The Agreement of a Two- and a Three-Dimensional Speckle-Tracking Global Longitudinal Strain
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Echocardiography
2.3. Statistics
2.4. Reproducibility Analysis
3. Results
4. Discussion
4.1. Previous Studies
4.2. Current Study
4.3. Implications
4.4. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Amundsen, B.H.; Helle-Valle, T.; Edvardsen, T.; Torp, H.; Crosby, J.; Lyseggen, E.; Støylen, A.; Ihlen, H.; Lima, J.A.; Smiseth, O.A.; et al. Noninvasive myocardial strain measurement by speckle tracking echocardiography: Validation against sonomicrometry and tagged magnetic resonance imaging. J. Am. Coll. Cardiol. 2006, 47, 789–793. [Google Scholar] [CrossRef] [PubMed]
- Curigliano, G.; Lenihan, D.; Fradley, M.; Ganatra, S.; Barac, A.; Blaes, A.; Herrmann, J.; Porter, C.; Lyon, A.R.; Lancellotti, P.; et al. Management of cardiac disease in cancer patients throughout oncological treatment: ESMO consensus recommendations. Ann. Oncol. 2020, 31, 171–190. [Google Scholar] [CrossRef] [PubMed]
- Argyle, R.A.; Ray, S.G. Stress and strain: Double trouble or useful tool? Eur. J. Echocardiogr. 2009, 10, 716–722. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Farsalinos, K.E.; Daraban, A.M.; Ünlü, S.; Thomas, J.D.; Badano, L.P.; Voigt, J.U. Head-to-head comparison of global longitudinal strain measurements among nine different vendors:The EACVI/ASE inter-vendor comparison study. J. Am. Soc. Echocardiogr. 2015, 28, 1171–1181. [Google Scholar] [CrossRef] [PubMed]
