Ezrin Contributes to the Plasma Membrane Expression of PD–L1 in A2780 Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. siRNA Treatment
2.3. RNA Extraction and Real–Time Reverse Transcription–Polymerase Chain Reaction
2.4. Confocal Laser Scanning Microscopy Analysis
2.5. Protein Isolation and Western Blotting
2.6. Flow Cytometry
2.7. Statistical Analysis
3. Results
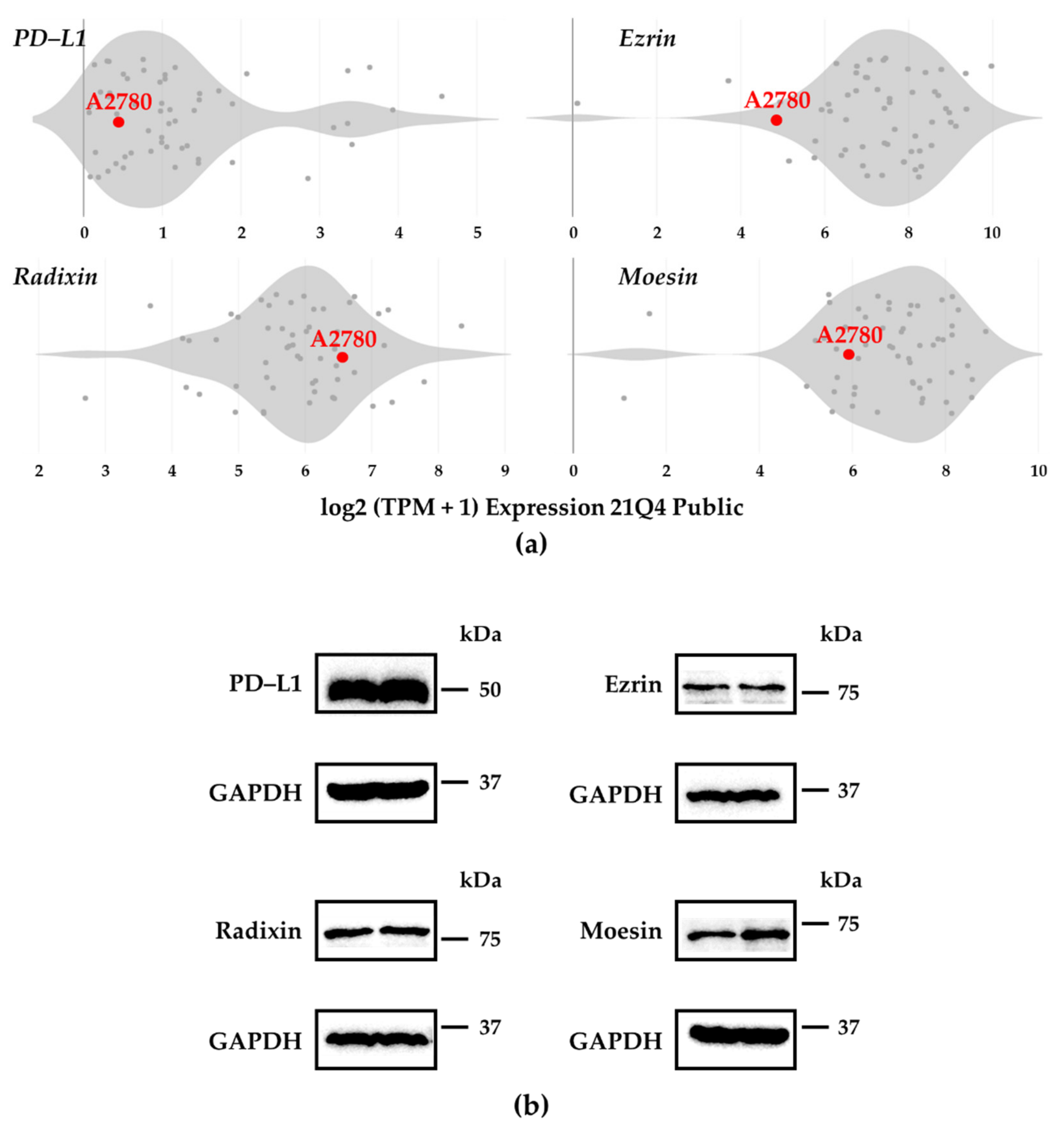
3.1. Expression Profiles of PD–L1 and ERMs in A2780 Cells
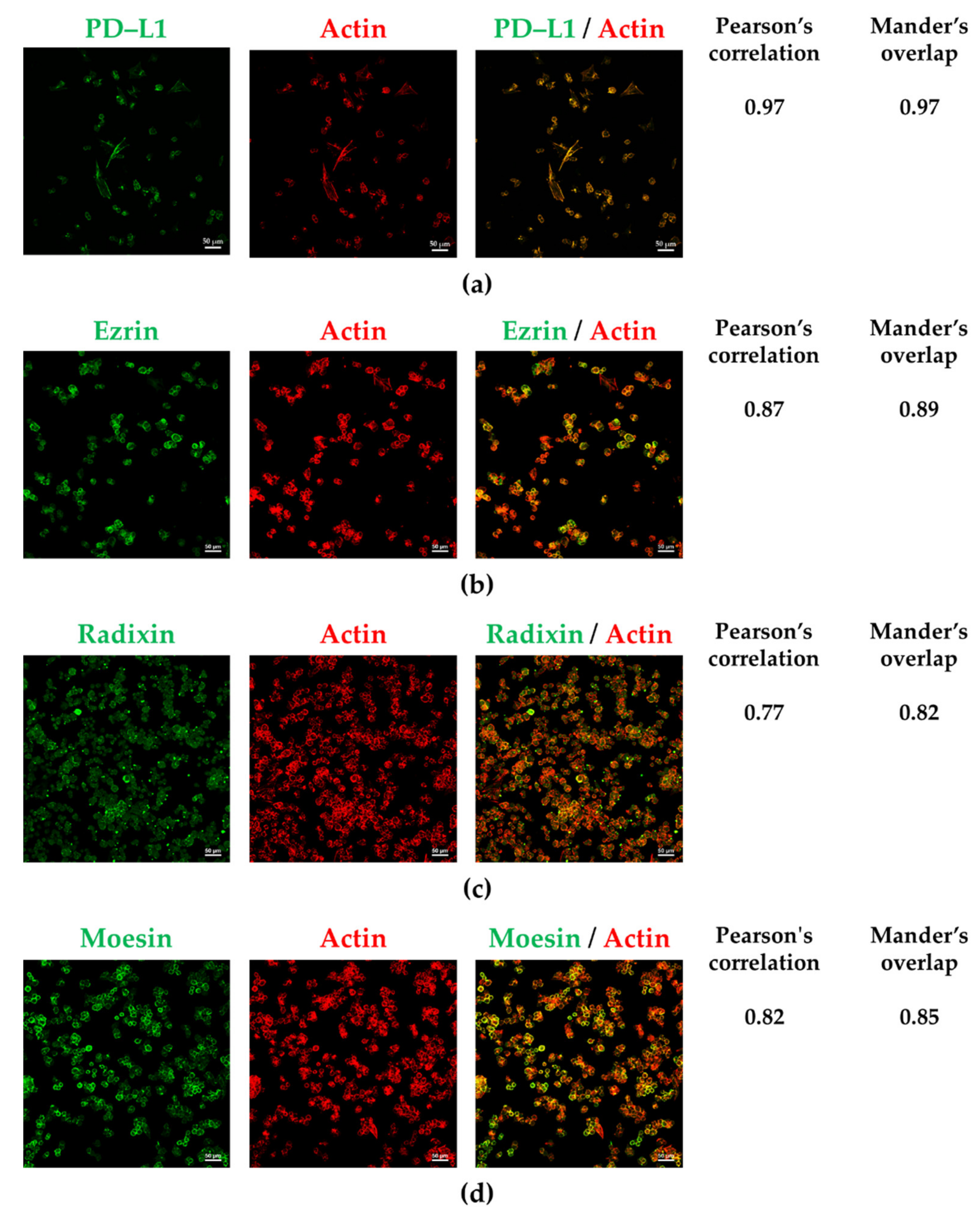
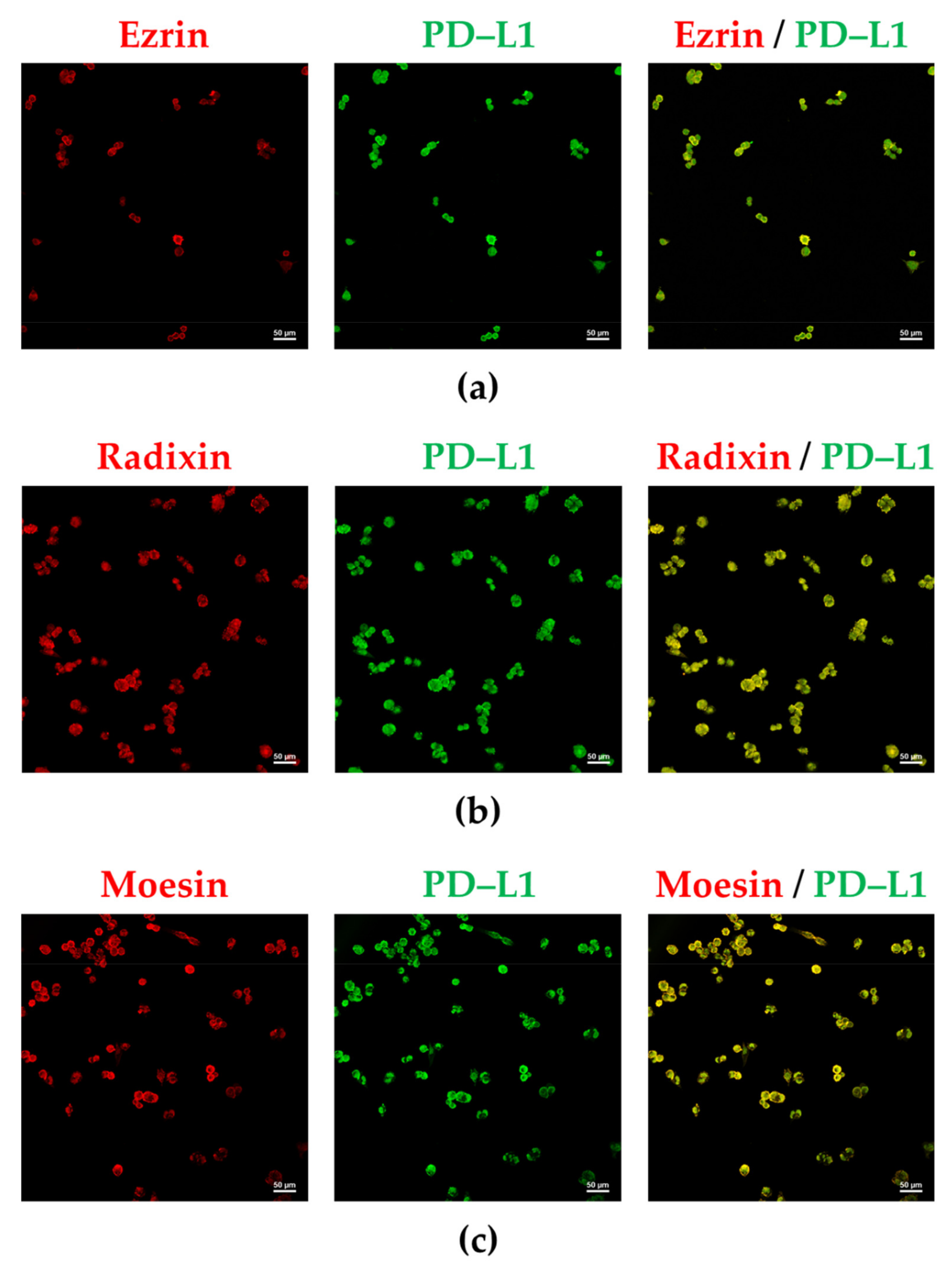
3.2. PD-L1 and ERM Were Localized in the Plasma Membrane of A2780 Cells
3.3. Influence of ERM Gene Suppression on the Expression of PD–L1 in A2780 Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, E.Y.; Kim, O.; Schilder, J.M.; Coffey, D.M.; Cho, C.H.; Bast, R.C., Jr. Cell Origins of High-Grade Serous Ovarian Cancer. Cancers 2018, 10, 433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ledermann, J.A.; Raja, F.A.; Fotopoulou, C.; Gonzalez-Martin, A.; Colombo, N.; Sessa, C.; Group, E.G.W. Newly diagnosed and relapsed epithelial ovarian carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2013, 24 (Suppl. 6), vi24–vi32. [Google Scholar] [CrossRef] [PubMed]
- Reid, B.M.; Permuth, J.B.; Sellers, T.A. Epidemiology of ovarian cancer: A review. Cancer Biol. Med. 2017, 14, 9–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohaegbulam, K.C.; Assal, A.; Lazar-Molnar, E.; Yao, Y.; Zang, X. Human cancer immunotherapy with antibodies to the PD-1 and PD-L1 pathway. Trends Mol. Med. 2015, 21, 24–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, L.; Chen, S.; Yang, L.; Li, Y. The role of PD-1 and PD-L1 in T-cell immune suppression in patients with hematological malignancies. J. Hematol. Oncol. 2013, 6, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, X.; Qin, B.; Zhang, Y.; Zhang, C.; Kahila, M.; Nowsheen, S.; Yin, P.; Yuan, J.; Pei, H.; Li, H.; et al. PD-L1 (B7-H1) Competes with the RNA Exosome to Regulate the DNA Damage Response and Can Be Targeted to Sensitize to Radiation or Chemotherapy. Mol. Cell 2019, 74, 1215–1226.e4. [Google Scholar] [CrossRef]
