Deep Learning with a Dataset Created Using Kanno Saitama Macro, a Self-Made Automatic Foveal Avascular Zone Extraction Program
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Optical Coherence Tomography Angiography
2.3. KSM (Modified Version) and Annotation Simplification
- (1)
- Noise processing
- (2)
- Area expansion
- (1)
- The interpolation processing setting was changed to “none” when enlarging/reducing the image.
- (2)
- Extraction was performed with “analyze particles” instead of the wand tool and the size of the extraction area was specified.
- (1)
- The folder containing the original image was loaded and displayed as a stack.
- (2)
- The FAZ was extracted from all the original images using the continuous method for every 5 images using ROI sets that specified the slices and the ROI sets were saved.
- (3)
- The entire window was selected and the “fill” command was used to suffuse all the original images with black (brightness value: 0). This image served as the background of the label image.
- (4)
- The ROI set saved in step 2 was loaded. The ROI for each slice was specified and the images were filled with white (luminance value: 255) (completion of label image).
- (5)
- The completed label images were saved one-by-one using the ROI sets specific to the slices.
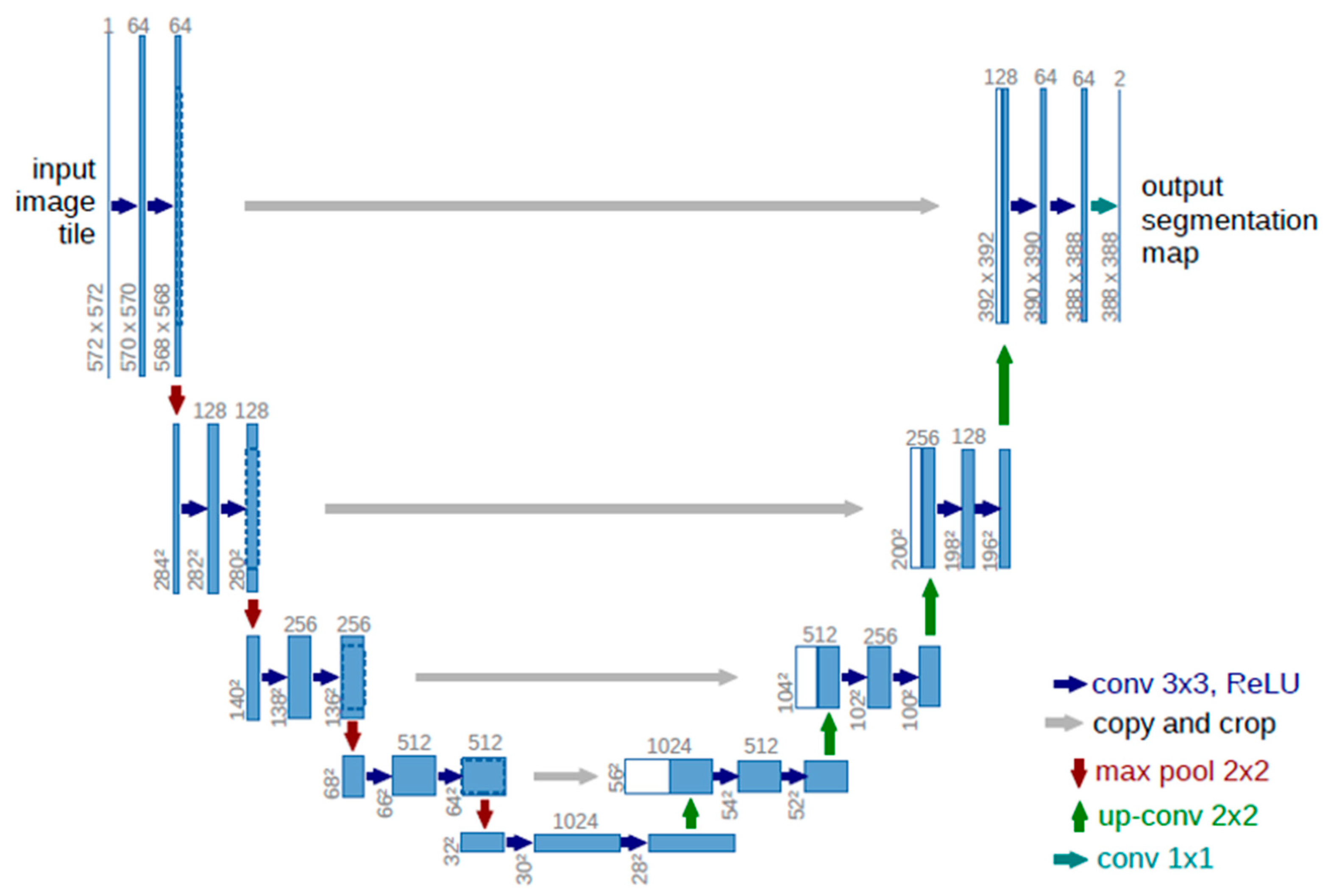
2.4. Deep Learning Network
2.5. The FAZ Extraction Method
2.5.1. The Manual Method (Examiner 1 and Examiner 2)
2.5.2. The Conventional Automatic Method (ARI)
2.5.3. Automatic Methods Using Deep Learning (U-Net)
2.6. Evaluation of the Extraction Accuracy
2.6.1. Coefficient of Variation and Correlation Coefficient of the Area
2.6.2. Measures of Similarity
Jaccard Similarity Coefficient
Dice Similarity Coefficient
2.7. Statistical Analysis
3. Results
3.1. Coefficient of Variation and Correlation Coefficient of the Area
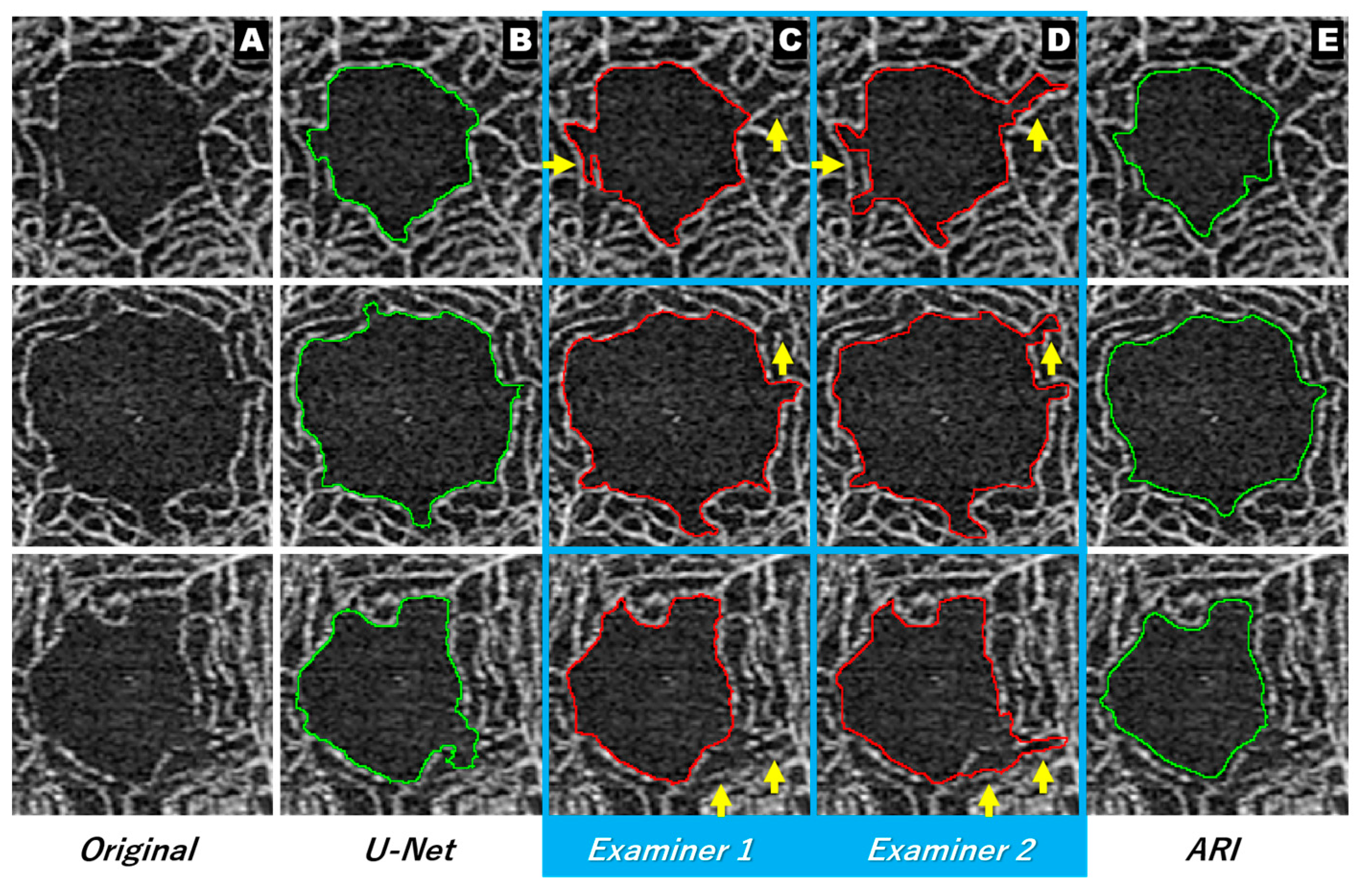
3.2. Two Types of Similarity, FN and FP (Excess or Deficiency of Extraction)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fujiwara, A.; Morizane, Y.; Hosokawa, M.; Kimura, S.; Shiode, Y.; Hirano, M.; Doi, S.; Toshima, S.; Takahashi, K.; Hosogi, M.; et al. Factors affecting foveal avascular zone in healthy eyes: An examination using swept-source optical coherence tomography angiography. PLoS ONE 2017, 12, e0188572. [Google Scholar] [CrossRef] [PubMed]