- Mirea, O.; Pagourelias, E.D.; Duchenne, J.; Bogaert, J.; Thomas, J.D.; Badano, L.P.; Voigt, J.U. Intervendor Differences in the Accuracy of detecting regional functional abnormalities: A report from the EACVI-ASE strain standardization task force. JACC Cardiovasc. Imaging 2018, 11, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Poyraz, E.; Tugba, K.O.; Güvenç, R.C.; Güvenç, T.S. Correlation and agreement between 2D and 3D speckle-tracking echocardiography for left ventricular volumetric, strain, and rotational parameters in healthy volunteers and in patients with mild mitral stenosis. Echocardiography 2019, 36, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Mor-Avi, V.; Lang, R.M.; Badano, L.P.; Belohlavek, M.; Cardim, N.M.; Derumeaux, G.; Galderisi, M.; Marwick, T.; Nagueh, S.F.; Sengupta, P.P.; et al. Current and evolving echocardiographic techniques for the quantitative evaluation of cardiac mechanics: ASE/EAE consensus statement on methodology and indications endorsed by the Japanese Society of Echocardiography. J. Am. Soc. Echocardiogr. 2011, 24, 277–313. [Google Scholar] [CrossRef] [PubMed]
- Edvardsen, T.; Gerber, B.L.; Garot, J.; Bluemke, D.A.; Lima, J.A.; Smiseth, O.A. Quantitative assessment of intrinsic regional myocardial deformation by Doppler strain rate echocardiography in humans: Validation against three-dimensional tagged magnetic resonance imaging. Circulation 2002, 106, 50–56. [Google Scholar] [CrossRef]
- Lumens, J.; Prinzen, F.W.; Delhaas, T. Longitudinal strain: “Think globally track locally”. JACC Cardiovasc. Imaging 2015, 8, 1360–1363. [Google Scholar] [CrossRef]
- Mazhari, R.; Omens, J.H.; Pavelec, R.S.; Covell, J.W.; McCulloch, A.D. Transmural distribution of threedimensional systolic strains in stunned myocardium. Circulation 2001, 104, 336–341. [Google Scholar] [CrossRef][Green Version]