- Rossi, J.F.; Ceballos, P.; Lu, Z.Y. Immune precision medicine for cancer: A novel insight based on the efficiency of immune effector cells. Cancer Commun. 2019, 39, 34. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Zhao, Z.; Qin, X.; Zhang, M.; Du, Q.; Li, Z.; Luan, Y. A Checkpoint-Regulatable Immune Niche Created by Injectable Hydrogel for Tumor Therapy. Adv. Funct. Mater. 2021, 31, 2104630. [Google Scholar] [CrossRef]
- Xue, C.; Zhu, D.; Chen, L.; Xu, Y.; Xu, B.; Zhang, D.; Jiang, J. Expression and prognostic value of PD-L1 and PD-L2 in ovarian cancer. Transl. Cancer Res. 2019, 8, 111–119. [Google Scholar] [CrossRef]
- Yarchoan, M.; Johnson, B.A., 3rd; Lutz, E.R.; Laheru, D.A.; Jaffee, E.M. Targeting neoantigens to augment antitumour immunity. Nat. Rev. Cancer 2017, 17, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Nixon, N.A.; Blais, N.; Ernst, S.; Kollmannsberger, C.; Bebb, G.; Butler, M.; Smylie, M.; Verma, S. Current landscape of immunotherapy in the treatment of solid tumours, with future opportunities and challenges. Curr. Oncol. 2018, 25, e373–e384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamanishi, J.; Mandai, M.; Ikeda, T.; Minami, M.; Kawaguchi, A.; Murayama, T.; Kanai, M.; Mori, Y.; Matsumoto, S.; Chikuma, S.; et al. Safety and Antitumor Activity of Anti-PD-1 Antibody, Nivolumab, in Patients With Platinum-Resistant Ovarian Cancer. J. Clin. Oncol. 2015, 33, 4015–4022. [Google Scholar] [CrossRef] [PubMed]
- Disis, M.L.; Taylor, M.H.; Kelly, K.; Beck, J.T.; Gordon, M.; Moore, K.M.; Patel, M.R.; Chaves, J.; Park, H.; Mita, A.C.; et al. Efficacy and Safety of Avelumab for Patients With Recurrent or Refractory Ovarian Cancer: Phase 1b Results From the JAVELIN Solid Tumor Trial. JAMA Oncol. 2019, 5, 393–401. [Google Scholar] [CrossRef] [Green Version]
- Borella, F.; Ghisoni, E.; Giannone, G.; Cosma, S.; Benedetto, C.; Valabrega, G.; Katsaros, D. Immune Checkpoint Inhibitors in Epithelial Ovarian Cancer: An Overview on Efficacy and Future Perspectives. Diagnostics 2020, 10, 146. [Google Scholar] [CrossRef] [Green Version]
