Microbiome and MicroRNA or Long Non-Coding RNA—Two Modern Approaches to Understanding Pancreatic Ductal Adenocarcinoma
Abstract
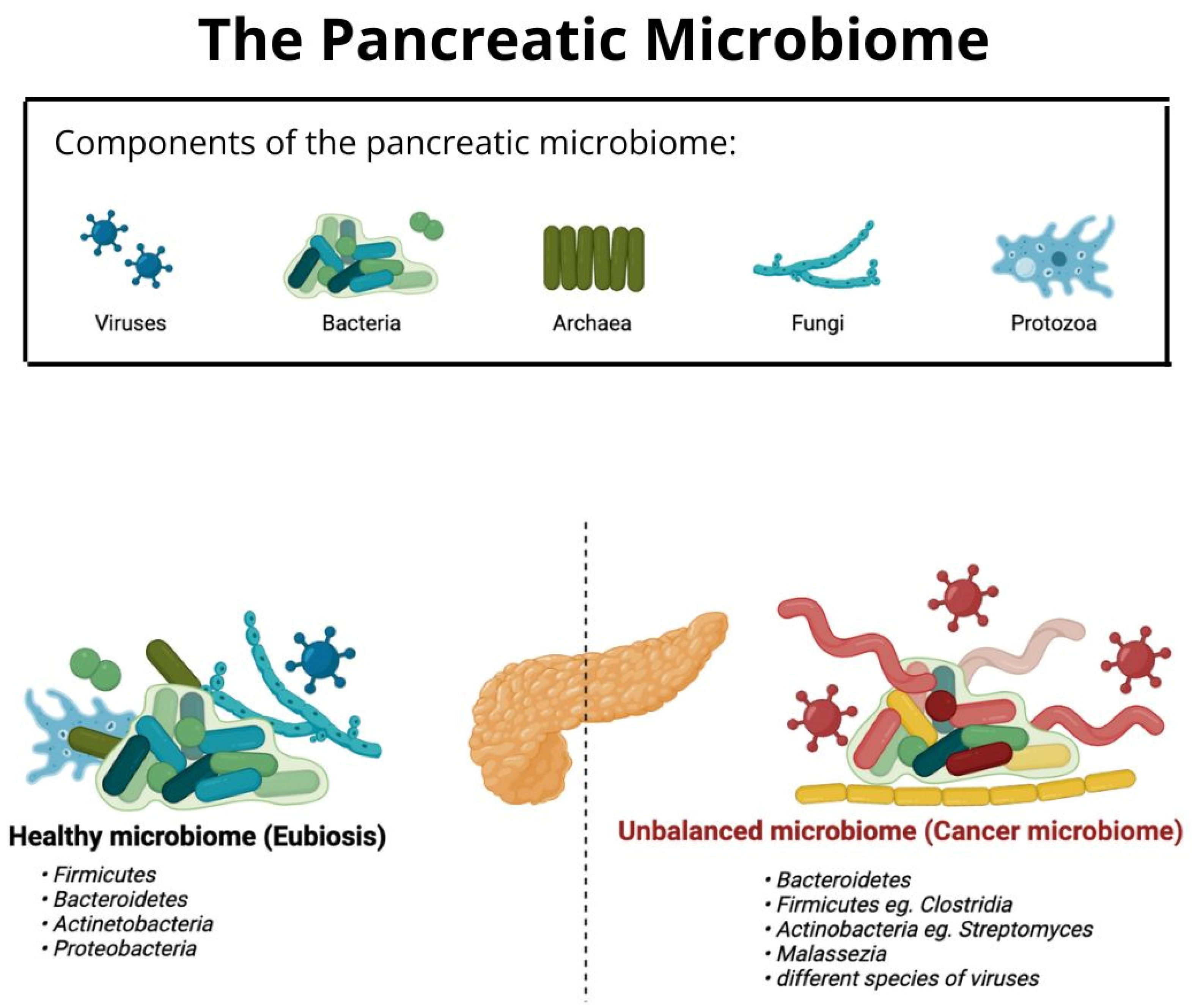
:1. Introduction
2. Evolution of Pancreatic Adenocarcinoma—What Occurs in Cells before Cancer?
3. Genetic Alterations in PanIN, MCN, and IPMNs
4. Genetic Alterations in PDAC and Their Consequences
5. Molecular Aspects of PDAC Metastases
6. miRNA and lncRNA Mechanisms of Intervention in PDAC Progression
7. Cross-Talk between Chosen ncRNA Types and Microbiome in PDAC
8. Conclusions and Knowledge Gaps
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Daniluk, J.; Daniluk, U.; Rogalski, P.; Dabrowski, A.; Swidnicka-Siergiejko, A. Microbiome—Friend or Foe of Pancreatic Cancer? J. Clin. Med. 2021, 10, 5624. [Google Scholar] [CrossRef] [PubMed]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Ryan, D.P.; Hong, T.S.; Bardeesy, N. Pancreatic Adenocarcinoma. N. Engl. J. Med. 2014, 371, 1039–1049. [Google Scholar] [CrossRef]
- Watson, M.D.; Miller-Ocuin, J.L.; Driedger, M.R.; Beckman, M.J.; McKillop, I.H.; Baker, E.H.; Martinie, J.B.; Vrochides, D.; Iannitti, D.A.; Ocuin, L.M. Factors Associated with Treatment and Survival of Early Stage Pancreatic Cancer in the Era of Modern Chemotherapy: An Analysis of the National Cancer Database. J. Pancreat. Cancer 2020, 6, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Mahesh, G.; Biswas, R. MicroRNA-155: A Master Regulator of Inflammation. J. Interf. Cytokine Res. 2019, 39, 321–330. [Google Scholar] [CrossRef]
- Wang, S.; Zheng, Y.; Yang, F.; Zhu, L.; Zhu, X.-Q.; Wang, Z.-F.; Wu, X.-L.; Zhou, C.-H.; Yan, J.-Y.; Hu, B.-Y.; et al. The molecular biology of pancreatic adenocarcinoma: Translational challenges and clinical perspectives. Signal Transduct. Target. Ther. 2021, 6, 249. [Google Scholar] [CrossRef]
- Wang, Q.; Ding, H.; Dong, G.; Xu, L.; Jiang, F.; Mao, Q. Bi-direction effects between microbiome and MiRNAs in carcinogenesis. J. Cancer Res. Clin. Oncol. 2021, 147, 1299–1305. [Google Scholar] [CrossRef]
- Elinav, E.; Garrett, W.S.; Trinchieri, G.; Wargo, J. The cancer microbiome. Nat. Rev. Cancer 2019, 19, 371–376. [Google Scholar] [CrossRef]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef]
