Exploring the Role of CD74 and D-Dopachrome Tautomerase in COVID-19: Insights from Transcriptomic and Serum Analyses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Transcriptomic Study
2.2. Patients
2.2.1. COVID-19 Patients and Healthy Donors
2.2.2. Clinical Characteristics
2.2.3. Blood Samples
2.2.4. Enzyme-Linked Immunosorbent Assay (ELISA) for Soluble Serum CD74 and D-DT
2.2.5. Flow Cytometry Analysis of Th1, Th2, and Th17 Cytokines
2.3. Statistical Analysis
3. Results
3.1. Transcriptomic Study
3.2. Patients
3.2.1. COVID-19 Patients Distribution
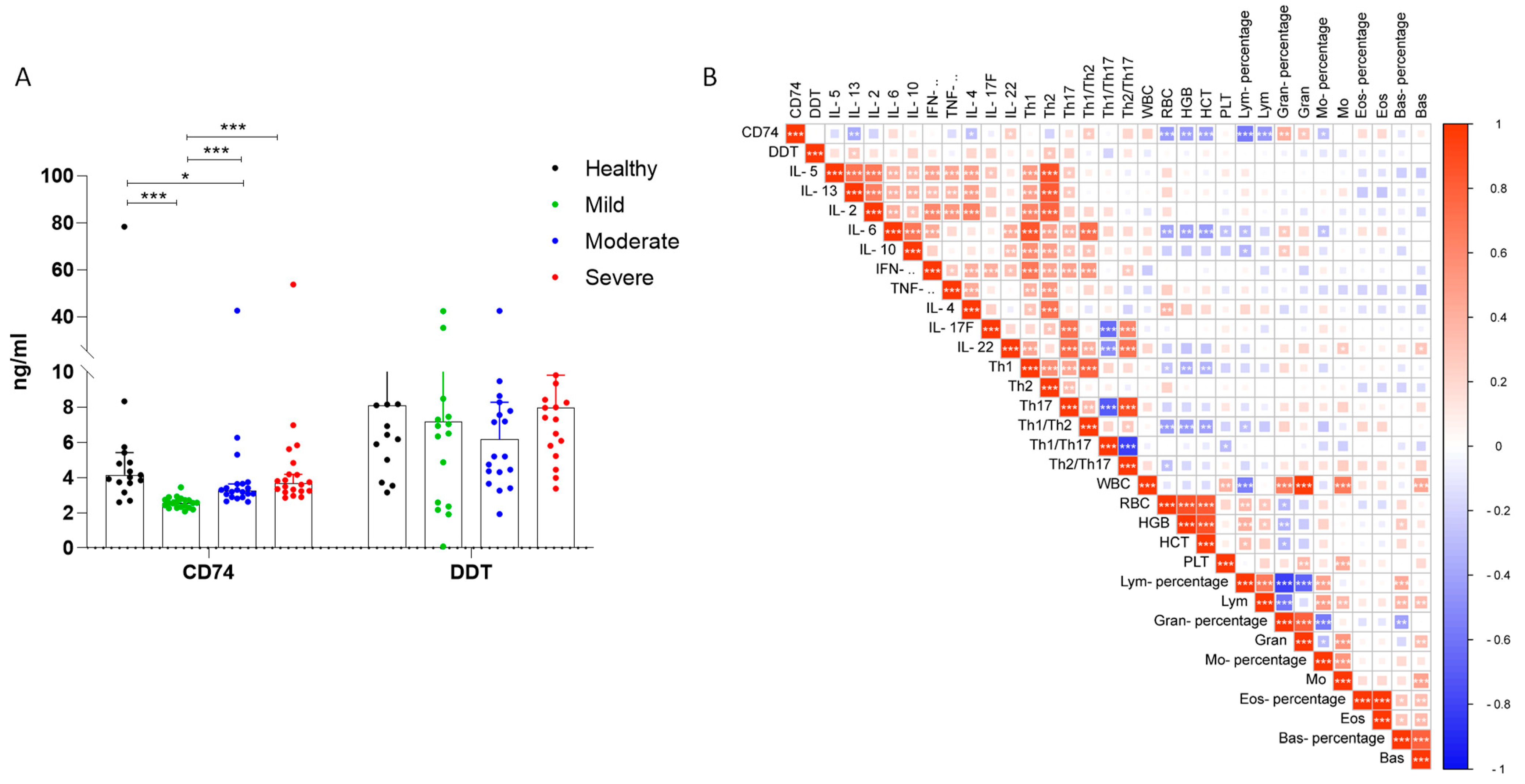
3.2.2. Serum CD74 and D-DT Levels in COVID-19 Patients
3.2.3. Analysis of CD74 and D-DT Levels in Recovered Cases and Lethal Cases from Severe COVID-19 Patients
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Latorre, D. Autoimmunity and SARS-CoV-2 infection: Unraveling the link in neurological disorders. Eur. J. Immunol. 2022, 52, 1561–1571. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, E.; Petralia, M.C.; Basile, M.S.; Bramanti, A.; Bramanti, P.; Nicoletti, F.; Spandidos, D.A.; Shoenfeld, Y.; Fagone, P. Transcriptomic analysis of COVID-19 lungs and bronchoalveolar lavage fluid samples reveals predominant B cell activation responses to infection. Int. J. Mol. Med. 2020, 46, 1266–1273. [Google Scholar] [CrossRef] [PubMed]
- Dharra, R.; Sharma, A.K.; Datta, S. Emerging aspects of cytokine storm in COVID-19: The role of proinflammatory cytokines and therapeutic prospects. Cytokine 2023, 169, 156287. [Google Scholar] [CrossRef] [PubMed]
- Maison, D.P.; Deng, Y.; Gerschenson, M. SARS-CoV-2 and the host-immune response. Front. Immunol. 2023, 14, 1195871. [Google Scholar] [CrossRef]
- Du, T.; Gao, C.; Lu, S.; Liu, Q.; Yu, W.; Li, W.; Sun, Y.Q.; Tang, C.; Wang, J.; Gao, J.; et al. Differential Transcriptomic Landscapes of SARS-CoV-2 Variants in Multiple Organs from Infected Rhesus Macaques. Genom. Proteom. Bioinform. 2023, in press. [CrossRef]
- Xu, J.; Li, X.-X.; Yuan, N.; Li, C.; Yang, J.-G.; Cheng, L.-M.; Lu, Z.-X.; Hou, H.-Y.; Zhang, B.; Hu, H.; et al. T cell receptor β repertoires in patients with COVID-19 reveal disease severity signatures. Front. Immunol. 2023, 14, 1190844. [Google Scholar] [CrossRef]
- Taylor, L. COVID-19: True global death toll from pandemic is almost 15 million, says WHO. BMJ 2022, 377, o1144. [Google Scholar] [CrossRef]
- Martonik, D.; Parfieniuk-Kowerda, A.; Starosz, A.; Grubczak, K.; Moniuszko, M.; Flisiak, R. Effect of antiviral and immunomodulatory treatment on a cytokine profile in patients with COVID-19. Front. Immunol. 2023, 14, 1222170. [Google Scholar] [CrossRef]
- Günther, S.; Fagone, P.; Jalce, G.; Atanasov, A.G.; Guignabert, C.; Nicoletti, F. Role of MIF and D-DT in immune-inflammatory, autoimmune, and chronic respiratory diseases: From pathogenic factors to therapeutic targets. Drug Discov. Today 2019, 24, 428–439. [Google Scholar] [CrossRef]
- Toldi, J.; Kelava, L.; Marton, S.; Muhl, D.; Kustan, P.; Feher, Z.; Maar, K.; Garai, J.; Pakai, E.; Garami, A. Distinct patterns of serum and urine macrophage migration inhibitory factor kinetics predict death in sepsis: A prospective, observational clinical study. Sci. Rep. 2023, 13, 588. [Google Scholar] [CrossRef]