- Weberpals, J.I.; Pugh, T.J.; Marco-Casanova, P.; Goss, G.D.; Andrews Wright, N.; Rath, P.; Torchia, J.; Fortuna, A.; Jones, G.N.; Roudier, M.P.; et al. Tumor genomic, transcriptomic, and immune profiling characterizes differential response to first-line platinum chemotherapy in high grade serous ovarian cancer. Cancer Med. 2021, 10, 3045–3058. [Google Scholar] [CrossRef]
- Corvigno, S.; Burks, J.K.; Hu, W.; Zhong, Y.; Jennings, N.B.; Fleming, N.D.; Westin, S.N.; Fellman, B.; Liu, J.; Sood, A.K. Immune microenvironment composition in high-grade serous ovarian cancers based on BRCA mutational status. J. Cancer Res. Clin. Oncol. 2021, 147, 3545–3555. [Google Scholar] [CrossRef]
- Wang, Y.N.; Lee, H.H.; Hsu, J.L.; Yu, D.; Hung, M.C. The impact of PD-L1 N-linked glycosylation on cancer therapy and clinical diagnosis. J. Biomed. Sci. 2020, 27, 77. [Google Scholar] [CrossRef]
- Li, S.M.; Zhou, J.; Wang, Y.; Nie, R.C.; Chen, J.W.; Xie, D. Recent Findings in the Posttranslational Modifications of PD-L1. J. Oncol. 2020, 2020, 5497015. [Google Scholar] [CrossRef]
- Cha, J.H.; Chan, L.C.; Li, C.W.; Hsu, J.L.; Hung, M.C. Mechanisms Controlling PD-L1 Expression in Cancer. Mol. Cell 2019, 76, 359–370. [Google Scholar] [CrossRef]
- Mezzadra, R.; Sun, C.; Jae, L.T.; Gomez-Eerland, R.; de Vries, E.; Wu, W.; Logtenberg, M.E.W.; Slagter, M.; Rozeman, E.A.; Hofland, I.; et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 2017, 549, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Burr, M.L.; Sparbier, C.E.; Chan, Y.C.; Williamson, J.C.; Woods, K.; Beavis, P.A.; Lam, E.Y.N.; Henderson, M.A.; Bell, C.C.; Stolzenburg, S.; et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 2017, 549, 101–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogihara, T.; Mizoi, K.; Kamioka, H.; Yano, K. Physiological Roles of ERM Proteins and Transcriptional Regulators in Supporting Membrane Expression of Efflux Transporters as Factors of Drug Resistance in Cancer. Cancers 2020, 12, 3352. [Google Scholar] [CrossRef]