- Ciloglu, E.; Unal, F.; Sukgen, E.A.; Koçluk, Y. Evaluation of Foveal Avascular Zone and Capillary Plexuses in Diabetic Patients by Optical Coherence Tomography Angiography. Korean J. Ophthalmol. 2019, 33, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Balaratnasingam, C.; Inoue, M.; Ahn, S.; McCann, J.; Dhrami-Gavazi, E.; Yannuzzi, L.A.; Freund, K.B. Visual Acuity Is Correlated with the Area of the Foveal Avascular Zone in Diabetic Retinopathy and Retinal Vein Occlusion. Ophthalmology 2016, 123, 2352–2367. [Google Scholar] [CrossRef]
- Shiihara, H.; Terasaki, H.; Sonoda, S.; Kakiuchi, N.; Yamaji, H.; Yamaoka, S.; Uno, T.; Watanabe, M.; Sakamoto, T. Association of foveal avascular zone with the metamorphopsia in epiretinal membrane. Sci. Rep. 2020, 10, 170–192. [Google Scholar] [CrossRef]
- Tsuboi, K.; Fukutomi, A.; Sasajima, H.; Ishida, Y.; Kusaba, K.; Kataoka, T.; Kamei, M. Visual Acuity Recovery After Macular Hole Closure Associated with Foveal Avascular Zone Change. Transl. Vis. Sci. Technol. 2020, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Jauregui, R.; Park, K.S.; Duong, J.K.; Mahajan, V.; Tsang, S.H. Quantitative progression of retinitis pigmentosa by optical coherence tomography angiography. Sci. Rep. 2018, 8, 13130. [Google Scholar] [CrossRef]
- Shoji, T.; Kanno, J.; Weinreb, R.N.; Yoshikawa, Y.; Mine, I.; Ishii, H.; Ibuki, H.; Shinoda, K. OCT angiography measured changes in the foveal avascular zone area after glaucoma surgery. Br. J. Ophthalmol. 2022, 106, 80–86. [Google Scholar] [CrossRef]
- Araki, S.; Miki, A.; Goto, K.; Yamashita, T.; Yoneda, T.; Haruishi, K.; Ieki, Y.; Kiryu, J.; Maehara, G.; Yaoeda, K. Foveal avascular zone and macular vessel density after correction for magnification error in unilateral amblyopia using optical coherence tomography angiography. BMC Ophthalmol. 2019, 19, 171. [Google Scholar] [CrossRef]
- Guo, M.; Zhao, M.; Cheong, A.M.Y.; Dai, H.; Lam, A.K.C.; Zhou, Y. Automatic quantification of superficial foveal avascular zone in optical coherence tomography angiography implemented with deep learning. Vis. Comput. Ind. Biomed. Art 2019, 2, 21. [Google Scholar] [CrossRef]
- Mirshahi, R.; Anvari, P.; Riazi-Esfahani, H.; Sardarinia, M.; Naseripour, M.; Falavarjani, K.G. Foveal avascular zone segmentation in optical coherence tomography angiography images using a deep learning approach. Sci. Rep. 2021, 11, 1031. [Google Scholar] [CrossRef]
