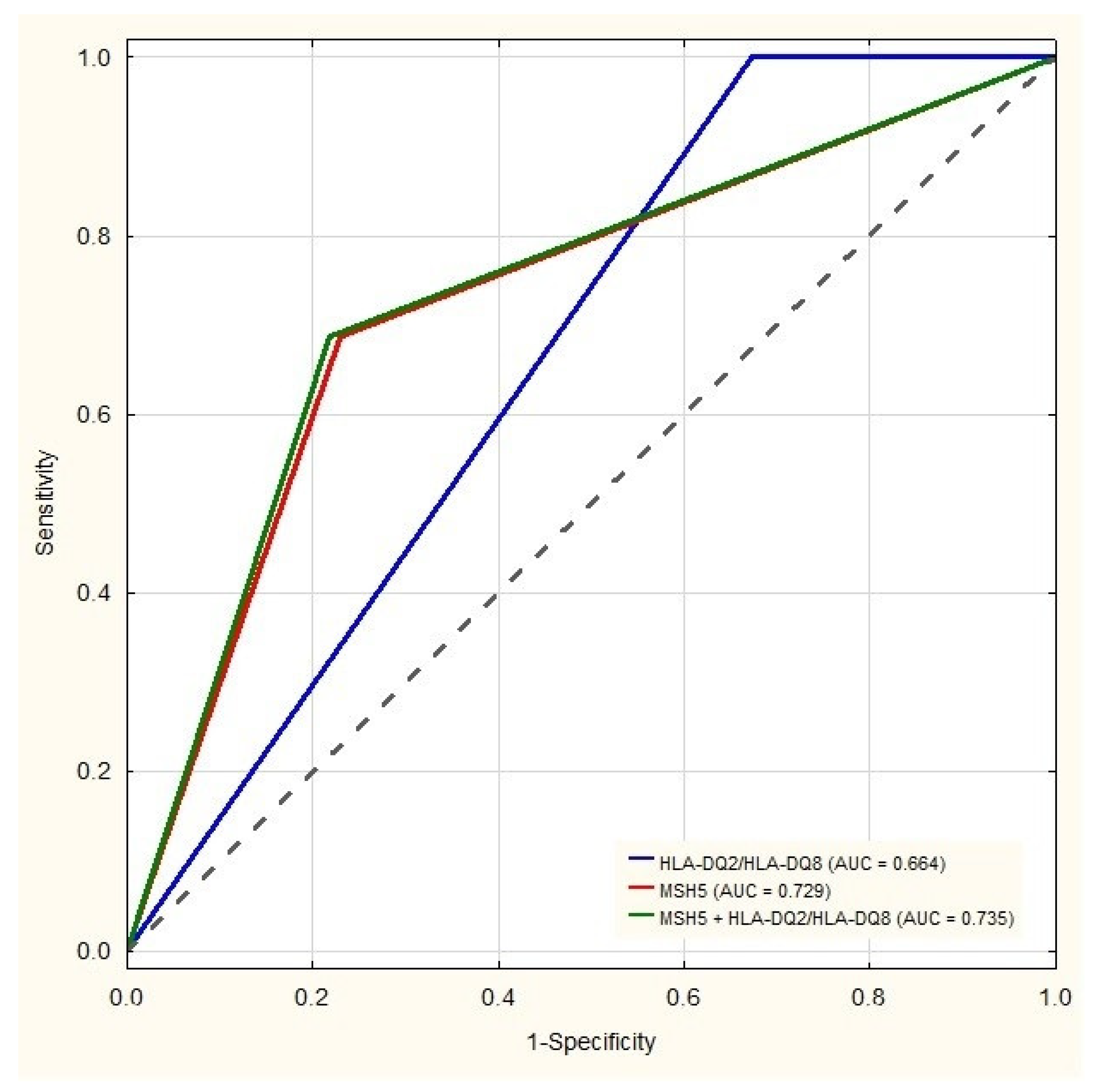
| T1D | T1D + CD | CD | Healthy Controls | |
|---|---|---|---|---|
| Total number | 228 | 20 | 287 | 551 |
| Median age (in years) | 9 | 5 * | 4 * | 28 |
| Age range (in years) | 9/12–18 | 2–11 | 1–17 | 18–68 |
| Female number (%) | 126 (55.3%) | 11 (55.0%) | 185 (64.5%) | 403 (73.1%) |
| Male number (%) | 102 (47.3%) | 9 (45.0%) | 102 (35.5%) | 148 (26.9%) |
| SNP | Assay ID * | Haplotype | Region/Gene | Allele/MAF ** | Genotype |
|---|---|---|---|---|---|
| rs7454108 | c_298171179_10 | DQ8 | N/A | C = 0.1018 | (T;T) |
| (C;T) | |||||
| (C;C) | |||||
| rs2187668 | c_58662585_10 | DQ2.5 | HLA-DQA1: Intron Variant | T = 0.1194 | (T;T) |
| (C;T) | |||||
| (C;C) | |||||
| rs2395182 | c_11409965_10 | DQ2.2 | HLA-DRA: 500B Downstream Variant | G = 0.2075 | (T;T) |
| (G;T) | |||||
| (G;G) | |||||
| rs7775228 | c_29315313_10 | DQ2.2 | N/A | C = 0.1287 | (T;T) |
| (C;T) | |||||
| (C;C) | |||||
| rs3130484 | C__30535385_10 | MSH5 | MSH5: Intron Variant | C = 0.0990 | (T;T) |
| (C;T) | |||||
| (C;C) |
| Haplotype | T1D n = 228 | T1D + CD n = 20 | Statistics T1D vs. T1D + CD | CD n = 287 | Statistics T1D vs. CD | Controls n = 551 | Statistics T1D vs. Controls | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n (%) | n(%) | p | OR (95% CI) | p # | n (%) | OR (95% CI) | p # | n (%) | OR (95% CI) | p # | |
| HLA-DQ8 | 128 (56.1%) | 13 (60%) | 0.49 | 1.4 (0.5–4.5) | 0.54 | 49 (17.1%) | 6.2 (4.1–9.5) | 5.98 × 10−20 | 98 (17.79%) | 5.90 (4.15–8.44) | 3.72 × 10−24 |
| HLA-DQ2.5 | 99 (43.4%) | 16 (80%) | 0.002 | 5.2 (1.6–22.0) | 0.04 | 234 (81.53%) | 5.7 (3.8–8.7) | 1.10 × 10−18 | 136 (24.68%) | 2.34 (1.67–3.28) | 5.05 × 10−7 |
| HLA-DQ2.2 | 38 (16.7%) | 2 (10%) | 0.75 | 1.8 (0.4–16.6) | 0.79 | 112 (39.0%) | 3.2 (2.1–5.0) | 3.05 × 10−8 | 142 (25.77%) | 0.58 (0.38–0.87) | 0.008 |