- Kawaguchi, K.; Yoshida, S.; Hatano, R.; Asano, S. Pathophysiological Roles of Ezrin/Radixin/Moesin Proteins. Biol. Pharm. Bull. 2017, 40, 381–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asp, N.; Kvalvaag, A.; Sandvig, K.; Pust, S. Regulation of ErbB2 localization and function in breast cancer cells by ERM proteins. Oncotarget 2016, 7, 25443–25460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clucas, J.; Valderrama, F. ERM proteins in cancer progression. J. Cell Sci. 2014, 127, 267–275. [Google Scholar] [CrossRef] [Green Version]
- Tameishi, M.; Kobori, T.; Tanaka, C.; Urashima, Y.; Ito, T.; Obata, T. Contribution of Ezrin on the Cell Surface Plasma Membrane Localization of Programmed Cell Death Ligand-1 in Human Choriocarcinoma JEG-3 Cells. Pharmaceuticals 2021, 14, 963. [Google Scholar] [CrossRef]
- Tanaka, C.; Kobori, T.; Tameishi, M.; Urashima, Y.; Ito, T.; Obata, T. Ezrin Modulates the Cell Surface Expression of Programmed Cell Death Ligand-1 in Human Cervical Adenocarcinoma Cells. Molecules 2021, 26, 5648. [Google Scholar] [CrossRef]
- Kobori, T.; Tanaka, C.; Tameishi, M.; Urashima, Y.; Ito, T.; Obata, T. Role of Ezrin/Radixin/Moesin in the Surface Localization of Programmed Cell Death Ligand-1 in Human Colon Adenocarcinoma LS180 Cells. Pharmaceuticals 2021, 14, 864. [Google Scholar] [CrossRef]
- Kobori, T.; Doukuni, R.; Ishikawa, H.; Ito, Y.; Okada, R.; Tanaka, C.; Tameishi, M.; Urashima, Y.; Ito, T.; Obata, T. Ezrin and Radixin Differentially Modulate Cell Surface Expression of Programmed Death Ligand-1 in Human Pancreatic Ductal Adenocarcinoma KP-2 Cells. Immuno 2022, 2, 68–84. [Google Scholar] [CrossRef]
- Tanaka, C.; Kobori, T.; Okada, R.; Doukuni, R.; Tameishi, M.; Urashima, Y.; Ito, T.; Takagaki, N.; Obata, T. Ezrin Regulates the Cell Surface Localization of PD-L1 in HEC-151 Cells. J. Clin. Med. 2022, 11, 2226. [Google Scholar] [CrossRef] [PubMed]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehar, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Ghandi, M.; Huang, F.W.; Jane-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R., 