Pathophysiology of Rejection in Kidney Transplantation
Abstract
1. Introduction
2. Evaluation of the Risk of Rejection
2.1. HLA Typing
2.2. Donor-Specific Antibodies
2.3. Other Factors
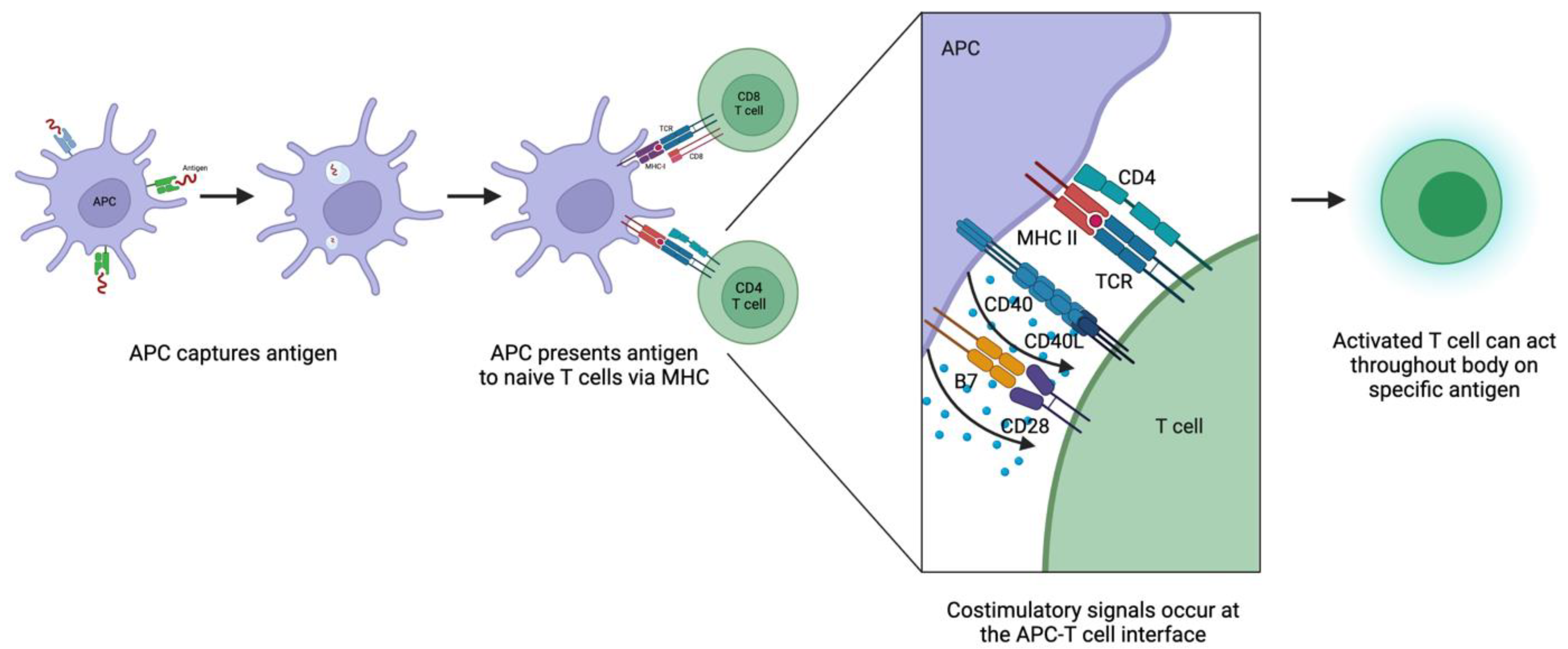
3. Mechanisms for Priming T Cells
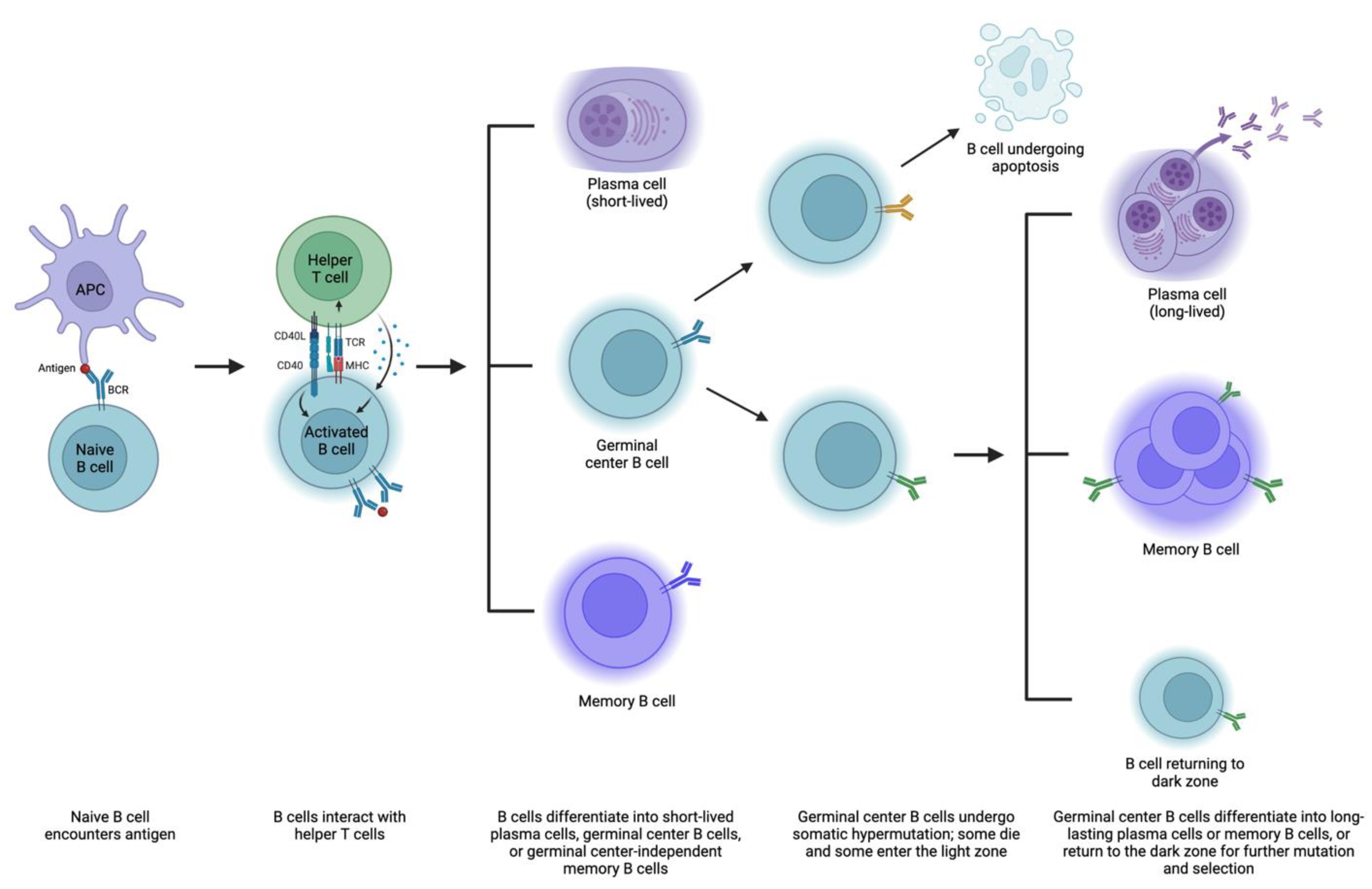
4. Mechanisms of Antibody Formation
5. Epidemiology of T-Cell-Mediated Rejection and Antibody-Mediated Rejection
6. Pathophysiology of T-Cell-Mediated Rejection
7. Pathophysiology of Antibody-Mediated Rejection
8. Limitations to Current Definitions of Rejection
9. Outcomes and Advances in Understanding of Pathophysiology of Rejection
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- United Network for Organ Sharing. Data and Trends. 2023. Available online: https://unos.org/data/ (accessed on 14 April 2023).
- Wolfe, R.A.; Ashby, V.B.; Milford, E.L.; Ojo, A.O.; Ettenger, R.E.; Agodoa, L.Y.C.; Held, P.J.; Port, F.K. Comparison of Mortality in All Patients on Dialysis, Patients on Dialysis Awaiting Transplantation, and Recipients of a First Cadaveric Transplant. N. Engl. J. Med. 1999, 341, 1725–1730. [Google Scholar] [CrossRef]
- Barker, C.F.; Markmann, J.F. Historical overview of transplantation. Cold Spring Harb. Perspect. Med. 2013, 3, a014977. [Google Scholar] [CrossRef] [PubMed]
- Pilch, N.A.; Bowman, L.J.; Taber, D.J. Immunosuppression trends in solid organ transplantation: The future of individualization, monitoring, and management. Pharmacotherapy 2021, 41, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Bestard, O.; Thaunat, O.; Bellini, M.I.; Böhmig, G.A.; Budde, K.; Claas, F.; Couzi, L.; Furian, L.; Heemann, U.; Mamode, N.; et al. Alloimmune Risk Stratification for Kidney Transplant Rejection. Transpl. Int. 2022, 35, 10138. [Google Scholar] [CrossRef]
- Dendrou, C.A.; Petersen, J.; Rossjohn, J.; Fugger, L. HLA variation and disease. Nat. Rev. Immunol. 2018, 18, 325–339. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.; Soormally, A.R.; Hayhurst, J.D.; Marsh, S.G.E. The IPD-IMGT/HLA Database—New developments in reporting HLA variation. Hum. Immunol. 2016, 77, 233–237. [Google Scholar] [CrossRef]
- Williams, R.C.; Opelz, G.; McGarvey, C.J.; Weil, E.J.; Chakkera, H.A. The Risk of Transplant Failure With HLA Mismatch in First Adult Kidney Allografts From Deceased Donors. Transplantation 2016, 100, 1094–1102. [Google Scholar] [CrossRef] [PubMed]
- Opelz, G.; Döhler, B. Effect of human leukocyte antigen compatibility on kidney graft survival: Comparative analysis of two decades. Transplantation 2007, 84, 137–143. [Google Scholar] [CrossRef]