- Ferreira, P.T.M.; Oliveira-Scussel, A.C.M.; Sousa, R.A.P.; Gomes, B.Q.; Félix, J.E.; Silva, R.J.; Millian, I.B.; Assunção, T.S.F.; Teixeira, S.C.; Gomes, M.d.L.M.; et al. Macrophage Migration Inhibitory Factor contributes to drive phenotypic and functional macrophages activation in response to Toxoplasma gondii infection. Immunobiology 2023, 228, 152357. [Google Scholar] [CrossRef]
- Huang, G.; Ma, L.; Shen, L.; Lei, Y.; Guo, L.; Deng, Y.; Ding, Y. MIF/SCL3A2 depletion inhibits the proliferation and metastasis of colorectal cancer cells via the AKT/GSK-3β pathway and cell iron death. J. Cell. Mol. Med. 2022, 26, 3410–3422. [Google Scholar] [CrossRef]
- Garcia-Gerique, L.; García, M.; Garrido-Garcia, A.; Gómez-González, S.; Torrebadell, M.; Prada, E.; Pascual-Pasto, G.; Muñoz, O.; Perez-Jaume, S.; Lemos, I.; et al. MIF/CXCR4 signaling axis contributes to survival, invasion, and drug resistance of metastatic neuroblastoma cells in the bone marrow microenvironment. BMC Cancer 2022, 22, 669. [Google Scholar] [CrossRef]
- Zan, C.; Yang, B.; Brandhofer, M.; El Bounkari, O.; Bernhagen, J. D-dopachrome tautomerase in cardiovascular and inflammatory diseases—A new kid on the block or just another MIF? FASEB J. 2022, 36, e22601. [Google Scholar] [CrossRef] [PubMed]
- Barthelmess, R.M.; Stijlemans, B.; Van Ginderachter, J.A. Hallmarks of Cancer Affected by the MIF Cytokine Family. Cancers 2023, 15, 395. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Cheng, R.; Zhang, D.; Guo, Y.; Li, F.; Li, Y.; Li, Y.; Bai, X.; Mo, J.; Huang, C. MIF promotes cell invasion by the LRP1-uPAR interaction in pancreatic cancer cells. Front. Oncol. 2023, 12, 1028070. [Google Scholar] [CrossRef]
- Zhu, G.-Q.; Tang, Z.; Huang, R.; Qu, W.-F.; Fang, Y.; Yang, R.; Tao, C.-Y.; Gao, J.; Wu, X.-L.; Sun, H.-X.; et al. CD36+ cancer-associated fibroblasts provide immunosuppressive microenvironment for hepatocellular carcinoma via secretion of macrophage migration inhibitory factor. Cell Discov. 2023, 9, 25. [Google Scholar] [CrossRef]
- Fang, T.; Liu, L.; Song, D.; Huang, D. The role of MIF in periodontitis: A potential pathogenic driver, biomarker, and therapeutic target. Oral Dis. 2023. [Google Scholar] [CrossRef] [PubMed]
- Huth, S.; Huth, L.; Heise, R.; Marquardt, Y.; Lopopolo, L.; Piecychna, M.; Boor, P.; Fingerle-Rowson, G.; Kapurniotu, A.; Yazdi, A.S.; et al. Macrophage migration inhibitory factor (MIF) and its homolog D-dopachrome tautomerase (D-DT) are significant promotors of UVB-but not chemically induced non-melanoma skin cancer. Sci. Rep. 2023, 13, 11611. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Li, R.; Song, S.; Ma, L.; Xue, H. The Role of MIF-173G/C Gene Polymorphism in the Susceptibility of Autoimmune Diseases. Mediat. Inflamm. 2020, 2020, 7825072. [Google Scholar] [CrossRef]
- Gupta, P.; Joshi, N.; Uprety, S.; Dogra, S.; De, D.; Handa, S.; Minz, R.W.; Singh, S.; Chhabra, S. Association of MIF gene polymorphisms with pemphigus vulgaris: A case-control study with comprehensive review of the literature. Int. J. Clin. Exp. Pathol. 2021, 14, 1080–1089. [Google Scholar] [PubMed]