3rd; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Q.; Zhang, Y.; Wang, Z.; Ding, J.; Song, Y.; Zhao, W. Cisplatin-mediated down-regulation of miR-145 contributes to up-regulation of PD-L1 via the c-Myc transcription factor in cisplatin-resistant ovarian carcinoma cells. Clin. Exp. Immunol. 2020, 200, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Carbotti, G.; Barisione, G.; Airoldi, I.; Mezzanzanica, D.; Bagnoli, M.; Ferrero, S.; Petretto, A.; Fabbi, M.; Ferrini, S. IL-27 induces the expression of IDO and PD-L1 in human cancer cells. Oncotarget 2015, 6, 43267–43280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, Y.; Zheng, W.; Liu, J.; Tang, Q.; Wang, S.S.; Yang, X.S. MiR-34a-5p/PD-L1 axis regulates cisplatin chemoresistance of ovarian cancer cells. Neoplasma 2020, 67, 93–101. [Google Scholar] [CrossRef]
- Chin, Y.T.; Wei, P.L.; Ho, Y.; Nana, A.W.; Changou, C.A.; Chen, Y.R.; Yang, Y.S.; Hsieh, M.T.; Hercbergs, A.; Davis, P.J.; et al. Thyroxine inhibits resveratrol-caused apoptosis by PD-L1 in ovarian cancer cells. Endocr. Relat. Cancer 2018, 25, 533–545. [Google Scholar] [CrossRef] [Green Version]
- Li, M.J.; Xiong, D.; Huang, H.; Wen, Z.Y. Ezrin Promotes the Proliferation, Migration, and Invasion of Ovarian Cancer Cells. Biomed. Environ. Sci. 2021, 34, 139–151. [Google Scholar] [CrossRef]
- Horwitz, V.; Davidson, B.; Stern, D.; Trope, C.G.; Tavor Re’em, T.; Reich, R. Ezrin Is Associated with Disease Progression in Ovarian Carcinoma. PLoS ONE 2016, 11, e0162502. [Google Scholar] [CrossRef] [Green Version]
- Konecny, G.E.; Glas, R.; Dering, J.; Manivong, K.; Qi, J.; Finn, R.S.; Yang, G.R.; Hong, K.L.; Ginther, C.; Winterhoff, B.; et al. Activity of the multikinase inhibitor dasatinib against ovarian cancer cells. Br. J. Cancer 2009, 101, 1699–1708. [Google Scholar] [CrossRef] [Green Version]