| HLA-DQ2 | 128 (65.1%) | 17 (85%) | 0.02 | 4.4 (1.2–24.1) | 0.06 | 276 (96.2%) | 19.5 (10.0–41.7) | 2.34 × 10−28 | 263 (47.73%) | 1.40 (1.02–1.94) | 0.04 |
| HLA-DQ2.2/HLA-DQ8 * | 156 (68.4%) | 15 (70%) | 1 | 1.1 (0.4–3.6) | 1 | 161 (55.1%) | 1.8 (182–2.6) | 0.002 | 222 (40.29%) | 3.23 (2.31–4.56) | 137 × 10−12 |
| HLA-DQ2.5/HLA-DQ8 * | 173 (75.9%) | 20 (100%) | 0.009 | 6.2 (0.8–215.2) | 0.05 | 248 (86.4%) | 2.0 (1.2–3.2) | 0.004 | 223 (40.47%) | 473 (330–6.85) | 2.28 × 10−19 |
| HLA-DQ2/HLA-DQ8 * | 192 (84.2%) | 20 (100%) | 0.09 | 3.7 (0.5–131.4) | 0.14 | 287 (100%) | 53.5 (6.7–1787.0) | 6.31 × 10−14 | 332 (60.25%) | 3.54 (2.36–5.42) | 5.03 × 10−11 |
| Genotype | T1D n = 228 | T1D + CD n = 20 | Statistics T1D vs. T1D + CD | CD n = 287 | Statistics T1D vs. CD | Controls n = 551 | Statistics T1D vs. Controls | Statistics CD vs. Controls | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n (%) | n(%) | p | OR (95%CI) | p # | n (%) | OR (95%CI) | p # | n (%) | OR (95%CI) | p # | OR (95%CI) | p # | |
| MSH5 | 81 (35.5%) | 12 (60%) | 0.05 | 2.7 (1.0–8.0) | 0.09 | 199 (69.3%) | 4.1 (2.8–6.0) | 4.15 × 10−14 | 98 (17.8%) | 2.54 (1.8–3.7) | 3.01 × 10−7 | 10.4 (7.4–14.8) | 3.87 × 10−49 |
| MSH5 + HLA-DQ8 | 113 (49.6%) | 13 (65%) | 0.39 | 1.7 (0.5–5.0) | 0.47 | 256 (89.2%) | 2.3 (1.4–4.0) | 0.001 | 226 (41.0%) | 17.08 (7.8–42.7) | 4.11 × 10−18 | 7.3 (3.2–18.8) | 7.22 × 10−8 |
| MSH5 + HLA-DQ2.5 | 102 (44.7%) | 16 (80%) | 0.03 | 2.9 (1.0–8.5) | 0.06 | 234 (81.5%) | 4.3 (3.0–6.4) | 3.99 × 10−15 | 150 (27.2%) | 2.89 (2.0–4.2) | 2.08 × 10−8 | 12.5 (8.8–18.0) | 2.03 × 10−54 |
| MSH5 + HLA-DQ2.2 | 131 (57.5%) | 17 (85%) | 0.45 | 1.9 (0.0–17.3) | 0.52 | 276 (96.2%) | 10.1 (4.2–29.3) | 6.94 × 10−11 | 272 (49.4%) | 1.04 (0.3–2.9) | 1 | 10.5 (5.6–20.8) | 1.28 × 10−17 |