- Su, X.; Zenios, S.A.; Chakkera, H.; Milford, E.L.; Chertow, G.M. Diminishing significance of HLA matching in kidney transplantation. Am. J. Transplant. 2004, 4, 1501–1508. [Google Scholar] [CrossRef]
- Wissing, K.M.; Fomegné, G.; Broeders, N.; Ghisdal, L.; Hoang, A.D.; Mikhalski, D.; Donckier, V.; Vereerstraeten, P.; Abramowicz, D. HLA mismatches remain risk factors for acute kidney allograft rejection in patients receiving quadruple immunosuppression with anti-interleukin-2 receptor antibodies. Transplantation 2008, 85, 411–416. [Google Scholar] [CrossRef]
- Beckingham, I.J.; Dennis, M.J.; Bishop, M.C.; Blamey, R.W.; Smith, S.J.; Nicholson, M.L. Effect of human leucocyte antigen matching on the incidence of acute rejection in renal transplantation. Br. J. Surg. 1994, 81, 574–577. [Google Scholar] [CrossRef] [PubMed]
- McKenna, R.M.; Lee, K.R.; Gough, J.C.; Jeffery, J.R.; Grimm, P.C.; Rush, D.N.; Nickerson, P. Matching for private or public HLA epitopes reduces acute rejection episodes and improves two-year renal allograft function. Transplantation 1998, 66, 38–43. [Google Scholar] [CrossRef]
- Lim, W.H.; Chadban, S.J.; Clayton, P.; Budgeon, C.A.; Murray, K.; Campbell, S.B.; Cohney, S.; Russ, G.R.; McDonald, S.P. Human leukocyte antigen mismatches associated with increased risk of rejection, graft failure, and death independent of initial immunosuppression in renal transplant recipients. Clin. Transplant. 2012, 26, E428–E437. [Google Scholar] [CrossRef]
- Opelz, G.; Döhler, B. Association of HLA mismatch with death with a functioning graft after kidney transplantation: A collaborative transplant study report. Am. J. Transplant. 2012, 12, 3031–3038. [Google Scholar] [CrossRef]
- Opelz, G.; Döhler, B. Impact of HLA mismatching on incidence of posttransplant non-hodgkin lymphoma after kidney transplantation. Transplantation 2010, 89, 567–572. [Google Scholar] [CrossRef]
- Opelz, G. Correlation of HLA matching with kidney graft survival in patients with or without cyclosporine treatment. Transplantation 1985, 40, 240–243. [Google Scholar] [CrossRef]
- Doxiadis, I.I.N.; de Fijter, J.W.; Mallat, M.J.K.; Haasnoot, G.W.; Ringers, J.; Persijn, G.G.; Claas, F.H.J. Simpler and Equitable Allocation of Kidneys From Postmortem Donors Primarily Based on Full HLA-DR Compatibility. Transplantation 2007, 83, 1207–1213. [Google Scholar] [CrossRef] [PubMed]
- European Bioinformatics Institute. IPD-IMGT/HLA. 2023. Available online: https://www.ebi.ac.uk/ipd/imgt/hla/ (accessed on 14 April 2023).