- Kobel, M.; Gradhand, E.; Zeng, K.; Schmitt, W.D.; Kriese, K.; Lantzsch, T.; Wolters, M.; Dittmer, J.; Strauss, H.G.; Thomssen, C.; et al. Ezrin promotes ovarian carcinoma cell invasion and its retained expression predicts poor prognosis in ovarian carcinoma. Int. J. Gynecol. Pathol. 2006, 25, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Fadiel, A.; Edusa, V.; Chen, Z.; So, J.; Sakamoto, H.; Fishman, D.A.; Naftolin, F. Estradiol-induced ezrin overexpression in ovarian cancer: A new signaling domain for estrogen. Cancer Lett. 2005, 220, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Fadiel, A.; Feng, Y.; Ohtani, K.; Rutherford, T.; Naftolin, F. Ovarian epithelial carcinoma tyrosine phosphorylation, cell proliferation, and ezrin translocation are stimulated by interleukin 1alpha and epidermal growth factor. Cancer 2001, 92, 3068–3075. [Google Scholar] [CrossRef]
- Nishizuka, S.; Chen, S.T.; Gwadry, F.G.; Alexander, J.; Major, S.M.; Scherf, U.; Reinhold, W.C.; Waltham, M.; Charboneau, L.; Young, L.; et al. Diagnostic markers that distinguish colon and ovarian adenocarcinomas: Identification by genomic, proteomic, and tissue array profiling. Cancer Res. 2003, 63, 5243–5250. [Google Scholar] [PubMed]
- Wan, T.; Sun, H.; Mao, Z.; Zhang, L.; Chen, X.; Shi, Y.; Shang, Y. Vitamin D deficiency inhibits microRNA-196b-5p which regulates ovarian granulosa cell hormone synthesis, proliferation, and apoptosis by targeting RDX and LRRC17. Ann. Transl. Med. 2021, 9, 1775. [Google Scholar] [CrossRef]
- Hu, X.; Lin, Z.; Wang, Z.; Zhou, Q. Emerging role of PD-L1 modification in cancer immunotherapy. Am. J. Cancer Res. 2021, 11, 3832–3840. [Google Scholar]
- Meng, F.; Su, Y.; Xu, B. Rho-associated protein kinase-dependent moesin phosphorylation is required for PD-L1 stabilization in breast cancer. Mol. Oncol. 2020, 14, 2701–2712. [Google Scholar] [CrossRef]
- Chan, L.C.; Li, C.W.; Xia, W.; Hsu, J.M.; Lee, H.H.; Cha, J.H.; Wang, H.L.; Yang, W.H.; Yen, E.Y.; Chang, W.C.; et al. IL-6/JAK1 pathway drives PD-L1 Y112 phosphorylation to promote cancer immune evasion. J. Clin. Invest. 2019, 129, 3324–3338. [Google Scholar] [CrossRef] [Green Version]
- Yi, M.; Niu, M.; Xu, L.; Luo, S.; Wu, K. Regulation of PD-L1 expression in the tumor microenvironment. J. Hematol. Oncol. 2021, 14, 10. [Google Scholar] [CrossRef]