| MSH5 + HLA-DQ2 | 163 (71.5%) | 19 (95%) | 0.03 | 2.9 (1.0–8.5) | 0.06 | 219 (76.3%) | 4.3 (3.0–6.4) | 3.99 × 10−15 | 188 (34.1%) | 2.70 (1.9–3.9) | 1.00 × 10−7 | 11.7 (8.3–16.7) | 1.74 × 10−52 |
| MSH5 + HLA-DQ2.2/ HLA-DQ8 * | 175 (76.8%) | 20 (100%) | 0.27 | 1.8 (0.6–5.2) | 0.35 | 248 (86.4%) | 1.4 (0.9–2.1) | 0.13 | 235 (42.65%) | 7.08 (4.1–12.6) | 5.81 × 10−14 | 9.8 (5.9–16.9) | 9.59 × 10−24 |
| MSH5 + HLA-DQ2.5/ HLA-DQ8 * | 185 (81.1%) | 19 (95%) | 0.03 | 2.8 (1.0–8.3) | 0.06 | 287 (100%) | 4.3 (2.9–6.3) | 1.10 × 10−14 | 235 (42.65%) | 2.94 (2.0–4.3) | 1.32 × 10−8 | 12.5 (8.8–18.0) | 2.03 × 10−54 |
| MSH + HLA-DQ2/ HLA-DQ8 * | 194 (85.1%) | 20 (100%) | 0.03 | 2.8 (1.0–8.3) | 0.06 | 287 (100%) | 4.3 (2.9–6.3) | 1.10 × 10−14 | 339 (61.52%) | 2.68 (1.85–3.87) | 1.07 × 10−7 | 11.4 (8.0–16.2) | 7.39 × 10−52 |
| Haplotypes/MSH5 | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | ACC (%) |
|---|---|---|---|---|---|
| HLA-DQ2 | 95.8 | 49.8 | 42.9 | 96.8 | 62.8 |
| HLA-DQ2.2 | 77.2 | 76.9 | 56.8 | 89.5 | 77.0 |
| HLA-DQ2.5 | 41.7 | 69.8 | 35.3 | 75.2 | 61.9 |
| HLA-DQ8 | 20.2 | 71.0 | 21.5 | 69.3 | 56.6 |
| HLA-DQ2.2/HLA DQ8 | 85.3 | 51.5 | 40.9 | 89.9 | 61.0 |
| HLA-DQ2.5/HLA-DQ8 | 58.0 | 49.2 | 31.0 | 74.8 | 51.7 |
| HLA-DQ2/HLA-DQ8 | 100 | 32.7 | 36.9 | 100 | 51.7 |
| MSH5 | 68.7 | 77.0 | 74.7 | 54.1 | 86.2 |
| MSH5 + HLA-DQ2.2 | 65.1 | 77.4 | 90.9 | 77.6 | 85.3 |
| MSH5 + HLA-DQ2.5 | 21.8 | 79.2 | 29.3 | 72.0 | 63.0 |
| MSH5 + HLA-DQ8 | 11.1 | 93.1 | 38.6 | 72.6 | 69.9 |
| MSH5 + HLA-DQ2.2/HLA-DQ8 | 67.1 | 90.5 | 73.6 | 87.5 | 83.9 |
| MSH5 + HLA-DQ2.5/HLA-DQ8 | 30.9 | 78.8 | 36.5 | 74.3 | 65.3 |