- Thiele, M.; Donnelly, S.C.; Mitchell, R.A. OxMIF: A druggable isoform of macrophage migration inhibitory factor in cancer and inflammatory diseases. J. Immunother. Cancer 2022, 10, e005475. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.T.; Beck, J.; Lue, H.; Fünfzig, H.; Kleemann, R.; Koolwijk, P.; Kapurniotu, A.; Bernhagen, J. A 16-Residue Peptide Fragment of Macrophage Migration Inhibitory Factor, MIF-(50–65), Exhibits Redox Activity and Has MIF-like Biological Functions. J. Biol. Chem. 2003, 278, 33654–33671. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.; VanPatten, S.; Deen, N.S.; Al-Abed, Y.; Morand, E.F. Rediscovering MIF: New Tricks for an Old Cytokine. Trends Immunol. 2019, 40, 447–462. [Google Scholar] [CrossRef] [PubMed]
- Kang, I.; Bucala, R. The immunobiology of MIF: Function, genetics and prospects for precision medicine. Nat. Rev. Rheumatol. 2019, 15, 427–437. [Google Scholar] [CrossRef]
- Merk, M.; Mitchell, R.A.; Endres, S.; Bucala, R. D-dopachrome tautomerase (D-DT or MIF-2): Doubling the MIF cytokine family. Cytokine 2012, 59, 10–17. [Google Scholar] [CrossRef] [Green Version]
- Tilstam, P.V.; Pantouris, G.; Corman, M.; Andreoli, M.; Mahboubi, K.; Davis, G.; Du, X.; Leng, L.; Lolis, E.; Bucala, R. A selective small-molecule inhibitor of macrophage migration inhibitory factor-2 (MIF-2), a MIF cytokine superfamily member, inhibits MIF-2 biological activity. J. Biol. Chem. 2019, 294, 18522–18531. [Google Scholar] [CrossRef] [Green Version]
- Aksakal, A.; Kerget, B.; Kerget, F.; Aşkın, S. Evaluation of the relationship between macrophage migration inhibitory factor level and clinical course in patients with COVID-19 pneumonia. J. Med. Virol. 2021, 93, 6519–6524. [Google Scholar] [CrossRef]
- Bleilevens, C.; Soppert, J.; Hoffmann, A.; Breuer, T.; Bernhagen, J.; Martin, L.; Stiehler, L.; Marx, G.; Dreher, M.; Stoppe, C.; et al. Macrophage Migration Inhibitory Factor (MIF) Plasma Concentration in Critically Ill COVID-19 Patients: A Prospective Observational Study. Diagnostics 2021, 11, 332. [Google Scholar] [CrossRef]
- Dheir, H.; Yaylaci, S.; Sipahi, S.; Genc, A.C.; Cekic, D.; Tuncer, F.B.; Cokluk, E.; Kocayigit, H.; Genc, A.B.; Salihi, S.; et al. Does Macrophage Migration Inhibitory Factor predict the prognosis of COVID-19 disease? J. Infect. Dev. Ctries. 2021, 15, 398–403. [Google Scholar] [CrossRef]
- Del Valle, D.M.; Kim-Schulze, S.; Huang, H.-H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020, 26, 1636–1643. [Google Scholar] [CrossRef] [PubMed]
- Langnau, C.; Janing, H.; Kocaman, H.; Gekeler, S.; Günter, M.; Petersen-Uribe, Á.; Jaeger, P.; Koch, B.; Kreisselmeier, K.-P.; Castor, T.; et al. Recovery of systemic hyperinflammation in patients with severe SARS-CoV-2 infection. Biomarkers 2023, 28, 97–110. [Google Scholar] [CrossRef]
- Arunachalam, P.S.; Wimmers, F.; Mok, C.K.P.; Perera, R.A.P.M.; Scott, M.; Hagan, T.; Sigal, N.; Feng, Y.; Bristow, L.; Tsang, O.T.-Y.; et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 2020, 369, 1210–1220. [Google Scholar] [CrossRef] [PubMed]
- Al Mahi, N.; Najafabadi, M.F.; Pilarczyk, M.; Kouril, M.; Medvedovic, M. GREIN: An Interactive Web Platform for Re-analyzing GEO RNA-seq Data. Sci. Rep. 2019, 9, 7580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, J.J.; Fan, W.; Par-Young, J.; Piecychna, M.; Leng, L.; Israni-Winger, K.; Qing, H.; Gu, J.; Zhao, H.; Schulz, W.L.; et al. MIF is a common genetic determinant of COVID-19 symptomatic infection and severity. QJM 2023, 116, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Rajasekaran, D.; Zierow, S.; Syed, M.; Bucala, R.; Bhandari, V.; Lolis, E.J. Targeting distinct tautomerase sites of D-DT and MIF with a single molecule for inhibition of neutrophil lung recruitment. FASEB J. 2014, 28, 4961–4971. [Google Scholar] [CrossRef] [Green Version]