- Lumens, J.; Delhaas, T.; Arts, T.; Cowan, B.R.; Young, A.A. Impaired subendocardial contractile myofiber function in asymptomatic aged humans, as detected using MRI. Am. J. Physiol. Heart Circ. Physiol. 2006, 291, H1573–H1579. [Google Scholar] [CrossRef]
- Karlsen, S.; Dahlslett, T.; Grenne, B.; Sjøli, B.; Smiseth, O.; Edvardsen, T.; Brunvand, H. Global longitudinal strain is more reproducible measure of left ventricular function than ejection fraction regardless of echocardiographic training. Cardiovasc. Ultrasound 2019, 17, 18. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.h.L.; Gonçalves, A.; Shah, A.M.; Cheng, S.; Kitzman, D.; Scott, S.D. Age and gender-related influences on left ventricular mechanics in elderly individuals free of prevalent heart failure: The atherosclerosis risk in communities study. Circ. Cardiovasc. Imaging 2017, 10, e004510. [Google Scholar] [CrossRef]
- Yodwut, C.; Weinert, L.; Klas, B.; Lang, R.M.; Mor-Avi, V. Effects of frame rate on three-dimensiona speckle tracking-based measurements of myocardial deformation. J. Am. Soc. Echocardiogr. 2012, 25, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Altman, M.; Bergerot, C.; Aussoleil, A.; Davidsen, E.S.; Sibellas, F.; Ovize, M.; Bonnefoy-Cudraz, E.; Thibault, H.; Derumeaux, G. Assesment of left ventricular systolic function by deformation imaging derived from speckle tracking: A comparison between 2D and 3D echo modalities. Eur. Heart J. Cardiovasc. Imaging 2014, 15, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Trache, T.; Stöbe, S.; Tarr, A.; Pfeiffer, D.; Hagendorff, A. The agreement between 3D, standard 2D and triplane 2D speckle tracking: Effects of image quality and 3D volume rate. Echo Res. Pract. 2014, 1, 71–83. [Google Scholar] [CrossRef][Green Version]