- Takamatsu, H.; Espinoza, J.L.; Lu, X.; Qi, Z.; Okawa, K.; Nakao, S. Anti-moesin antibodies in the serum of patients with aplastic anemia stimulate peripheral blood mononuclear cells to secrete TNF-alpha and IFN-gamma. J. Immunol. 2009, 182, 703–710. [Google Scholar] [CrossRef]
- Suzuki, K.; Nagao, T.; Itabashi, M.; Hamano, Y.; Sugamata, R.; Yamazaki, Y.; Yumura, W.; Tsukita, S.; Wang, P.C.; Nakayama, T.; et al. A novel autoantibody against moesin in the serum of patients with MPO-ANCA-associated vasculitis. Nephrol. Dial. Transplant. 2014, 29, 1168–1177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lall, N.; Henley–Smith, C.J.; De Canha, M.N.; Oosthuizen, C.B.; Berrington, D. Viability Reagent, PrestoBlue, in Comparison with Other Available Reagents, Utilized in Cytotoxicity and Antimicrobial Assays. Int. J. Microbiol. 2013, 2013, 420601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boncler, M.; Rozalski, M.; Krajewska, U.; Podsedek, A.; Watala, C. Comparison of PrestoBlue and MTT assays of cellular viability in the assessment of anti–proliferative effects of plant extracts on human endothelial cells. J. Pharmacol. Toxicol. Methods 2014, 69, 9–16. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tameishi, M.; Ishikawa, H.; Tanaka, C.; Kobori, T.; Urashima, Y.; Ito, T.; Obata, T. Ezrin Contributes to the Plasma Membrane Expression of PD–L1 in A2780 Cells. J. Clin. Med. 2022, 11, 2457. https://doi.org/10.3390/jcm11092457
Tameishi M, Ishikawa H, Tanaka C, Kobori T, Urashima Y, Ito T, Obata T. Ezrin Contributes to the Plasma Membrane Expression of PD–L1 in A2780 Cells. Journal of Clinical Medicine. 2022; 11(9):2457. https://doi.org/10.3390/jcm11092457
Chicago/Turabian StyleTameishi, Mayuka, Honami Ishikawa, Chihiro Tanaka, Takuro Kobori, Yoko Urashima, Takuya Ito, and Tokio Obata. 2022. "Ezrin Contributes to the Plasma Membrane Expression of PD–L1 in A2780 Cells" Journal of Clinical Medicine 11, no. 9: 2457. https://doi.org/10.3390/jcm11092457