- Keene, J.D. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc. Natl. Acad. Sci. USA 2001, 98, 7018–7024. [Google Scholar] [CrossRef]
- Gibellini, L.; De Biasi, S.; Meschiari, M.; Gozzi, L.; Paolini, A.; Borella, R.; Mattioli, M.; Tartaro, D.L.; Fidanza, L.; Neroni, A.; et al. Plasma Cytokine Atlas Reveals the Importance of TH2 Polarization and Interferons in Predicting COVID-19 Severity and Survival. Front. Immunol. 2022, 13, 842150. [Google Scholar] [CrossRef]
- Pons, M.J.; Ymaña, B.; Mayanga-Herrera, A.; Sáenz, Y.; Alvarez-Erviti, L.; Tapia-Rojas, S.; Gamarra, R.; Blanco, A.B.; Moncunill, G.; Ugarte-Gil, M.F. Cytokine Profiles Associated With Worse Prognosis in a Hospitalized Peruvian COVID-19 Cohort. Front. Immunol. 2021, 12, 700921. [Google Scholar] [CrossRef]
- Montazersaheb, S.; Khatibi, S.M.H.; Hejazi, M.S.; Tarhriz, V.; Farjami, A.; Sorbeni, F.G.; Farahzadi, R.; Ghasemnejad, T. COVID-19 infection: An overview on cytokine storm and related interventions. Virol. J. 2022, 19, 92. [Google Scholar] [CrossRef]
- Donlan, A.N.; Sutherland, T.E.; Marie, C.; Preissner, S.; Bradley, B.T.; Carpenter, R.M.; Sturek, J.M.; Ma, J.Z.; Moreau, G.B.; Donowitz, J.R.; et al. IL-13 is a driver of COVID-19 severity. J. Clin. Investig. 2021, 6, e150107. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, F.; Mancuso, G.; Cusumano, V.; Di Marco, R.; Zaccone, P.; Bendtzen, K.; Teti, G. Prevention of endotoxin-induced lethality in neonatal mice by interleukin-13. Eur. J. Immunol. 1997, 27, 1580–1583. [Google Scholar] [CrossRef] [PubMed]
- Baumhofer, J.M.; Beinhauer, B.G.; Wang, J.E.; Brandmeier, H.; Geissler, K.; Losert, U.; Philip, R.; Aversa, G.; Rogy, M.A. Gene Transfer with IL-4 and IL-13 Improves Survival in Lethal Endotoxemia in the Mouse and Ameliorates Peritoneal Macrophages Immune Competence. Eur. J. Immunol. 1998, 28, 610–615. [Google Scholar] [CrossRef]
- Muchamuel, T.; Menon, S.; Pisacane, P.; Howard, M.C.; A Cockayne, D. IL-13 protects mice from lipopolysaccharide-induced lethal endotoxemia: Correlation with down-modulation of TNF-alpha, IFN-gamma, and IL-12 production. J. Immunol. 1997, 158, 2898–2903. [Google Scholar] [CrossRef] [PubMed]
- Szabo, S.; Zayachkivska, O.; Hussain, A.; Muller, V. What is really ‘Long COVID’? Inflammopharmacology 2023, 31, 551–557. [Google Scholar] [CrossRef]
- Cavalli, E.; Mazzon, E.; Basile, M.S.; Mangano, K.; Di Marco, R.; Bramanti, P.; Nicoletti, F.; Fagone, P.; Petralia, M.C. Upregulated Expression of Macrophage Migration Inhibitory Factor, Its Analogue D-Dopachrome Tautomerase, and the CD44 Receptor in Peripheral CD4 T Cells from Clinically Isolated Syndrome Patients with Rapid Conversion to Clinical Defined Multiple Sclerosis. Medicina 2019, 55, 667. [Google Scholar] [CrossRef] [Green Version]
- Fagone, P.; Mazzon, E.; Cavalli, E.; Bramanti, A.; Petralia, M.C.; Mangano, K.; Al-Abed, Y.; Bramati, P.; Nicoletti, F. Contribution of the macrophage migration inhibitory factor superfamily of cytokines in the pathogenesis of preclinical and human multiple sclerosis: In silico and in vivo evidences. J. Neuroimmunol. 2018, 322, 46–56. [Google Scholar] [CrossRef]