- Obokata, M.; Nagata, Y.; Wu, V.C.; Kado, Y.; Kurabayashi, M.; Otsuji, Y.; Takeuchi, M. Direct comparison of cardiac magnetic resonance feature tracking and 2D/3D echoardiography speckle tracking for evaluation of global left ventricular strain. Eur. Heart J. Cardiovasc. Imaging 2016, 17, 525–532. [Google Scholar] [CrossRef]
- Dillikar, M.V.; Venkateshvaran, A.; Barooah, B.; Varyani, R.; Kini, P.; Dash, P.K.; Sola, S. Three dimensional versus two dimensional strain for assessment of myocardial function: A case series. J. Indian Acad. Echocrdiogr. Cardiovsc. Imaging 2017, 1, 18–23. [Google Scholar]
- Mirea, O.; Pagourelias, E.D.; Duchenne, J.; Bogaert, J.; Thomas, J.D.; Badano, L.P.; Voigt, J.U. Variability and reproducibility of segmental longitudinal strain measurement: A report from the EACVI-ASE strain standardization task force. JACC Cardiovasc. Imaging 2018, 11, 15–24. [Google Scholar] [CrossRef]
- Pedrizzetti, G.; Claus, P.; Kilner, P.J.; Nagel, E. Principles of cardiovascular magnetic resonance feature tracking and echocardiographic speckle tracking for informed clinical use. J. Cardiovasc. Magn. Reson. 2016, 18, 51. [Google Scholar] [CrossRef]
- Patrianakos, A.P.; Zacharaki, A.A.; Kalogerakis, A.; Solidakis, G.; Parthenakis, F.I.; Vardas, P.E. Two-dimensional global and segmental longitudinal strain: Are the results from software in different high-end ultrasound systems comparable? Echo Res. Pract. 2015, 2, 29–39. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Muraru, D.; Niero, A.; Rodriguez-Zanella Cherata, D.; Badano, L. Three-dimensional speckle-tracking echocardiography- benefits and limitations of integrating myocardial mechanics with three-dimensional imaging. Cardiovasc. Diagn Ther. 2018, 8, 101–117. [Google Scholar] [CrossRef] [PubMed]