- Gimferrer, I. HLA Testing for Solid Organ Transplantation. Pearls Lab. Med. 2019. Available online: https://www.aacc.org/-/media/Files/Transcripts/Pearls-of-Laboratory-Medicine/2019/Transcript/HLA-Testing-for-Solid-Organ-Transplantation-Gimferrer-Transcript.pdf?la=en&hash=97FBAD0198D35DEC45186CF34706D26D40724498 (accessed on 14 April 2023).
- Dunckley, H. HLA Typing by SSO and SSP Methods. In Immunogenetics: Methods and Applications in Clinical Practice; Christiansen, F.T., Tait, B.D., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 9–25. [Google Scholar]
- Mohan, S.; Palanisamy, A.; Tsapepas, D.; Tanriover, B.; Crew, R.J.; Dube, G.; Ratner, L.E.; Cohen, D.J.; Radhakrishnan, J. Donor-specific antibodies adversely affect kidney allograft outcomes. J. Am. Soc. Nephrol. 2012, 23, 2061–2071. [Google Scholar] [CrossRef]
- Patel, R.; Terasaki, P.I. Significance of the positive crossmatch test in kidney transplantation. N. Engl. J. Med. 1969, 280, 735–739. [Google Scholar] [CrossRef]
- Higgins, R.; Lowe, D.; Daga, S.; Hathaway, M.; Williams, C.; Lam, F.T.; Kashi, H.; Tan, L.C.; Imray, C.; Fletcher, S.; et al. Pregnancy-induced HLA antibodies respond more vigorously after renal transplantation than antibodies induced by prior transplantation. Hum. Immunol. 2015, 76, 546–552. [Google Scholar] [CrossRef] [PubMed]
- Gebel, H.M.; Bray, R.A.; Nickerson, P. Pre-Transplant Assessment of Donor-Reactive, HLA-Specific Antibodies in Renal Transplantation: Contraindication vs. Risk. Am. J. Transplant. 2003, 3, 1488–1500. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.M.; Pancoska, C.; Mulgaonkar, S.; Weng, F.L. Renal transplantation in patients with pre-transplant donor-specific antibodies and negative flow cytometry crossmatches. Am. J. Transplant. 2007, 7, 2371–2377. [Google Scholar] [CrossRef]
- Ho, E.K.; Vasilescu, E.R.; Colovai, A.I.; Stokes, M.B.; Hallar, M.; Markowitz, G.S.; D’Agati, V.D.; Cohen, D.J.; Ratner, L.E.; Suciu-Foca, N. Sensitivity, specificity and clinical relevance of different cross-matching assays in deceased-donor renal transplantation. Transpl. Immunol. 2008, 20, 61–67. [Google Scholar] [CrossRef]
- Aubert, V.; Venetz, J.P.; Pantaleo, G.; Pascual, M. Low levels of human leukocyte antigen donor-specific antibodies detected by solid phase assay before transplantation are frequently clinically irrelevant. Hum. Immunol. 2009, 70, 580–583. [Google Scholar] [CrossRef]
- Graff, R.J.; Buchanan, P.M.; Dzebisashvili, N.; Schnitzler, M.A.; Tuttle-Newhall, J.; Xiao, H.; Schadde, E.; Gheorghian, A.; Lentine, K.L. The clinical importance of flow cytometry crossmatch in the context of CDC crossmatch results. Transplant. Proc. 2010, 42, 3471–3474. [Google Scholar] [CrossRef] [PubMed]
- Lentine, K.L.; Graff, R.J.; Xiao, H.; Modanlou, K.A.; Salvalaggio, P.R.; Brennan, D.C.; Pinsky, B.W.; Burroughs, T.E.; Schnitzler, M.A. Flow cytometry crossmatch before kidney transplantation in contemporary practice: Target cell utilization, results patterns, and associated long-term graft survival. Clin. Transplant. 2008, 253–266. [Google Scholar]
- Antibody Mediated Rejection in Organ Transplantation; Thermo Fisher Scientific Inc.: West Hills, CA, USA, 2019; Available online: https://www.thermofisher.com/us/en/home/life-science/antibodies.html?ef_id=CjwKCAjwyqWkBhBMEiwAp2yUFm6FETJ0AMjk_tXUlG9QCohRX9tXuP8PkLCakwLxljAhlGmYHERdzRoCqoIQAvD_BwE:G:s&s_kwcid=AL!3652!3!598837518400!p!!g!!thermofisher%20antibodies!2081760689!79794335227&cid=bid_pca_aup_r01_co_cp1359_pjt0000_bid00000_0se_gaw_bt_pur_con&gclid=CjwKCAjwyqWkBhBMEiwAp2yUFm6FETJ0AMjk_tXUlG9QCohRX9tXuP8PkLCakwLxljAhlGmYHERdzRoCqoIQAvD_BwE (accessed on 14 April 2023).