- Benedek, G.; Meza-Romero, R.; Jordan, K.; Zhang, Y.; Nguyen, H.; Kent, G.; Li, J.; Siu, E.; Frazer, J.; Piecychna, M.; et al. MIF and D-DT are potential disease severity modifiers in male MS subjects. Proc. Natl. Acad. Sci. USA 2017, 114, E8421–E8429. [Google Scholar] [CrossRef]
- Han, Z.; Qu, J.; Zhao, J.; Zou, X. Genetic Variant rs755622 Regulates Expression of the Multiple Sclerosis Severity Modifier D-Dopachrome Tautomerase in a Sex-Specific Way. BioMed Res. Int. 2018, 2018, 8285653. [Google Scholar] [CrossRef] [Green Version]
- Nicoletti, F.; Créange, A.; Orlikowski, D.; Bolgert, F.; Mangano, K.; Metz, C.; Di Marco, R.; Al Abed, Y. Macrophage migration inhibitory factor (MIF) seems crucially involved in Guillain–Barré syndrome and experimental allergic neuritis. J. Neuroimmunol. 2005, 168, 168–174. [Google Scholar] [CrossRef]
- Laudanski, K.; Hajj, J.; Restrepo, M.; Siddiq, K.; Okeke, T.; Rader, D.J. Dynamic Changes in Central and Peripheral Neuro-Injury vs. Neuroprotective Serum Markers in COVID-19 Are Modulated by Different Types of Anti-Viral Treatments but Do Not Affect the Incidence of Late and Early Strokes. Biomedicines 2021, 9, 1791. [Google Scholar] [CrossRef] [PubMed]


| A. | ||||||||||
| ID Sample | Age | Gender | WBC | RBC | HGB g/L | HCT | PLT | Lym% | Lym# | Gran% |
| 2 | 51 | woman | 4.63 | 5.6 | 133 | 41.8 | 280 | 40.4 | 1.87 | 48.4 |
| 5 | 44 | woman | 10.04 | 4.49 | 135 | 39.7 | 241 | 28.3 | 2.84 | 63.1 |
| 6 | 45 | man | 8.61 | 5.66 | 159 | 46.7 | 265 | 27.1 | 2.33 | 63.3 |
| 8 | 49 | man | 9.73 | 5.08 | 152 | 44.8 | 289 | 28.7 | 2.79 | 61.7 |
| 12 | 72 | man | 6.13 | 5.18 | 169 | 48.1 | 106 | 18.6 | 1.14 | 77.3 |
| 14 | 52 | man | 9.01 | 5.4 | 163 | 48.2 | 376 | 9.2 | 0.83 | 75.8 |
| 15 | 61 | man | 5.24 | 5.11 | 161 | 45.9 | 219 | 27.9 | 1.46 | 63.5 |
| 40 | 40 | man | 8.13 | 5.3 | 148 | 44.3 | 261 | 9.1 | 0.74 | 86.5 |
| 46 | 39 | man | 2.55 | 4.74 | 139 | 40.9 | 185 | 24.7 | 0.63 | 67.5 |
| 48 | 75 | man | 14.98 | 3.52 | 108 | 33.3 | 262 | 13.8 | 2.07 | 81.4 |
| 67 | 57 | man | 5.68 | 4.86 | 146 | 42.3 | 91 | 31.3 | 1.78 | 56.5 |
| 75 | 47 | woman | 6.55 | 5.66 | 150 | 45.6 | 236 | 25.3 | 1.66 | 68.5 |
| 76 | 50 | man | 6.32 | 4.18 | 153 | 44.2 | 204 | 16.9 | 1.07 | 72.8 |
| 78 | 80 | man | 14.95 | 4.19 | 141 | 40.7 | 440 | 14.4 | 2.15 | 86.8 |
| 85 | 75 | man | 6.23 | 5.68 | 165 | 48.3 | 174 | 39.5 | 2.46 | 48.4 |
| 92 | 48 | man | 3.9 | 4.76 | 146 | 42.1 | 195 | 28.7 | 1.12 | 61.8 |
| 93 | 83 | woman | 3.94 | 4.51 | 140 | 4 | 138 | 32 | 1.26 | 59.8 |
| 94 | 70 | man | 10.68 | 5.07 | 135 | 41.6 | 219 | 5.3 | 0.57 | 90 |
| 96 | 48 | woman | 7.66 | 3.91 | 109 | 34.2 | 354 | 17.2 | 1.32 | 73.1 |
| 99 | 25 | woman | 5.24 | 5.05 | 144 | 44.5 | 173 | 16.6 | 0.87 | 80.3 |
| B. | ||||||||||
| ID Sample | Age | Gran# | Mo% | Mo# | Eo% | Eo# | Ba% | Ba# | SO2% | Fever |
| 2 | 51 | 2.24 | 11 | 0.51 | 0 | 0 | 0.2 | 0.01 | 96% | 38.6 |
| 5 | 44 | 6.34 | 8.2 | 0.82 | 0.2 | 0.02 | 0.2 | 0.02 | 98% | 37.6 |
| 6 | 45 | 5.45 | 8.1 | 0.7 | 1.3 | 0.11 | 0.2 | 0.02 | 95% | 37.3 |
| 8 | 49 | 6 | 8.4 | 0.82 | 0.9 | 0.09 | 0.9 | 0.09 | 98% | 37.7 |
| 12 | 72 | 4.74 | 3.9 | 0.24 | 0 | 0 | 0.2 | 0.01 | 95% | 37.4 |
| 14 | 52 | 6.83 | 14.9 | 1.34 | 0 | 0 | 0.1 | 0.01 | 96% | 37.9 |
| 15 | 61 | 3.33 | 8.2 | 0.43 | 0.2 | 0.01 | 0.2 | 0.01 | 97% | 37.8 |