- Castel, A.L.; Menet, A.; Ennezat, P.V.; Delelis, F.; Le Goffic, C.; Binda, C.; Guerbaai, R.A.; Levy, F.; Graux, P.; Tribouilloy, C.; et al. Global longitudinal strain software upgrade: Implications for intervendor consistency and longitudinal imaging studies. Arch. Cardiovasc. Dis. 2016, 109, 22–30. [Google Scholar] [CrossRef]
- Stokke, T.M.; Hasselberg, N.E.; Smedsrud, M.K.; Sarvari, S.I.; Haugaa, K.H.; Smiseth, O.A.; Edvardsen, T.; Remme, E.W. Geometry as a confounder when assessing ventricular systolic function: Comparison between ejection fraction and strain. J. Am. Coll. Cardiol. 2017, 70, 942–954. [Google Scholar] [CrossRef] [PubMed]

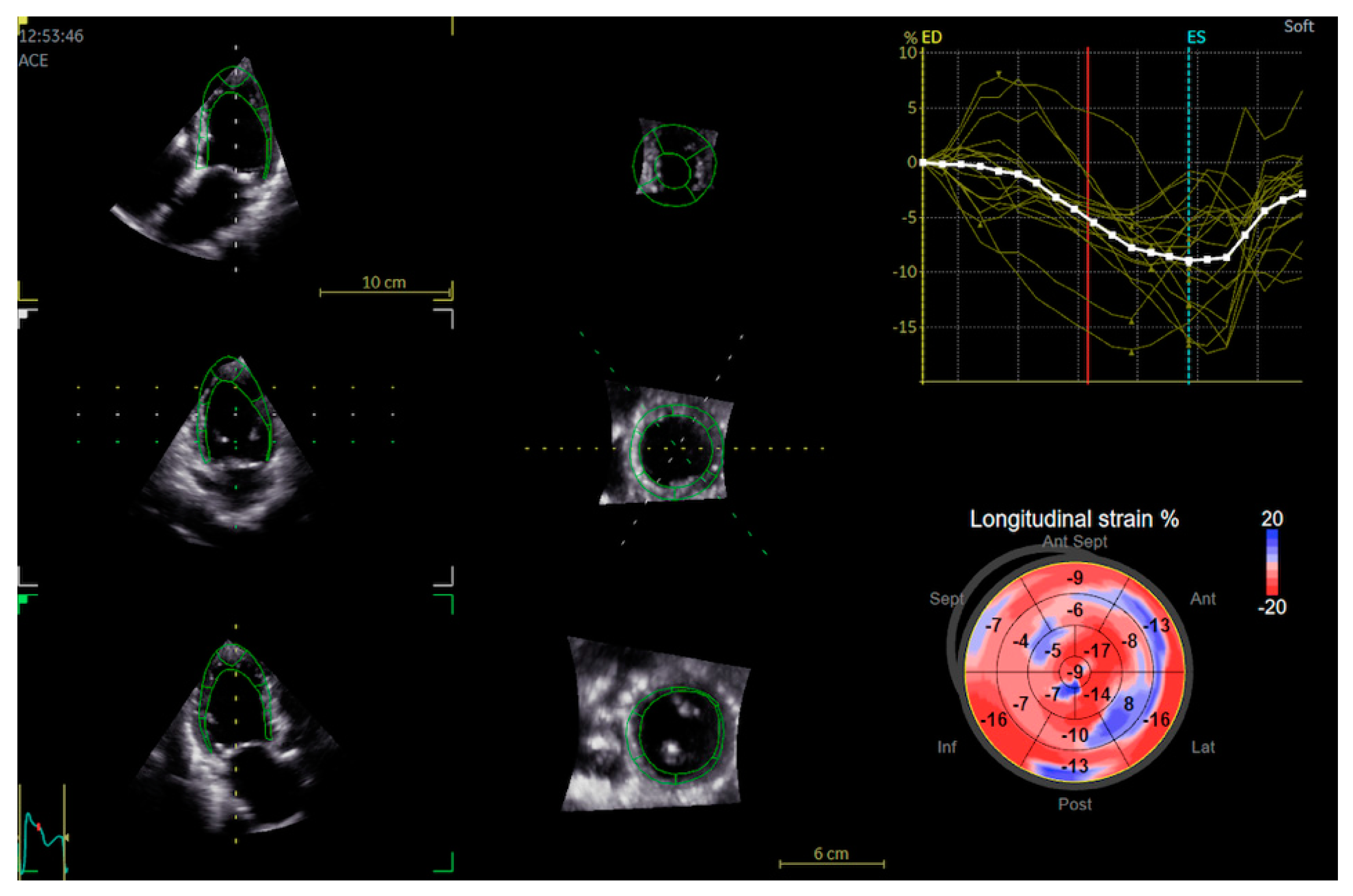
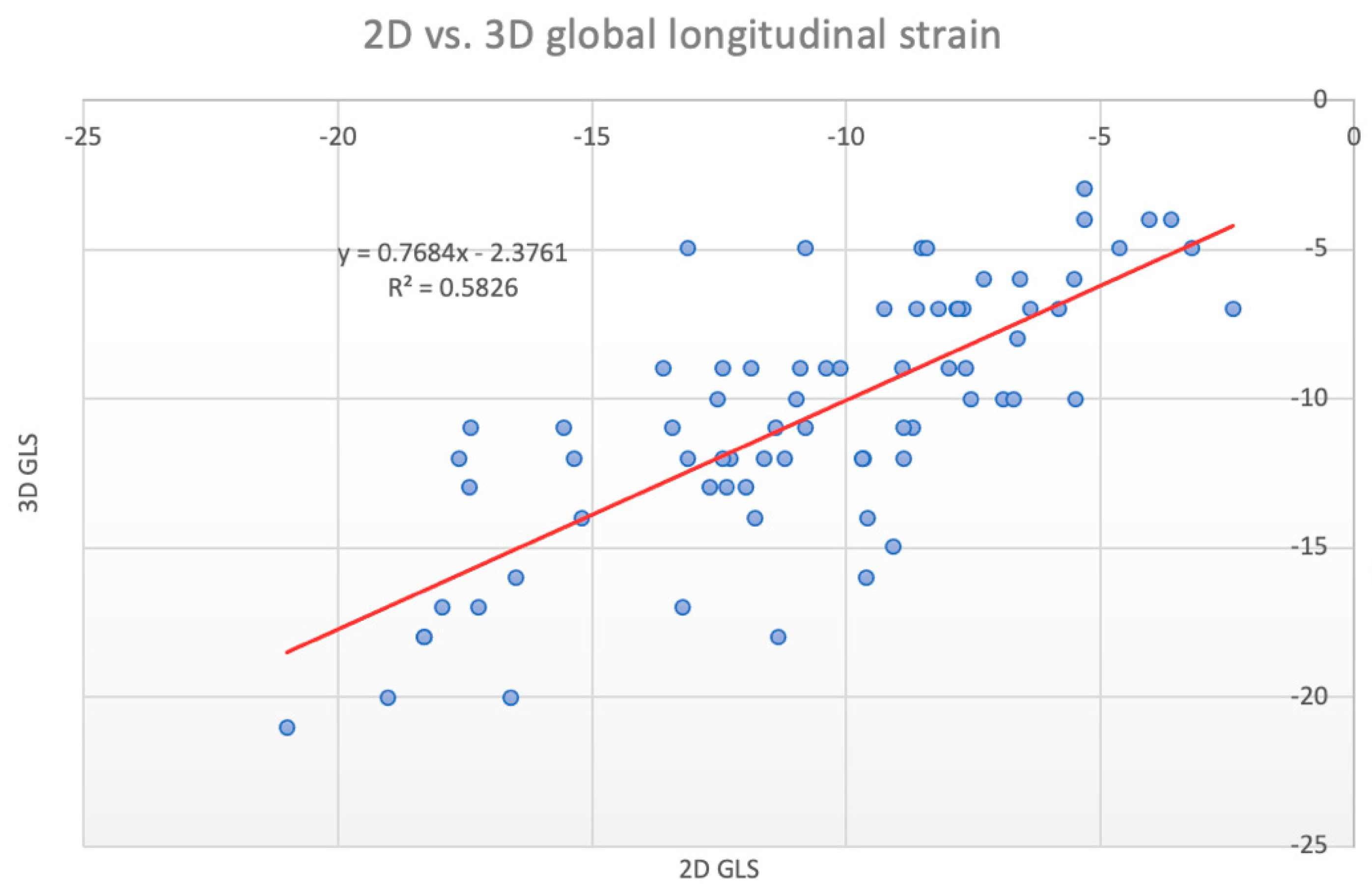


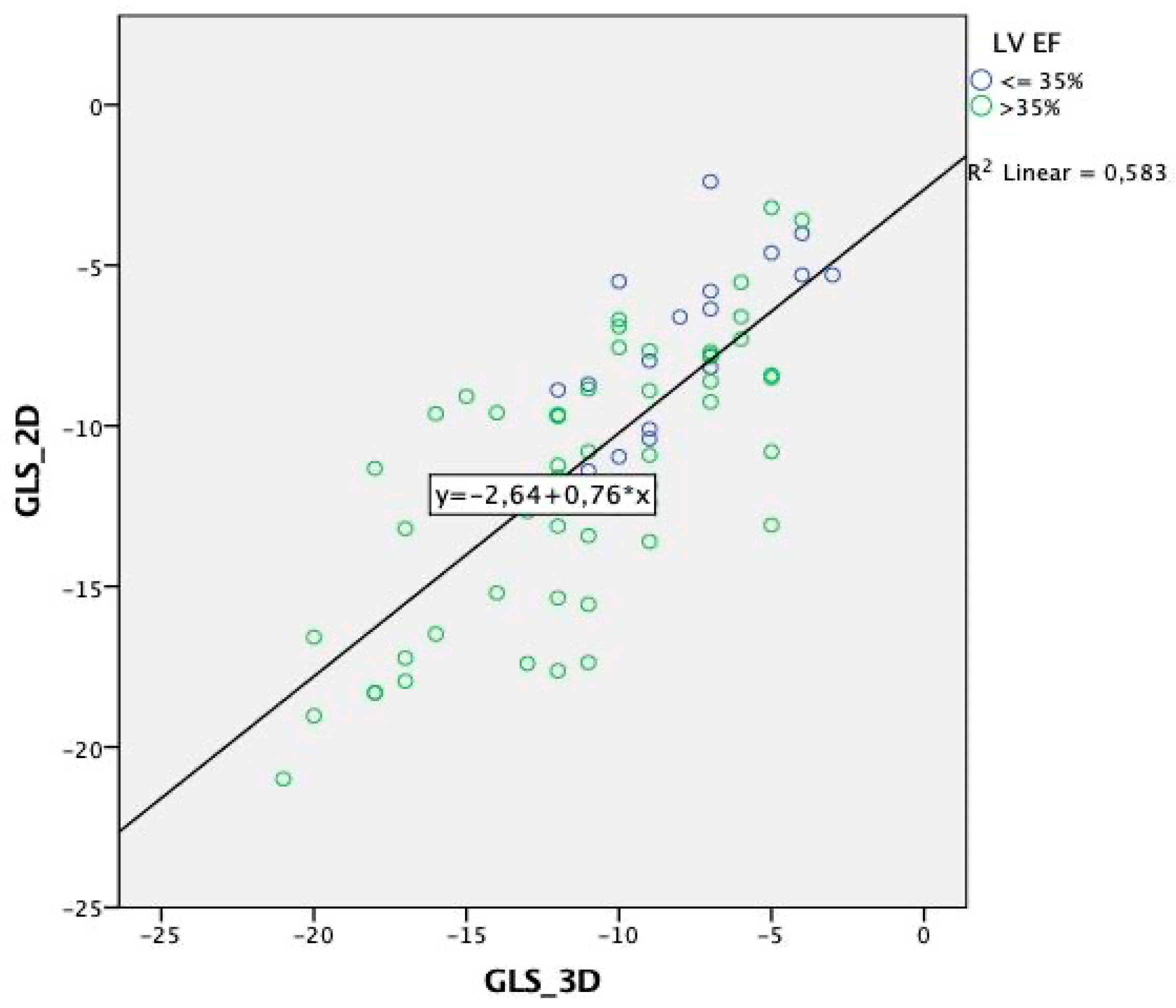
| Total Population | Males | Females | p Value | |
|---|---|---|---|---|
| N = 90 | N = 53 | N = 37 | ||
| Age (years) | 73.2 ± 11.2 | 70.5 ± 12.3 | 76.7 ± 8.3 | 0.681 |
| Males (%) | 59 | - | - | |
| Body weight (kg) | 85.1 ± 18.4 | 91.4 ± 16.5 | 77 ± 17.7 | 0.882 |
| Body height (cm) | 169.1 ± 9.9 | 175.5 ± 6.4 | 160.7 ± 7 | 0.068 |
| Body mass index (kg/m2) | 29.6 ± 5.4 | 29.5 ± 4.6 | 29.8 ± 6.4 | 0.92 |
| LV EF | 48 ± 12.7 | 45 ± 12.9 | 51 ± 11.8 | 0.039 |
| Coronary artery disease (%) | 46.5 | 49 | 43.2 | 0.269 |
| Hyperlipoproteinemia (%) | 52.6 | 55.1 | 48.6 | 0.726 |
| Myocardial infarction (%) | 27.9 | 32.7 | 21.6 | 0.744 |
| Peripheral arterial disease (%) | 4.7 | 8.2 | 2.7 | 0.660 |
| Hypertension (%) | 79.1 | 73 | 86.5 | 0.371 |
| Heart failure (%) | 8.1 | 18.0 | 10.8 | 0.021 |
| Diabetes mellitus (%) | 25.6 | 24.5 | 27 | 0.732 |
| Previous stroke/TIA (%) | 9.3 | 10.2 | 8.1 | 0.585 |