- Lefaucheur, C.; Loupy, A.; Hill, G.S.; Andrade, J.; Nochy, D.; Antoine, C.; Gautreau, C.; Charron, D.; Glotz, D.; Suberbielle-Boissel, C. Preexisting donor-specific HLA antibodies predict outcome in kidney transplantation. J. Am. Soc. Nephrol. 2010, 21, 1398–1406. [Google Scholar] [CrossRef]
- Tambur, A.R.; Herrera, N.D.; Haarberg, K.M.; Cusick, M.F.; Gordon, R.A.; Leventhal, J.R.; Friedewald, J.J.; Glotz, D. Assessing Antibody Strength: Comparison of MFI, C1q, and Titer Information. Am. J. Transplant. 2015, 15, 2421–2430. [Google Scholar] [CrossRef] [PubMed]
- Viglietti, D.; Bouatou, Y.; Kheav, V.D.; Aubert, O.; Suberbielle-Boissel, C.; Glotz, D.; Legendre, C.; Taupin, J.-L.; Zeevi, A.; Loupy, A.; et al. Complement-binding anti-HLA antibodies are independent predictors of response to treatment in kidney recipients with antibody-mediated rejection. Kidney Int. 2018, 94, 773–787. [Google Scholar] [CrossRef]
- Wan, S.S.; Chadban, S.J.; Watson, N.; Wyburn, K. Development and outcomes of de novo donor-specific antibodies in low, moderate, and high immunological risk kidney transplant recipients. Am. J. Transplant. 2020, 20, 1351–1364. [Google Scholar] [CrossRef] [PubMed]
- Wiebe, C.; Gibson, I.W.; Blydt-Hansen, T.D.; Pochinco, D.; Birk, P.E.; Ho, J.; Karpinski, M.; Goldberg, A.; Storsley, L.; Rush, D.N.; et al. Rates and determinants of progression to graft failure in kidney allograft recipients with de novo donor-specific antibody. Am. J. Transplant. 2015, 15, 2921–2930. [Google Scholar] [CrossRef] [PubMed]
- Wiebe, C.; Gibson, I.W.; Blydt-Hansen, T.D.; Karpinski, M.; Ho, J.; Storsley, L.J.; Goldberg, A.; Birk, P.E.; Rush, D.N.; Nickerson, P.W. Evolution and Clinical Pathologic Correlations of De Novo Donor-Specific HLA Antibody Post Kidney Transplant. Am. J. Transplant. 2012, 12, 1157–1167. [Google Scholar] [CrossRef]
- Dieplinger, G.; Ditt, V.; Arns, W.; Huppertz, A.; Kisner, T.; Hellmich, M.; Bauerfeind, U.; Stippel, D.L. Impact of de novo donor-specific HLA antibodies detected by Luminex solid-phase assay after transplantation in a group of 88 consecutive living-donor renal transplantations. Transpl. Int. 2014, 27, 60–68. [Google Scholar] [CrossRef]
- Ginevri, F.; Nocera, A.; Comoli, P.; Innocente, A.; Cioni, M.; Parodi, A.; Fontana, I.; Magnasco, A.; Nocco, A.; Tagliamacco, A.; et al. Posttransplant de novo donor-specific hla antibodies identify pediatric kidney recipients at risk for late antibody-mediated rejection. Am. J. Transplant. 2012, 12, 3355–3362. [Google Scholar] [CrossRef]
- Lefaucheur, C.; Louis, K.; Morris, A.B.; Taupin, J.-L.; Nickerson, P.; Tambur, A.R.; Gebel, H.M.; Reed, E.F.; Kobashigawa, J.A.; Chandraker, A.; et al. Clinical recommendations for posttransplant assessment of anti–HLA (Human Leukocyte Antigen) donor-specific antibodies: A Sensitization in Transplantation: Assessment of Risk consensus document. Am. J. Transplant. 2023, 23, 115–132. [Google Scholar] [CrossRef]
- Eckardt, K.-U.; Kasiske, B.L.; Zeier, M.G. Special Issue: KDIGO Clinical Practice Guideline for the Care of Kidney Transplant Recipients. Am. J. Transplant. 2009, 9, S1–S155. [Google Scholar] [CrossRef]
- Oweira, H.; Ramouz, A.; Ghamarnejad, O.; Khajeh, E.; Ali-Hasan-Al-Saegh, S.; Nikbakhsh, R.; Reißfelder, C.; Rahbari, N.; Mehrabi, A.; Sadeghi, M. Risk Factors of Rejection in Renal Transplant Recipients: A Narrative Review. J. Clin. Med. 2022, 11, 1392. [Google Scholar] [CrossRef]
- Hoegy, D.; Bleyzac, N.; Robinson, P.; Bertrand, Y.; Dussart, C.; Janoly-Dumenil, A. Medication adherence in pediatric transplantation and assessment methods: A systematic review. Patient Prefer. Adherence 2019, 13, 705–719. [Google Scholar] [CrossRef]
- Dharnidharka, V.R.; Lamb, K.E.; Zheng, J.; Schechtman, K.B.; Meier-Kriesche, H.-U. Across all solid organs, adolescent age recipients have worse transplant organ survival than younger age children: A US national registry analysis. Pediatr. Transplant. 2015, 19, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Foster, B.J.; Dahhou, M.; Zhang, X.; Platt, R.W.; Samuel, S.M.; Hanley, J.A. Association between age and graft failure rates in young kidney transplant recipients. Transplantation 2011, 92, 1237–1243. [Google Scholar] [CrossRef] [PubMed]
- Noppakun, K.; Cosio, F.G.; Dean, P.G.; Taler, S.J.; Wauters, R.; Grande, J.P. Living Donor Age and Kidney Transplant Outcomes. Am. J. Transplant. 2011, 11, 1279–1286. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.W.; Bennett, L.E.; Breen, T.J. Effect of donor age on outcome of kidney transplantation. A two-year analysis of transplants reported to the United Network for Organ Sharing Registry. Transplantation 1994, 57, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Fuggle, S.V.; Allen, J.E.; Johnson, R.J.; Collett, D.; Mason, P.D.; Dudley, C.; Rudge, C.J.; Bradley, J.A.; Watson, C.J. Factors affecting graft and patient survival after live donor kidney transplantation in the UK. Transplantation 2010, 89, 694–701. [Google Scholar] [CrossRef] [PubMed]