| 40 | 40 | 7.03 | 4.4 | 0.36 | 0 | 0 | 0 | 0 | 97% | 38 |
| 46 | 39 | 1.72 | 7.8 | 0.2 | 0 | 0 | 0 | 0 | 96% | 37.7 |
| 48 | 75 | 12.19 | 4.1 | 0.62 | 0.4 | 0.06 | 0.3 | 0.04 | 97% | 37.2 |
| 67 | 57 | 3.21 | 12 | 0.68 | 0 | 0 | 0.2 | 0.01 | 97% | 38 |
| 75 | 47 | 4.49 | 6 | 0.39 | 0 | 0 | 0.2 | 0.01 | 96% | 38.6 |
| 76 | 50 | 4.6 | 9.5 | 0.6 | 0.5 | 0.03 | 0.3 | 0.02 | 97% | 37.3 |
| 78 | 80 | 12.97 | 8.1 | 1.21 | 0.3 | 0.05 | 0.4 | 0.06 | 98% | 37.8 |
| 85 | 75 | 3.02 | 11.6 | 0.72 | 0 | 0 | 0.5 | 0.03 | 96% | 37.8 |
| 92 | 48 | 1.12 | 9 | 0.35 | 0 | 0 | 0.5 | 0.02 | 96% | 37.9 |
| 93 | 83 | 2.36 | 7.9 | 0.31 | 0 | 0 | 0.3 | 0.01 | 97% | 37 |
| 94 | 70 | 9.61 | 4.6 | 0.49 | 0 | 0 | 0.1 | 0.01 | 96% | 37.4 |
| 96 | 48 | 5.6 | 9.7 | 0.74 | 0 | 0 | 0 | 0 | 95% | 37.6 |
| 99 | 25 | 4.21 | 2.9 | 0.15 | 0 | 0 | 0.2 | 0.01 | 98% | 37.8 |
| A. | ||||||||||
| ID Sample | Age | Gender | WBC | RBC | HGB g/L | HCT | PLT | Lym% | Lym# | Gran% |
| 1 | 58 | woman | 7.63 | 4.32 | 122 | 37.1 | 197 | 8 | 0.61 | 86.9 |
| 3 | 58 | woman | 10.28 | 3.97 | 119 | 34.6 | 295 | 10.1 | 1.04 | 86.3 |
| 4 | 77 | man | 8.23 | 2.83 | 95 | 28.3 | 170 | 1.8 | 0.15 | 94.2 |
| 11 | 58 | woman | 5.44 | 4.36 | 132 | 39.2 | 241 | 27.8 | 1.51 | 63.9 |
| 18 | 66 | man | 16.93 | 5.11 | 151 | 44.4 | 400 | 4.3 | 0.72 | 88.7 |
| 20 | 73 | man | 5.08 | 3.78 | 120 | 35.8 | 247 | 12 | 0.61 | 80.7 |
| 23 | 45 | man | 8.62 | 4.94 | 142 | 39.8 | 229 | 13.5 | 1.16 | 83 |
| 24 | 68 | man | 7.85 | 4.83 | 168 | 46.9 | 176 | 16.1 | 0.51 | 76.9 |
| 34 | 84 | man | 11.1 | 3.58 | 111 | 33.5 | 379 | 7.5 | 0.83 | 83.6 |
| 41 | 82 | woman | 11.32 | 3.34 | 99 | 31.4 | 210 | 9.2 | 1.04 | 84.6 |
| 43 | 42 | woman | 5.73 | 3.48 | 106 | 32.1 | 351 | 13.8 | 0.79 | 80.6 |
| 44 | 60 | woman | 6.59 | 4.9 | 136 | 40.1 | 328 | 20.6 | 1.36 | 76.5 |
| 45 | 44 | man | 7.92 | 4.38 | 137 | 38.4 | 220 | 16.5 | 1.31 | 80 |
| 47 | 67 | woman | 11.33 | 4.08 | 132 | 0.38 | 204 | 4.1 | 0.46 | 91.8 |
| 49 | 51 | man | 8.78 | 4.86 | 150 | 44.6 | 266 | 21.4 | 1.88 | 72.5 |
| 52 | 65 | woman | 5.97 | 4.07 | 120 | 35.8 | 364 | 22.9 | 1.37 | 66 |
| 56 | 76 | woman | 5.11 | 4.12 | 131 | 38.1 | 187 | 14.9 | 0.76 | 81.2 |
| 57 | 51 | man | 6.01 | 4.92 | 142 | 24.5 | 373 | 20.8 | 1.25 | 67.8 |
| 59 | 63 | woman | 5.89 | 3.92 | 124 | 35.8 | 161 | 17.3 | 0.33 | 76.4 |
| 60 | 66 | man | 22.06 | 4.86 | 139 | 40 | 416 | 4.1 | 0.9 | 92.2 |
| B. | ||||||||||
| ID Sample | Age | Gran# | Mo% | Mo# | Eo% | Eo# | Ba% | Ba# | SO2% | Fever |
| 1 | 58 | 6.63 | 5.1 | 0.39 | 0 | 0 | 0 | 0 | 88% | 39 |
| 3 | 58 | 8.87 | 3.5 | 0.36 | 0 | 0 | 0.1 | 0.01 | 90% | 39.5 |
| 4 | 77 | 7.75 | 4 | 0.33 | 0 | 0 | 0 | 0 | 94% | 39.2 |
| 11 | 58 | 3.48 | 7.9 | 0.43 | 0 | 0 | 0.4 | 0.02 | 92–93% | 38.2 |
| 18 | 66 | 15.03 | 6.6 | 1.12 | 0 | 0 | 0.4 | 0.06 | 92% | 38.4 |
| 20 | 73 | 4.1 | 7.1 | 0.36 | 0 | 0 | 0.2 | 0.01 | 91% | 38.5 |
| 23 | 45 | 7.15 | 3.2 | 0.28 | 0 | 0 | 0.3 | 0.03 | 85% | 38 |
| 24 | 68 | 6.04 | 6.5 | 0.51 | 0.1 | 0.01 | 0.4 | 0.03 | 90% | 38.3 |
| 34 | 84 | 9.28 | 7.4 | 0.82 | 1.4 | 0.16 | 0.1 | 0.01 | 90% | 39 |
| 41 | 82 | 9.58 | 6.2 | 0.7 | 0 | 0 | 0 | 0 | 93% | 38.7 |
| 43 | 42 | 4.62 | 5.6 | 0.32 | 0 | 0 | 0 | 0 | 88–92% | 39.4 |
| 44 | 60 | 5.04 | 2.7 | 0.18 | 0 | 0 | 0.2 | 0.01 | 90% | 39 |
| 45 | 44 | 6.33 | 16.5 | 0.26 | 0.1 | 0.01 | 0.1 | 0.01 | 88–89% | 37.9 |