| COPD (%) | 8.1 | 8.2 | 8.1 | 0.314 |
| DCM | 8.1 | 10.2 | 5.4 | 0.27 |
| HCM | 1.2 | 2 | 0 | N/A |
| CABG (%) | 16.3 | 24.5 | 5.4 | 0.027 |
| PM (%) | 38 | 32 | 48 | 0.656 |
| LINEAR REGRESSION + PEARSON’S AND SPEARMAN CORRELATION | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2D vs. 3D GLS | ALL | MALES | FEMALES | ||||||||||||||||||
| SLOPE ± SEM | R | p | r | p | ρ | p | SLOPE ± SEM | R | p | r | p | ρ | p | SLOPE ± SEM | R | p | ρ | p | r | p | |
| 0.77 ± 0.27 | 0.76 | <0.001 | 0.76 | <0.001 | 0.74 | <0.001 | 0.82 ± 0.35 | 0.78 | <0.001 | 0.78 | <0.001 | 0.75 | <0.001 | 0.69 ± 0.39 | 0.69 | <0.001 | 0.66 | <0.001 | 0.69 | <0.001 | |
| Seg.1 | 0.38 ± 0.39 | 0.27 | 0.019 | 0.27 | 0.02 | 0.32 | 0.005 | 0.36 ± 0.36 | 0.16 | 0.3 | 0.18 | 0.23 | 0.42 | 0.006 | 0.26 ± 0.57 | 0.15 | 0.41 | 0.23 | 0.218 | 0.16 | 0.40 |
| Seg.2 | 0.66 ± 0.52 | 0.55 | <0.001 | 0.55 | <0.001 | 0.50 | <0.001 | 0.79 ± 0.51 | 0.54 | <0.001 | 0.54 | <0.001 | 0.57 | <0.001 | 0.58 ± 1.03 | 0.58 | <0.001 | 0.47 | 0.009 | 0.58 | <0.001 |
| Seg.3 | 0.12 ± 0.59 | 0.18 | 0.123 | 0.20 | 0.09 | 0.23 | 0.053 | 0.29 ± 0.74 | 0.3 | 0.05 | 0.32 | 0.04 | 0.36 | 0.018 | 0.09 ± 0.91 | 0.07 | 0.715 | 0.06 | 0.740 | 0.08 | 0.682 |
| Seg.4 | 0.46 ± 0.60 | 0.35 | 0.002 | 0.35 | 0.002 | 0.35 | 0.002 | 0.46 ± 0.82 | 0.35 | 0.023 | 0.35 | 0.02 | 0.34 | 0.027 | 0.44 ± 089 | 0.33 | 0.075 | 0.30 | 0.104 | 0.32 | 0.086 |
| Seg.5 | 0.42 ± 0.68 | 0.318 | 0.006 | 0.30 | 0.009 | 0.35 | 0.002 | 0.38 ± 0.79 | 0.26 | 0.096 | 0.24 | 0.119 | 0.25 | 0.116 | 0.50 ± 1.19 | 0.40 | 0.03 | 0.46 | 0.011 | 0.39 | 0.03 |
| Seg.6 | 0.05 ± 0.60 | 0.068 | 0.569 | 0.04 | 0.733 | 0.12 | 0.327 | −0.01 ± 0.65 | 0.05 | 0.750 | 0.01 | 0.970 | 0.13 | 0.413 | 0.06 ± 1.11 | 0.06 | 0.745 | 0.06 | 0.754 | 0.06 | 0.753 |