- El-Khoury, B.; Yang, T.C. Reviewing Racial Disparities in Living Donor Kidney Transplantation: A Socioecological Approach. J. Racial Ethn. Health Disparities 2023. Epub ahead of print. [Google Scholar] [CrossRef]
- Beatty, P.G.; Mori, M.; Milford, E. Impact of racial genetic polymorphism on the probability of finding an HLA-matched donor. Transplantation 1995, 60, 778–783. [Google Scholar] [CrossRef]
- Dharnidharka, V.R.; Seifert, M.E. Kidney transplant results in children: Progress made, but blacks lag behind. Kidney Int. 2015, 87, 492–494. [Google Scholar] [CrossRef] [PubMed]
- Mannon, R.B. Delayed Graft Function: The AKI of Kidney Transplantation. Nephron 2018, 140, 94–98. [Google Scholar] [CrossRef]
- Dorr, C.R.; Oetting, W.S.; Jacobson, P.A.; Israni, A.K. Genetics of acute rejection after kidney transplantation. Transpl. Int. 2018, 31, 263–277. [Google Scholar] [CrossRef]
- Wahrmann, M.; Döhler, B.; Arnold, M.L.; Scherer, S.; Mayer, K.A.; Haindl, S.; Haslacher, H.; Böhmig, G.A.; Süsal, C. Functional Fc Gamma Receptor Gene Polymorphisms and Long-Term Kidney Allograft Survival. Front. Immunol. 2021, 12, 724331. [Google Scholar] [CrossRef]
- Arnold, M.L.; Kainz, A.; Hidalgo, L.G.; Eskandary, F.; Kozakowski, N.; Wahrmann, M.; Haslacher, H.; Oberbauer, R.; Heilos, A.; Spriewald, B.M.; et al. Functional Fc gamma receptor gene polymorphisms and donor-specific antibody-triggered microcirculation inflammation. Am. J. Transplant. 2018, 18, 2261–2273. [Google Scholar] [CrossRef]
- Delves, P.J.; Roitt, I.M. The Immune System. N. Engl. J. Med. 2000, 343, 37–49. [Google Scholar] [CrossRef]
- Callemeyn, J.; Lamarthée, B.; Koenig, A.; Koshy, P.; Thaunat, O.; Naesens, M. Allorecognition and the spectrum of kidney transplant rejection. Kidney Int. 2022, 101, 692–710. [Google Scholar] [CrossRef]
- Charles, A.; Janeway, J.; Travers, P.; Walport, M.; Shlomchik, M.J. The production of armed effector T cells. In Immunobiology: The Immune System in Health and Disease, 5th ed.; Garland Science: New York, NY, USA, 2001. [Google Scholar]
- Kambayashi, T.; Laufer, T.M. Atypical MHC class II-expressing antigen-presenting cells: Can anything replace a dendritic cell? Nat. Rev. Immunol. 2014, 14, 719–730. [Google Scholar] [CrossRef]
- Charles, A.; Janeway, J.; Travers, P.; Walport, M.; Shlomchik, M.J. Antigen recognition by T cells. In Immunobiology: The Immune System in Health and Disease, 5th ed.; Garland Science: New York, NY, USA, 2001. [Google Scholar]
- Sherman, L.A.; Chattopadhyay, S. The molecular basis of allorecognition. Annu. Rev. Immunol. 1993, 11, 385–402. [Google Scholar] [CrossRef] [PubMed]
- Matzinger, P.; Bevan, M.J. Hypothesis: Why do so many lymphocytes respond to major histocompatibility antigens? Cell Immunol. 1977, 29, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Herrera, O.B.; Golshayan, D.; Tibbott, R.; Salcido Ochoa, F.; James, M.J.; Marelli-Berg, F.M.; Lechler, R.I. A novel pathway of alloantigen presentation by dendritic cells. J. Immunol. 2004, 173, 4828–4837. [Google Scholar] [CrossRef]
- Li, X.C.; Turka, L.A. An update on regulatory T cells in transplant tolerance and rejection. Nat. Rev. Nephrol. 2010, 6, 577–583. [Google Scholar] [CrossRef]
- Ohkura, N.; Kitagawa, Y.; Sakaguchi, S. Development and Maintenance of Regulatory T cells. Immunity 2013, 38, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, J.; Burrows, P.D.; Wang, J.-Y. B Cell Development and Maturation. In B Cells in Immunity and Tolerance; Wang, J.-Y., Ed.; Springer: Singapore, 2020; pp. 1–22. [Google Scholar]
- Pillai, S.; Cariappa, A. The follicular versus marginal zone B lymphocyte cell fate decision. Nat. Rev. Immunol. 2009, 9, 767–777. [Google Scholar] [CrossRef]
- Akkaya, M.; Kwak, K.; Pierce, S.K. B cell memory: Building two walls of protection against pathogens. Nat. Rev. Immunol. 2020, 20, 229–238. [Google Scholar] [CrossRef]
- Chong, A.S. Mechanisms of organ transplant injury mediated by B cells and antibodies: Implications for antibody-mediated rejection. Am. J. Transplant. 2020, 20 (Suppl. 4), 23–32. [Google Scholar] [CrossRef] [PubMed]
- Steele, D.J.; Laufer, T.M.; Smiley, S.T.; Ando, Y.; Grusby, M.J.; Glimcher, L.H.; Auchincloss, H., Jr. Two levels of help for B cell alloantibody production. J. Exp. Med. 1996, 183, 699–703. [Google Scholar] [CrossRef]