| 47 | 67 | 10.41 | 4 | 0.45 | 0 | 0 | 0.1 | 0.01 | 93% | 38.3 |
| 49 | 51 | 6.36 | 6 | 0.53 | 0 | 0 | 0.1 | 0.01 | 91% | 39.4 |
| 52 | 65 | 3.93 | 9.5 | 0.57 | 1.3 | 0.08 | 0.3 | 0.02 | 93–94% | 38.2 |
| 56 | 76 | 4.15 | 3.9 | 0.2 | 0 | 0 | 0 | 0 | 90% | 39.5 |
| 57 | 51 | 4.07 | 10.3 | 0.62 | 0.08 | 0.05 | 0.3 | 0.02 | 94–95% | 38 |
| 59 | 63 | 4.5 | 5.6 | 0.33 | 0.2 | 0.01 | 0.5 | 0.03 | 94% | 38.6 |
| 60 | 66 | 20.34 | 3.5 | 0.77 | 0 | 0 | 0.2 | 0.05 | 92% | 38.5 |
| A. | ||||||||||
| ID Sample | Age | Gender | WBC | RBC | HGB g/L | HCT | PLT | Lym% | Lym# | Gran% |
| 7 | 64 | man | 9.69 | 4.57 | 125 | 38.1 | 209 | 12.1 | 1.17 | 71.2 |
| 9 | 53 | man | 6.65 | 3.55 | 120 | 35.5 | 85 | 6.8 | 0.45 | 90.7 |
| 22 | 77 | man | 2.6 | 3.76 | 118 | 38.8 | 26 | 6.5 | 0.17 | 59.6 |
| 26 | 50 | man | 2.67 | 4.54 | 138 | 42.9 | 161 | 3.2 | 0.94 | 58.1 |
| 27 | 74 | woman | 14.28 | 4.35 | 133 | 38.8 | 325 | 2.7 | 0.39 | 92.7 |
| 32 | 79 | woman | 32.19 | 1.29 | 40 | 11.4 | 96 | 2.5 | 0.79 | 93.9 |
| 36 | 48 | man | 7.42 | 4.84 | 147 | 44 | 330 | 10.1 | 0.74 | 86.7 |
| 37 | 75 | woman | 42.3 | 2.4 | 74 | 23.1 | 132 | 3.4 | 1.44 | 85.3 |
| 38 | 43 | man | 6.76 | 5.13 | 147 | 44.7 | 270 | 6.4 | 0.43 | 89.9 |
| 51 | 73 | man | 10.54 | 4.81 | 133 | 38.9 | 310 | 8.4 | 0.89 | 83.1 |
| 53 | 98 | man | 6.68 | 3.93 | 124 | 36.7 | 185 | 13 | 0.87 | 81.5 |
| 54 | 48 | woman | 6.79 | 4.61 | 136 | 40.3 | 227 | 16.5 | 1.12 | 75 |
| 63 | 79 | woman | 11.68 | 4.01 | 122 | 36.6 | 263 | 9.9 | 1.16 | 84.6 |
| 65 | 66 | woman | 10.5 | 4.37 | 128 | 38.6 | 320 | 7.3 | 0.77 | 89.5 |
| 66 | 72 | woman | 16.34 | 4.49 | 134 | 39.8 | 245 | 7.7 | 1.26 | 86.8 |
| 77 | 82 | woman | 16.17 | 4.67 | 127 | 38.2 | 272 | 3.6 | 0.58 | 89.8 |
| 80 | 49 | man | 10.6 | 5.21 | 152 | 45.2 | 226 | 8 | 0.85 | 86.8 |
| 84 | 64 | woman | 11.29 | 4.71 | 140 | 41.5 | 287 | 9.3 | 1.05 | 81.5 |
| 89 | 62 | man | 3.61 | 3.43 | 109 | 32 | 106 | 10.5 | 0.38 | 84 |
| 91 | 62 | woman | 9.7 | 4.39 | 123 | 35.7 | 233 | 3.7 | 0.36 | 89.9 |
| B. | ||||||||||
| ID Sample | Age | Gran# | Mo% | Mo# | Eo% | Eo# | Ba% | Ba# | SO2% | Fever |
| 7 | 64 | 6.89 | 4.4 | 0.43 | 0.6 | 0.06 | 0.4 | 0.04 | 82% | 39 |
| 9 | 53 | 6.04 | 2.3 | 0.15 | 0 | 0 | 0.2 | 0.01 | 86% | 38.4 |
| 22 | 77 | 1.55 | 2.7 | 0.07 | 30.8 | 0.8 | 0.4 | 0.01 | 79% | 39 |
| 26 | 50 | 1.55 | 6.7 | 0.18 | 0 | 0 | 0 | 0 | 86% | 38 |
| 27 | 74 | 13.23 | 4.5 | 0.64 | 0 | 0 | 0.1 | 0.02 | 81% | 39 |
| 32 | 79 | 30.25 | 3.3 | 0.73 | 1.2 | 0.38 | 0.1 | 0.04 | 87% | 39 |
| 36 | 48 | 6.43 | 3.2 | 0.24 | 0 | 0 | 0.1 | 0.01 | 83% | 39.5 |
| 37 | 75 | 36.08 | 11.1 | 4.69 | 0 | 0 | 0.2 | 0.09 | 82% | 39.6 |
| 38 | 43 | 6.08 | 1.5 | 0.1 | 2.1 | 0.14 | 0.1 | 0.01 | 87% | 39.4 |
| 51 | 73 | 8.75 | 8.3 | 0.88 | 0.1 | 0.01 | 0.1 | 0.01 | 88% | 38.9 |
| 53 | 98 | 5.44 | 4.5 | 0.3 | 0.9 | 0.06 | 0.1 | 0.01 | 80% | 38 |
| 54 | 48 | 5.09 | 8.2 | 0.56 | 0 | 0 | 0.3 | 0.02 | 82% | 38.5 |
| 63 | 79 | 9.87 | 4.3 | 0.5 | 0.9 | 0.11 | 0.3 | 0.04 | 63% | 37.5 |
| 65 | 66 | 9.39 | 3 | 0.32 | 0 | 0 | 0.2 | 0.02 | 65% | 39.3 |
| 66 | 72 | 14.18 | 5.4 | 0.88 | 0 | 0 | 0.1 | 0.02 | 77% | 37.9 |
| 77 | 82 | 14.53 | 6.4 | 1.03 | 0 | 0 | 0.2 | 0.03 | 80% | 38 |
| 80 | 49 | 9.2 | 5.2 | 0.55 | 0 | 0 | 0 | 0 | 83% | 40 |
| 84 | 64 | 9.21 | 7.2 | 0.81 | 1.9 | 0.21 | 0.1 | 0.01 | 86% | 39 |