| Seg.7 | 0.50 ± 0.43 | 0.44 | <0.001 | 0.44 | <0.001 | 0.48 | <0.001 | 0.43 ± 0.60 | 0.42 | 0.006 | 0.42 | 0.004 | 0.51 | 0.001 | 0.61 ± 0.57 | 0.432 | 0.019 | 0.49 | 0.007 | 0.43 | 0.016 |
| Seg.8 | 0.44 ± 0.48 | 0.47 | <0.001 | 0.47 | <0.001 | 0.56 | <0.001 | 0.59 ± 0.54 | 0.53 | <0.001 | 0.53 | <0.001 | 0.56 | <0.001 | 0.33 ± 0.88 | 0.42 | 0.024 | 0.51 | 0.004 | 0.41 | 0.02 |
| Seg.9 | 0.50 ± 0.47 | 0.41 | <0.001 | 0.41 | <0.001 | 0.33 | 0.005 | 0.57 ± 0.59 | 0.49 | 0.001 | 0.49 | 0.001 | 0.45 | 0.003 | 0.40 ± 0.76 | 0.32 | 0.086 | 0.22 | 0.251 | 0.31 | 0.09 |
| Seg.10 | 0.32 ± 0.52 | 0.27 | 0.02 | 0.27 | 0.02 | 0.35 | 0.002 | 0.59 ± 0.65 | 0.53 | <0.001 | 0.52 | <0.001 | 0.62 | <0.001 | −0.1 ± 0.85 | 0.04 | 0.850 | 0.631 | 0.091 | 0.05 | 0.794 |
| Seg.11 | 0.531 ± 0.528 | 0.45 | <0.001 | 0.45 | <0.001 | 0.50 | <0.001 | 0.63 ± 0.63 | 0.51 | <0.001 | 0.50 | <0.001 | 0.55 | <0.001 | 0.36 ± 0.91 | 0.36 | 0.05 | 0.31 | 0.1 | 0.37 | 0.04 |
| Seg.12 | 0.50 ± 0.52 | 0.45 | <0.001 | 0.45 | <0.001 | 0.39 | 0.001 | 0.60 ± 0.59 | 0.54 | <0.001 | 0.54 | <0.001 | 0.51 | 0.001 | 0.39 ± 0.92 | 0.36 | 0.05 | 0.28 | 0.229 | 0.37 | 0.04 |
| Seg.13 | 0.29 ± 0.65 | 0.48 | <0.001 | 0.48 | <0.001 | 0.58 | <0.001 | 0.37 ± 0.75 | 0.51 | <0.001 | 0.51 | <0.001 | 0.62 | <0.001 | 0.16 ± 1.15 | 0.38 | 0.05 | 0.49 | 0.007 | 0.37 | 0.04 |
| Seg.14 | 0.46 ± 0.66 | 0.52 | <0.001 | 0.52 | <0.001 | 0.50 | <0.001 | 0.56 ± 0.75 | 0.56 | <0.001 | 0.57 | <0.001 | 0.56 | <0.001 | 0.39 ± 1.18 | 0.49 | 0.007 | 0.43 | 0.017 | 0.49 | 0.006 |
| Seg.15 | 0.23 ± 0.61 | 0.32 | 0.007 | 0.26 | 0.03 | 0.31 | 0.007 | 0.28 ± 0.75 | 0.42 | 0.006 | 0.31 | 0.05 | 0.37 | 0.016 | 0.17 ± 1.01 | 0.23 | 0.220 | 0.19 | 0.376 | 0.2 | 0.291 |