- Díaz, M.; Novo, J.; Cutrín, P.; Gómez-Ulla, F.; Penedo, M.G.; Ortega, M. Automatic segmentation of the foveal avascular zone in ophthalmological OCT-A images. PLoS ONE 2019, 14, e0212364. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Simonett, J.M.; Wang, J.; Zhang, M.; Hwang, T.; Hagag, A.; Huang, D.; Li, D.; Jia, Y. Evaluation of Automatically Quantified Foveal Avascular Zone Metrics for Diagnosis of Diabetic Retinopathy Using Optical Coherence Tomography Angiography. Investig. Opthalmology Vis. Sci. 2018, 59, 2212–2221. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.Y.; Ng, D.S.; Lam, A.; Luk, F.; Wong, R.; Chan, C.; Mohamed, S.; Fong, A.; Lok, J.; Tso, T.; et al. Determinants of Quantitative Optical Coherence Tomography Angiography Metrics in Patients with Diabetes. Sci. Rep. 2017, 7, 2575. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Fang, D.; Li, C.; Cheung, C.Y.; Chen, H. Improved Automated Foveal Avascular Zone Measurement in Cirrus Optical Coherence Tomography Angiography Using the Level Sets Macro. Transl. Vis. Sci. Technol. 2020, 9, 20. [Google Scholar] [CrossRef]
- Ishii, H.; Shoji, T.; Yoshikawa, Y.; Kanno, J.; Ibuki, H.; Shinoda, K. Automated Measurement of the Foveal Avascular Zone in Swept-Source Optical Coherence Tomography Angiography Images. Transl. Vis. Sci. Technol. 2019, 8, 28. [Google Scholar] [CrossRef]
- Jimenez, G.; Racoceanu, D. Deep Learning for Semantic Segmentation vs. Classification in Computational Pathology: Application to Mitosis Analysis in Breast Cancer Grading. Front. Bioeng. Biotechnol. 2019, 7, 145. [Google Scholar] [CrossRef]
- Hollon, T.C.; Pandian, B.; Adapa, A.R.; Urias, E.; Save, A.V.; Khalsa, S.S.S.; Eichberg, D.G.; D’Amico, R.S.; Farooq, Z.U.; Lewis, S.; et al. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat. Med. 2020, 26, 52–58. [Google Scholar] [CrossRef]
- Bevilacqua, V.; Brunetti, A.; Cascarano, G.D.; Guerriero, A.; Pesce, F.; Moschetta, M.; Gesualdo, L. A comparison between two semantic deep learning frameworks for the autosomal dominant polycystic kidney disease segmentation based on magnetic resonance images. BMC Med. Inform. Decis. Mak. 2019, 19, 1244. [Google Scholar] [CrossRef]
- Casalegno, F.; Newton, T.; Daher, R.; Abdelaziz, M.; Lodi-Rizzini, A.; Schürmann, F.; Krejci, I.; Markram, H. Caries Detection with Near-Infrared Transillumination Using Deep Learning. J. Dent. Res. 2019, 98, 1227–1233. [Google Scholar] [CrossRef]