- Martin, A.; Chahwan, R.; Parsa, J.Y.; Scharff, M.D. Somatic Hypermutation: The Molecular Mechanisms Underlying the Production of Effective High-Affinity Antibodies. In Molecular Biology of B Cells, 2nd ed.; Alt, F.W., Honjo, T., Radbruch, A., Reth, M., Eds.; Academic Press: London, UK, 2015; pp. 363–388. [Google Scholar]
- Mayer, C.T.; Gazumyan, A.; Kara, E.E.; Gitlin, A.D.; Golijanin, J.; Viant, C.; Pai, J.; Oliveira, T.Y.; Wang, Q.; Escolano, A.; et al. The microanatomic segregation of selection by apoptosis in the germinal center. Science 2017, 358, eaao2602. [Google Scholar] [CrossRef] [PubMed]
- Nutt, S.L.; Hodgkin, P.D.; Tarlinton, D.M.; Corcoran, L.M. The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 2015, 15, 160–171. [Google Scholar] [CrossRef]
- Shapiro-Shelef, M.; Calame, K. Regulation of plasma-cell development. Nat. Rev. Immunol. 2005, 5, 230–242. [Google Scholar] [CrossRef]
- Gatto, D.; Brink, R. The germinal center reaction. J. Allergy Clin. Immunol. 2010, 126, 898–907. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Malisan, F.; de Bouteiller, O.; Guret, C.; Lebecque, S.; Banchereau, J.; Mills, F.C.; Max, E.E.; Martinez-Valdez, H. Within Germinal Centers, Isotype Switching of Immunoglobulin Genes Occurs after the Onset of Somatic Mutation. Immunity 1996, 4, 241–250. [Google Scholar] [CrossRef]
- Ochiai, K.; Maienschein-Cline, M.; Simonetti, G.; Chen, J.; Rosenthal, R.; Brink, R.; Chong, A.S.; Klein, U.; Dinner, A.R.; Singh, H.; et al. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity 2013, 38, 918–929. [Google Scholar] [CrossRef] [PubMed]
- Zotos, D.; Coquet, J.M.; Zhang, Y.; Light, A.; D’Costa, K.; Kallies, A.; Corcoran, L.M.; Godfrey, D.I.; Toellner, K.M.; Smyth, M.J.; et al. IL-21 regulates germinal center B cell differentiation and proliferation through a B cell-intrinsic mechanism. J. Exp. Med. 2010, 207, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.T.; Gonzalez, D.G.; Cote, C.M.; Kerfoot, S.M.; Deng, S.; Cheng, Y.; Magari, M.; Haberman, A.M. Germinal center B cell development has distinctly regulated stages completed by disengagement from T cell help. Elife 2017, 6, e19552. [Google Scholar] [CrossRef]
- Mohib, K.; Cherukuri, A.; Rothstein, D.M. Regulatory B cells and transplantation: Almost prime time? Curr. Opin. Organ. Transplant. 2018, 23, 524–532. [Google Scholar] [CrossRef] [PubMed]
- Lucas, J.G.; Co, J.P.; Nwaogwugwu, U.T.; Dosani, I.; Sureshkumar, K.K. Antibody-mediated rejection in kidney transplantation: An update. Expert Opin. Pharmacother. 2011, 12, 579–592. [Google Scholar] [CrossRef] [PubMed]
- Helanterä, I.; Mengel, M. Revisiting acute T cell–mediated rejection in kidney allografts. Am. J. Transplant. 2022, 22, 681–682. [Google Scholar] [CrossRef]
- Shapiro, R.; Young, J.B.; Milford, E.L.; Trotter, J.F.; Bustami, R.T.; Leichtman, A.B. Immunosuppression: Evolution in practice and trends, 1993–2003. Am. J. Transplant. 2005, 5, 874–886. [Google Scholar] [CrossRef]
- Lentine, K.L.; Smith, J.M.; Miller, J.M.; Bradbrook, K.; Larkin, L.; Weiss, S.; Handarova, D.K.; Temple, K.; Israni, A.K.; Snyder, J.J. OPTN/SRTR 2021 Annual Data Report: Kidney; The Scientific Registry of Transplant Recipients: Minneapolis, MN, USA, 2021.
- Gaston, R.S.; Cecka, J.M.; Kasiske, B.L.; Fieberg, A.M.; Leduc, R.; Cosio, F.C.; Gourishankar, S.; Grande, J.; Halloran, P.; Hunsicker, L.; et al. Evidence for antibody-mediated injury as a major determinant of late kidney allograft failure. Transplantation 2010, 90, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Stegall, M.D.; Park, W.D.; Larson, T.S.; Gloor, J.M.; Cornell, L.D.; Sethi, S.; Dean, P.G.; Prieto, M.; Amer, H.; Textor, S.; et al. The histology of solitary renal allografts at 1 and 5 years after transplantation. Am. J. Transplant. 2011, 11, 698–707. [Google Scholar] [CrossRef]
- Roberts, I.S.D.; Reddy, S.; Russell, C.; Davies, D.R.; Friend, P.J.; Handa, A.I.; Morris, P.J. Subclinical rejection and borderline changes in early protocol biopsy specimens after renal transplantation. Transplantation 2004, 77, 1194–1198. [Google Scholar] [CrossRef]
- Mengel, M.; Gwinner, W.; Schwarz, A.; Bajeski, R.; Franz, I.; Bröcker, V.; Becker, T.; Neipp, M.; Klempnauer, J.; Haller, H.; et al. Infiltrates in protocol biopsies from renal allografts. Am. J. Transplant. 2007, 7, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Moreso, F.; Ibernon, M.; Gomà, M.; Carrera, M.; Fulladosa, X.; Hueso, M.; Gil-Vernet, S.; Cruzado, J.M.; Torras, J.; Grinyó, J.M.; et al. Subclinical rejection associated with chronic allograft nephropathy in protocol biopsies as a risk factor for late graft loss. Am. J. Transplant. 2006, 6, 747–752. [Google Scholar] [CrossRef]