| 89 | 62 | 3.03 | 5.5 | 0.2 | 0 | 0 | 0 | 0 | 83% | 37.4 |
| 91 | 62 | 8.71 | 6.3 | 0.61 | 0.1 | 0.01 | 0.1 | 0.01 | 87% | 38 |
| C. | ||||||||||
| ID Sample | Age | CRP | Fibrinogen | D-Dimer | Urea | Creatinine | Ferritin | |||
| 7 | 64 | recovery | 201.2 | 6.86 | 1.32 | 8.1 | 124 | 2042 | ||
| 9 | 53 | recovery | 5.4 | 4.27 | 0.59 | 84 | 383 | |||
| 22 | 77 | exitus letalis | 226 | 5.24 | 9.94 | 11.5 | 303 | 1295 | ||
| 26 | 50 | recovery | 130 | 5.38 | 5.6 | 94 | 756 | |||
| 27 | 74 | recovery | 155.5 | 6.2 | 1.38 | 10.1 | 82 | 1297 | ||
| 32 | 79 | exitus letalis | 32.3 | 3.78 | 14.76 | 21.1 | 295 | 1122 | ||
| 36 | 48 | recovery | 69.4 | 5.88 | 0.39 | 9.0 | 71 | 1418 | ||
| 37 | 75 | exitus letalis | 205.9 | 8.0 | 1.99 | 478 | 424 | 1892 | ||
| 38 | 43 | recovery | 177.1 | 6.01 | 1.22 | 7.1 | 111 | 2324 | ||
| 51 | 73 | recovery | 40.7 | 7.35 | 0.7 | 5.3 | 87 | 237 | ||
| 53 | 98 | recovery | 245.5 | 5.1 | 0.73 | 3.2 | 82 | 439.0 | ||
| 54 | 48 | exitus letalis | 19.3 | 5.2 | 0.82 | 4.7 | 101 | 520 | ||
| 63 | 79 | exitus letalis | 229.8 | 6.07 | 2.19 | 13.1 | 138 | 604 | ||
| 65 | 66 | exitus letalis | 140 | 4.93 | 3.56 | 17.3 | 194 | 776 | ||
| 66 | 72 | recovery | 124.6 | 7.5 | 1.6 | 5.1 | 115 | 492 | ||
| 77 | 82 | recovery | 170 | 6.27 | 7.94 | 13.7 | 153 | 1266 | ||
| 80 | 49 | recovery | 60.8 | 5.38 | 1.03 | 4.4 | 102 | 259 | ||
| 84 | 64 | recovery | 104.9 | 6.17 | 0.50 | 5.4 | 63 | 405 | ||
| 89 | 62 | exitus letalis | 71.6 | 3.57 | 1.25 | 20.6 | 478 | 553 | ||
| 91 | 62 | exitus letalis | 183.2 | 5.31 | 0.86 | 24.7 | 236 | 597 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ralchev Ralchev, N.; Lyubenova Bradyanova, S.; Valerieva Doneva, Y.; Mihaylova, N.; Vikentieva Elefterova-Florova, E.; Ivanov Tchorbanov, A.; Munoz-Valle, J.F.; Petralia, M.C.; Checconi, P.; Nicoletti, F.; et al. Exploring the Role of CD74 and D-Dopachrome Tautomerase in COVID-19: Insights from Transcriptomic and Serum Analyses. J. Clin. Med. 2023, 12, 5037. https://doi.org/10.3390/jcm12155037
Ralchev Ralchev N, Lyubenova Bradyanova S, Valerieva Doneva Y, Mihaylova N, Vikentieva Elefterova-Florova E, Ivanov Tchorbanov A, Munoz-Valle JF, Petralia MC, Checconi P, Nicoletti F, et al. Exploring the Role of CD74 and D-Dopachrome Tautomerase in COVID-19: Insights from Transcriptomic and Serum Analyses. Journal of Clinical Medicine. 2023; 12(15):5037. https://doi.org/10.3390/jcm12155037
Chicago/Turabian StyleRalchev Ralchev, Nikola, Silviya Lyubenova Bradyanova, Yana Valerieva Doneva, Nikolina Mihaylova, Elena Vikentieva Elefterova-Florova, Andrey Ivanov Tchorbanov, José Francisco Munoz-Valle, Maria Cristina Petralia, Paola Checconi, Ferdinando Nicoletti, and et al. 2023. "Exploring the Role of CD74 and D-Dopachrome Tautomerase in COVID-19: Insights from Transcriptomic and Serum Analyses" Journal of Clinical Medicine 12, no. 15: 5037. https://doi.org/10.3390/jcm12155037
APA StyleRalchev Ralchev, N., Lyubenova Bradyanova, S., Valerieva Doneva, Y., Mihaylova, N., Vikentieva Elefterova-Florova, E., Ivanov Tchorbanov, A., Munoz-Valle, J. F., Petralia, M. C., Checconi, P., Nicoletti, F., & Fagone, P. (2023). Exploring the Role of CD74 and D-Dopachrome Tautomerase in COVID-19: Insights from Transcriptomic and Serum Analyses. Journal of Clinical Medicine, 12(15), 5037. https://doi.org/10.3390/jcm12155037