- Nemoto, T.; Futakami, N.; Yagi, M.; Kumabe, A.; Takeda, A.; Kunieda, E.; Shigematsu, N. Efficacy evaluation of 2D, 3D U-Net semantic segmentation and atlas-based segmentation of normal lungs excluding the trachea and main bronchi. J. Radiat. Res. 2020, 61, 257–264. [Google Scholar] [CrossRef]
- Bakr, S.; Gevaert, O.; Echegaray, S.; Ayers, K.; Zhou, M.; Shafiq, M.; Zheng, H.; Benson, J.A.; Zhang, W.; Leung, A.N.C.; et al. A radiogenomic dataset of non-small cell lung cancer. Sci. Data 2018, 5, 180202. [Google Scholar] [CrossRef] [PubMed]
- Bojikian, K.D.; Chen, C.-L.; Wen, J.C.; Zhang, Q.; Xin, C.; Gupta, D.; Mudumbai, R.C.; Johnstone, M.A.; Wang, R.K.; Chen, P.P. Optic Disc Perfusion in Primary Open Angle and Normal Tension Glaucoma Eyes Using Optical Coherence Tomography-Based Microangiography. PLoS ONE 2016, 11, e0154691. [Google Scholar] [CrossRef] [PubMed]
- Tambe, S.B.; Kulhare, D.; Nirmal, M.D.; Prajapati, G. Image Processing (IP) Through Erosion and Dilation Methods. Int. J. Emerg. Technol. Adv. Eng. 2013, 3, 285–289. [Google Scholar]
- Kumar, M.; Singh, S. Edge Detection and Denoising Medical Image Using Morphology. Int. J. Emerg. Technol. Adv. Eng. 2012, 2, 66–72. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Kou, C.; Li, W.; Liang, W.; Yu, Z.; Hao, J. Microaneurysms segmentation with a U-Net based on recurrent residual convolutional neural network. J. Med. Imaging 2019, 6, 025008. [Google Scholar] [CrossRef] [PubMed]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Moghimi, S.; Hosseini, H.; Riddle, J.; Lee, G.Y.; Bitrian, E.; Giaconi, J.; Caprioli, J.; Nouri-Mahdavi, K. Measurement of Optic Disc Size and Rim Area with Spectral-Domain OCT and Scanning Laser Ophthalmoscopy. Investig. Opthalmology Vis. Sci. 2012, 53, 4519–4530. [Google Scholar] [CrossRef]
- Belgacem, R.; Malek, I.T.; Trabelsi, H.; Jabri, I. A supervised machine learning algorithm SKVMs used for both classification and screening of glaucoma disease. New Front. Ophthalmol. 2018, 4, 1–27. [Google Scholar] [CrossRef]
- Bertels, J.; Eelbode, T.; Berman, M.; Vandermeulen, D.; Maes, F.; Bisschops, R.; Blaschko, M.B. Optimizing the Dice Score and Jaccard Index for Medical Image Segmentation: Theory and Practice. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2019; pp. 92–100. [Google Scholar] [CrossRef]