- Loupy, A.; Suberbielle-Boissel, C.; Hill, G.S.; Lefaucheur, C.; Anglicheau, D.; Zuber, J.; Martinez, F.; Thervet, E.; Méjean, A.; Charron, D.; et al. Outcome of subclinical antibody-mediated rejection in kidney transplant recipients with preformed donor-specific antibodies. Am. J. Transplant. 2009, 9, 2561–2570. [Google Scholar] [CrossRef]
- Loupy, A.; Haas, M.; Roufosse, C.; Naesens, M.; Adam, B.; Afrouzian, M.; Akalin, E.; Alachkar, N.; Bagnasco, S.; Becker, J.U.; et al. The Banff 2019 Kidney Meeting Report (I): Updates on and clarification of criteria for T cell- and antibody-mediated rejection. Am. J. Transplant. 2020, 20, 2318–2331. [Google Scholar] [CrossRef] [PubMed]
- Oellerich, M.; Sherwood, K.; Keown, P.; Schütz, E.; Beck, J.; Stegbauer, J.; Rump, L.C.; Walson, P.D. Liquid biopsies: Donor-derived cell-free DNA for the detection of kidney allograft injury. Nat. Rev. Nephrol. 2021, 17, 591–603. [Google Scholar] [CrossRef]
- Cornell, L.D.; Smith, R.N.; Colvin, R.B. Kidney transplantation: Mechanisms of rejection and acceptance. Annu. Rev. Pathol. 2008, 3, 189–220. [Google Scholar] [CrossRef]
- Colvin, R.B.; Nickeleit, V. Renal transplant pathology. In Heptinstall’s Pathology of the Kidney; Jennette, J.C., Olson, J.L., Schwartz, M.M., Silva, F.G., Eds.; Lippincott-Raven: Philadelphia, PA, USA, 2006; pp. 1347–1490. [Google Scholar]
- Justiz Vaillant, A.A.; Misra, S.; Fitzgerald, B.M. Acute Transplantation Rejection. In StatPearls; StatPearls Publishing LLC: Treasure Island, FL, USA, 2023. [Google Scholar]
- Hassanein, M.; Augustine, J.J. Chronic Kidney Transplant Rejection. In StatPearls; StatPearls Publishing LLC: Treasure Island, FL, USA, 2023. [Google Scholar]
- Colvin, R.B.; Smith, R.N. Antibody-mediated organ-allograft rejection. Nat. Rev. Immunol. 2005, 5, 807–817. [Google Scholar] [CrossRef]
- Kim, M.; Martin, S.T.; Townsend, K.R.; Gabardi, S. Antibody-Mediated Rejection in Kidney Transplantation: A Review of Pathophysiology, Diagnosis, and Treatment Options. Pharmacotherapy 2014, 34, 733–744. [Google Scholar] [CrossRef]
- Janeway, C.A., Jr.; Travers, P.; Walport, M.; Shlomchik, M.J. The complement system and innate immunity. In Immunobiology: The Immune System in Health and Disease, 5th ed.; Garland Science: New York, NY, USA, 2001. [Google Scholar]
- Walport, M.J. Complement. N. Engl. J. Med. 2001, 344, 1058–1066. [Google Scholar] [CrossRef]
- van den Elsen, J.M.; Martin, A.; Wong, V.; Clemenza, L.; Rose, D.R.; Isenman, D.E. X-ray crystal structure of the C4d fragment of human complement component C4. J. Mol. Biol. 2002, 322, 1103–1115. [Google Scholar] [CrossRef]
- Benzaquen, L.R.; Nicholson-Weller, A.; Halperin, J.A. Terminal complement proteins C5b-9 release basic fibroblast growth factor and platelet-derived growth factor from endothelial cells. J. Exp. Med. 1994, 179, 985–992. [Google Scholar] [CrossRef]
- Saadi, S.; Holzknecht, R.A.; Patte, C.P.; Stern, D.M.; Platt, J.L. Complement-mediated regulation of tissue factor activity in endothelium. J. Exp. Med. 1995, 182, 1807–1814. [Google Scholar] [CrossRef] [PubMed]
- Gómez Román, V.R.; Murray, J.C.; Weiner, L.M. Chapter 1—Antibody-Dependent Cellular Cytotoxicity (ADCC). In Antibody Fc; Ackerman, M.E., Nimmerjahn, F., Eds.; Academic Press: Boston, MA, USA, 2014; pp. 1–27. [Google Scholar]
- Jeong, H.J. Diagnosis of renal transplant rejection: Banff classification and beyond. Kidney Res. Clin. Pract. 2020, 39, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Haas, M.; Sis, B.; Racusen, L.C.; Solez, K.; Glotz, D.; Colvin, R.B.; Castro, M.C.; David, D.S.; David-Neto, E.; Bagnasco, S.M.; et al. Banff 2013 meeting report: Inclusion of c4d-negative antibody-mediated rejection and antibody-associated arterial lesions. Am. J. Transplant. 2014, 14, 272–283. [Google Scholar] [CrossRef]
- Dominy, K.M.; Willicombe, M.; Al Johani, T.; Beckwith, H.; Goodall, D.; Brookes, P.; Cook, H.T.; Cairns, T.; McLean, A.; Roufosse, C. Molecular Assessment of C4d-Positive Renal Transplant Biopsies Without Evidence of Rejection. Kidney Int. Rep. 2019, 4, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Senev, A.; Coemans, M.; Lerut, E.; Van Sandt, V.; Daniëls, L.; Kuypers, D.; Sprangers, B.; Emonds, M.P.; Naesens, M. Histological picture of antibody-mediated rejection without donor-specific anti-HLA antibodies: Clinical presentation and implications for outcome. Am. J. Transplant. 2019, 19, 763–780. [Google Scholar] [CrossRef]
- Delville, M.; Lamarthée, B.; Pagie, S.; See, S.B.; Rabant, M.; Burger, C.; Gatault, P.; Giral, M.; Thaunat, O.; Arzouk, N.; et al. Early Acute Microvascular Kidney Transplant Rejection in the Absence of Anti-HLA Antibodies Is Associated with Preformed IgG Antibodies against Diverse Glomerular Endothelial Cell Antigens. J. Am. Soc. Nephrol. 2019, 30, 692–709. [Google Scholar] [CrossRef] [PubMed]