| MSH5 + HLA-DQ2/HLA-DQ8 | 68.7 | 78.2 | 75.5 | 55.4 | 86.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wysocka-Mincewicz, M.; Groszek, A.; Ambrozkiewicz, F.; Paziewska, A.; Dąbrowska, M.; Rybak, A.; Konopka, E.; Ochocińska, A.; Żeber-Lubecka, N.; Karczmarski, J.; et al. Combination of HLA-DQ2/-DQ8 Haplotypes and a Single MSH5 Gene Variant in a Polish Population of Patients with Type 1 Diabetes as a First Line Screening for Celiac Disease? J. Clin. Med. 2022, 11, 2223. https://doi.org/10.3390/jcm11082223
Wysocka-Mincewicz M, Groszek A, Ambrozkiewicz F, Paziewska A, Dąbrowska M, Rybak A, Konopka E, Ochocińska A, Żeber-Lubecka N, Karczmarski J, et al. Combination of HLA-DQ2/-DQ8 Haplotypes and a Single MSH5 Gene Variant in a Polish Population of Patients with Type 1 Diabetes as a First Line Screening for Celiac Disease? Journal of Clinical Medicine. 2022; 11(8):2223. https://doi.org/10.3390/jcm11082223
Chicago/Turabian StyleWysocka-Mincewicz, Marta, Artur Groszek, Filip Ambrozkiewicz, Agnieszka Paziewska, Michalina Dąbrowska, Anna Rybak, Ewa Konopka, Agnieszka Ochocińska, Natalia Żeber-Lubecka, Jakub Karczmarski, and et al. 2022. "Combination of HLA-DQ2/-DQ8 Haplotypes and a Single MSH5 Gene Variant in a Polish Population of Patients with Type 1 Diabetes as a First Line Screening for Celiac Disease?" Journal of Clinical Medicine 11, no. 8: 2223. https://doi.org/10.3390/jcm11082223
APA StyleWysocka-Mincewicz, M., Groszek, A., Ambrozkiewicz, F., Paziewska, A., Dąbrowska, M., Rybak, A., Konopka, E., Ochocińska, A., Żeber-Lubecka, N., Karczmarski, J., Bierła, J. B., Trojanowska, I., Rogowska, A., Ostrowski, J., & Cukrowska, B. (2022). Combination of HLA-DQ2/-DQ8 Haplotypes and a Single MSH5 Gene Variant in a Polish Population of Patients with Type 1 Diabetes as a First Line Screening for Celiac Disease? Journal of Clinical Medicine, 11(8), 2223. https://doi.org/10.3390/jcm11082223