- Jaccard, P. The Distribution of the Flora in the Alpine Zone. New Phytol. 1912, 11, 37–50. [Google Scholar] [CrossRef]
- Dice, L.R. Measures of the Amount of Ecologic Association Between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Zou, K.H.; Warfield, S.; Bharatha, A.; Tempany, C.M.; Kaus, M.R.; Haker, S.J.; Wells, W.M., 3rd; Jolesz, F.A.; Kikinis, R. Statistical validation of image segmentation quality based on a spatial overlap index1: Scientific reports. Acad. Radiol. 2004, 11, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Hwang, T.S.; Dongye, C.; Wilson, D.J.; Huang, D.; Jia, Y. Automated Quantification of Nonperfusion in Three Retinal Plexuses Using Projection-Resolved Optical Coherence Tomography Angiography in Diabetic Retinopathy. Investig. Opthalmol. Vis. Sci. 2016, 57, 5101–5106. [Google Scholar] [CrossRef] [PubMed]




| Method | Area (Mean ± SD) (mm2) |
|---|---|
| Examiner 1 | 0.271 ± 0.086 |
| Examiner 2 | 0.265 ± 0.086 |
| U-Net | 0.265 ± 0.085 |
| ARI | 0.240 ± 0.081 |
| p-value * | <0.001 |
| Method | CV (Mean [95%CI]) (%) | rho | p-Value |
|---|---|---|---|
| Examiner 1 vs Examiner 2 | 1.61 (1.23–1.98) | 0.995 | <0.001 * |
| Examiner 1 vs U-Net | 1.35 (0.95–1.75) | 0.994 | <0.001 * |
| Examiner 2 vs U-Net | 1.01 (0.73–1.29) | 0.995 | <0.001 * |
| Examiner 1 vs ARI | 6.35 (5.68–7.02) | 0.987 | <0.001 * |
| Examiner 2 vs ARI | 4.99 (4.33–5.65) | 0.987 | <0.001 * |
| <0.001 † |
| Jaccard (95%CI) | DSC (95%CI) | |
|---|---|---|
| Examiner 1 vs Examiner 2 | 0.931 (0.923–0.940) | 0.964 (0.959–0.969) |
| Examiner 1 vs U-Net | 0.951 (0.943–0.959) | 0.975 (0.971–0.979) |
| Examiner 2 vs U-Net | 0.933 (0.924–0.942) | 0.965 (0.960–0.970) |
| Examiner 1 vs ARI | 0.875 (0.864–0.887) | 0.933 (0.926–0.940) |
| Examiner 2 vs ARI | 0.894 (0.881–0.906) | 0.944 (0.936–0.951) |
| p value * | <0.001 | <0.001 |
| Mean FN (%) | Mean FP (%) | p-Value * | |
|---|---|---|---|
| Examiner 1 vs Examiner 2 | 4.87 | 2.21 | <0.001 |
| Examiner 1 vs U-Net | 3.65 | 1.34 | <0.001 |
| Examiner 2 vs U-Net | 3.3 | 3.66 | 0.128 |
| Examiner 1 vs ARI | 12.22 | 0.35 | <0.001 |
| Examiner 2 vs ARI | 10.07 | 0.68 | <0.001 |
| Study Using the Jaccard Similarity Coefficient | ||||||
| Author | Imaging Device | n | Slab | Method | Maximum Mean of Jaccard Similarity Coefficient | Area Correlation Coefficient * |
| Diaz et al. [11] | TOPCON DRI OCT Triton | 144 | SRL | Second observer | 0.83 | 0.93 |