| Seg.16 | 0.14 ± 0.56 | 0.19 | 0.103 | 0.18 | 0.129 | 0.135 | 0.257 | 0.33 ± 0.75 | 0.45 | 0.003 | 0.42 | 0.006 | 0.36 | 0.018 | −0.15 ± 0.76 | 0.17 | 0.359 | 0.25 | 0.190 | 0.17 | 0.361 |
| Seg.17 | 0.27 ± 0.49 | 0.3 | 0.01 | 0.29 | 0.01 | 0.26 | 0.028 | 0.42 ± 0.66 | 0.47 | 0.002 | 0.44 | 0.003 | 0.40 | 0.773 | 0.03 ± 0.71 | 0.04 | 0.830 | 0.05 | 0.773 | 0.03 | 0.877 |
| N | R | p Value | |
|---|---|---|---|
| LV EF < 35% | 17 | 0.699 | 0.002 |
| LV EF > 35% | 73 | 0.727 | <0.001 |
| VP > 50% | 41 | 0.62 | <0.001 |
| VP < 50% | 49 | 0.8 | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Plášek, J.; Rychlý, T.; Drieniková, D.; Cisovský, O.; Grézl, T.; Homza, M.; Václavík, J. The Agreement of a Two- and a Three-Dimensional Speckle-Tracking Global Longitudinal Strain. J. Clin. Med. 2022, 11, 2402. https://doi.org/10.3390/jcm11092402
Plášek J, Rychlý T, Drieniková D, Cisovský O, Grézl T, Homza M, Václavík J. The Agreement of a Two- and a Three-Dimensional Speckle-Tracking Global Longitudinal Strain. Journal of Clinical Medicine. 2022; 11(9):2402. https://doi.org/10.3390/jcm11092402
Chicago/Turabian StylePlášek, Jiří, Tomáš Rychlý, Diana Drieniková, Ondřej Cisovský, Tomáš Grézl, Miroslav Homza, and Jan Václavík. 2022. "The Agreement of a Two- and a Three-Dimensional Speckle-Tracking Global Longitudinal Strain" Journal of Clinical Medicine 11, no. 9: 2402. https://doi.org/10.3390/jcm11092402
APA StylePlášek, J., Rychlý, T., Drieniková, D., Cisovský, O., Grézl, T., Homza, M., & Václavík, J. (2022). The Agreement of a Two- and a Three-Dimensional Speckle-Tracking Global Longitudinal Strain. Journal of Clinical Medicine, 11(9), 2402. https://doi.org/10.3390/jcm11092402