- Callemeyn, J.; Lerut, E.; de Loor, H.; Arijs, I.; Thaunat, O.; Koenig, A.; Meas-Yedid, V.; Olivo-Marin, J.-C.; Halloran, P.; Chang, J.; et al. Transcriptional Changes in Kidney Allografts with Histology of Antibody-Mediated Rejection without Anti-HLA Donor-Specific Antibodies. J. Am. Soc. Nephrol. 2020, 31, 2168–2183. [Google Scholar] [CrossRef]
- Sablik, K.A.; Clahsen-van Groningen, M.C.; Looman, C.W.N.; Damman, J.; Roelen, D.L.; van Agteren, M.; Betjes, M.G.H. Chronic-active antibody-mediated rejection with or without donor-specific antibodies has similar histomorphology and clinical outcome—A retrospective study. Transpl. Int. 2018, 31, 900–908. [Google Scholar] [CrossRef]
- Koenig, A.; Mezaache, S.; Callemeyn, J.; Barba, T.; Mathias, V.; Sicard, A.; Charreau, B.; Rabeyrin, M.; Dijoud, F.; Picard, C.; et al. Missing Self-Induced Activation of NK Cells Combines with Non-Complement-Fixing Donor-Specific Antibodies to Accelerate Kidney Transplant Loss in Chronic Antibody-Mediated Rejection. J. Am. Soc. Nephrol. 2021, 32, 479–494. [Google Scholar] [CrossRef]
- Reindl-Schwaighofer, R.; Heinzel, A.; Kainz, A.; van Setten, J.; Jelencsics, K.; Hu, K.; Loza, B.L.; Kammer, M.; Heinze, G.; Hruba, P.; et al. Contribution of non-HLA incompatibility between donor and recipient to kidney allograft survival: Genome-wide analysis in a prospective cohort. Lancet 2019, 393, 910–917. [Google Scholar] [CrossRef] [PubMed]
- Schinstock, C.A.; Sapir-Pichhadze, R.; Naesens, M.; Batal, I.; Bagnasco, S.; Bow, L.; Campbell, P.; Clahsen-van Groningen, M.C.; Cooper, M.; Cozzi, E.; et al. Banff survey on antibody-mediated rejection clinical practices in kidney transplantation: Diagnostic misinterpretation has potential therapeutic implications. Am. J. Transplant. 2019, 19, 123–131. [Google Scholar] [CrossRef]
- Yoo, D.; Goutaudier, V.; Divard, G.; Gueguen, J.; Astor, B.C.; Aubert, O.; Raynaud, M.; Demir, Z.; Hogan, J.; Weng, P.; et al. An automated histological classification system for precision diagnostics of kidney allografts. Nat. Med. 2023, 29, 1211–1220. [Google Scholar] [CrossRef]
- Tran, T.H.; Döhler, B.; Heinold, A.; Scherer, S.; Ruhenstroth, A.; Opelz, G. Deleterious impact of mismatching for human leukocyte antigen-C in presensitized recipients of kidney transplants. Transplantation 2011, 92, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, K.; Vögeler, U.; Albrecht, K.H.; Eigler, F.W.; Buchholz, B.; Grosse-Wilde, H. HLA-DP antibodies in patients awaiting renal transplantation. Transpl. Int. 1995, 8, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Goral, S.; Prak, E.L.; Kearns, J.; Bloom, R.D.; Pierce, E.; Doyle, A.; Grossman, R.; Naji, A.; Kamoun, M. Preformed donor-directed anti-HLA-DP antibodies may be an impediment to successful kidney transplantation. Nephrol. Dial. Transplant. 2008, 23, 390–392. [Google Scholar] [CrossRef] [PubMed]
- Sypek, M.P.; Hughes, P. HLA Eplet Mismatches in Kidney Transplantation: More Than Just Adding Things Up. Kidney Int. Rep. 2021, 6, 1500–1502. [Google Scholar] [CrossRef]
- Kishikawa, H.; Kinoshita, T.; Hashimoto, M.; Fukae, S.; Taniguchi, A.; Yamanaka, K.; Nakagawa, M.; Nishimura, K. Class II HLA Eplet Mismatch Is a Risk Factor for De Novo Donor-Specific Antibody Development and Antibody-mediated Rejection in Kidney Transplantation Recipients. Transplant. Proc. 2018, 50, 2388–2391. [Google Scholar] [CrossRef]
- Duquesnoy, R.J. HLAMatchmaker: A molecularly based algorithm for histocompatibility determination. I. Description of the algorithm. Hum. Immunol. 2002, 63, 339–352. [Google Scholar] [CrossRef]
- Sawitzki, B.; Harden, P.N.; Reinke, P.; Moreau, A.; Hutchinson, J.A.; Game, D.S.; Tang, Q.; Guinan, E.C.; Battaglia, M.; Burlingham, W.J.; et al. Regulatory cell therapy in kidney transplantation (The ONE Study): A harmonised design and analysis of seven non-randomised, single-arm, phase 1/2A trials. Lancet 2020, 395, 1627–1639. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tamargo, C.L.; Kant, S. Pathophysiology of Rejection in Kidney Transplantation. J. Clin. Med. 2023, 12, 4130. https://doi.org/10.3390/jcm12124130
Tamargo CL, Kant S. Pathophysiology of Rejection in Kidney Transplantation. Journal of Clinical Medicine. 2023; 12(12):4130. https://doi.org/10.3390/jcm12124130
Chicago/Turabian StyleTamargo, Christina L., and Sam Kant. 2023. "Pathophysiology of Rejection in Kidney Transplantation" Journal of Clinical Medicine 12, no. 12: 4130. https://doi.org/10.3390/jcm12124130
APA StyleTamargo, C. L., & Kant, S. (2023). Pathophysiology of Rejection in Kidney Transplantation. Journal of Clinical Medicine, 12(12), 4130. https://doi.org/10.3390/jcm12124130