| System | 0.82 | 0.90 | ||||
| Zhang et al. [34] | Optovue RTVue-XR | 22 | SRL | Automated Detection | 0.85 | |
| Lu et al. [12] | Optovue RTVue-XR | 19 | Inner Retinal | GGVF snake algorithm | 0.87 | |
| Current study | Zeiss PLEX Elite 9000 | 40 | SRL | Second observer | 0.931 | 0.995 |
| Typical U-Net (KSM Datasets) | 0.951 | 0.994 | ||||
| ARI | 0.894 | 0.987 | ||||
| Study Using the Dice Similarity Coefficient | ||||||
| Author | Imaging Device | n | Slab | Method | Maximum Mean of Dice Similarity Coefficient | Area Correlation Coefficient |
| Lin et al. [14] | Zeiss Cirrus HD-OCT 5000 | 34 | SRL | Second observer | 0.931 | |
| Level-sets macro | 0.924 | |||||
| Unadjusted KSM | 0.910 | |||||
| Guo et al. [9] | Zeiss Cirrus HD-OCT 5000 | 45 | SRL | Improved U-Net (Manual Datasets) | 0.976 | 0.997 |
| Mirshahi et al. [10] | RTVue XR 100 Avanti | 10 | Inner Retinal | Mask R-CNN (Manual Datasets) | 0.974 | 0.995 |
| Current study | Zeiss PLEX Elite 9000 | 40 | SRL | Second observer | 0.964 | 0.995 |
| Typical U-Net (KSM Datasets) | 0.975 | 0.994 | ||||
| ARI | 0.944 | 0.987 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanno, J.; Shoji, T.; Ishii, H.; Ibuki, H.; Yoshikawa, Y.; Sasaki, T.; Shinoda, K. Deep Learning with a Dataset Created Using Kanno Saitama Macro, a Self-Made Automatic Foveal Avascular Zone Extraction Program. J. Clin. Med. 2023, 12, 183. https://doi.org/10.3390/jcm12010183
Kanno J, Shoji T, Ishii H, Ibuki H, Yoshikawa Y, Sasaki T, Shinoda K. Deep Learning with a Dataset Created Using Kanno Saitama Macro, a Self-Made Automatic Foveal Avascular Zone Extraction Program. Journal of Clinical Medicine. 2023; 12(1):183. https://doi.org/10.3390/jcm12010183
Chicago/Turabian StyleKanno, Junji, Takuhei Shoji, Hirokazu Ishii, Hisashi Ibuki, Yuji Yoshikawa, Takanori Sasaki, and Kei Shinoda. 2023. "Deep Learning with a Dataset Created Using Kanno Saitama Macro, a Self-Made Automatic Foveal Avascular Zone Extraction Program" Journal of Clinical Medicine 12, no. 1: 183. https://doi.org/10.3390/jcm12010183
APA StyleKanno, J., Shoji, T., Ishii, H., Ibuki, H., Yoshikawa, Y., Sasaki, T., & Shinoda, K. (2023). Deep Learning with a Dataset Created Using Kanno Saitama Macro, a Self-Made Automatic Foveal Avascular Zone Extraction Program. Journal of Clinical Medicine, 12(1), 183. https://doi.org/10.3390/jcm12010183