- Li, J.J.; Zhu, M.; Kashyap, P.C.; Chia, N.; Tran, N.H.; McWilliams, R.R.; Bekaii-Saab, T.S.; Ma, W.W. The role of microbiome in pancreatic cancer. Cancer Metastasis Rev. 2021, 40, 777–789. [Google Scholar] [CrossRef]
- Kunej, T.; Obsteter, J.; Pogacar, Z.; Horvat, S.; Calin, G.A. The decalog of long non-coding RNA involvement in cancer diagnosis and monitoring. Crit. Rev. Clin. Lab. Sci. 2014, 51, 344–357. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Huang, S. Role of non-coding RNA in pancreatic cancer (Review). Oncol. Lett. 2019, 18, 3963–3973. [Google Scholar] [CrossRef]
- Yonemori, K.; Kurahara, H.; Maemura, K.; Natsugoe, S. MicroRNA in pancreatic cancer. J. Hum. Genet. 2016, 62, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhi, X.; Gao, Y.; Ta, N.; Jiang, H.; Zheng, J. LncRNAs in pancreatic cancer. Oncotarget 2016, 7, 57379–57390. [Google Scholar] [CrossRef]
- Singh, M.; Maitra, A. Precursor Lesions of Pancreatic Cancer: Molecular Pathology and Clinical Implications. Pancreatology 2007, 7, 9–19. [Google Scholar] [CrossRef]
- Distler, M.; Aust, D.; Weitz, J.; Pilarsky, C.; Grützmann, R. Precursor lesions for sporadic pancreatic cancer: PanIN, IPMN, and MCN. BioMed Res. Int. 2014, 2014, 474905. [Google Scholar] [CrossRef]
- Meeker, A.K.; De Marzo, A.M. Recent advances in telomere biology: Implications for human cancer. Curr. Opin. Oncol. 2004, 16, 32–38. [Google Scholar] [CrossRef]
- Moskaluk, C.; Hruban, R.; Kern, S. P16 and k-ras gene mutations in the intraductal precursors of human pancreatic adenocarci-noma. Cancer Res. 1997, 57, 2140–2143. [Google Scholar] [PubMed]
- Kaino, M.; Kondoh, S.; Okita, S.; Hatano, S.; Shiraishi, K.; Kaino, S.; Okita, K. Detection of K-ras and p53 gene mutations in pancreatic juice for the diagnosis of intraductal papillary mucinous tumors. Pancreas 1999, 18, 294–299. [Google Scholar] [CrossRef]
- Yoshizawa, K.; Nagai, H.; Sakurai, S.; Hironaka, M.; Morinaga, S.; Saitoh, K.; Fukayama, M. Clonality and K- ras mutation analyses of epithelia in intraductal papillary mucinous tumor and mucinous cystic tumor of the pancreas. Virchows Arch. 2002, 441, 437–443. [Google Scholar] [CrossRef]
- Caldas, C.; Hahn, S.A.; da Costa, L.T.; Redston, M.S.; Schutte, M.; Seymour, A.B.; Weinstein, C.L.; Hruban, R.H.; Yeo, C.J.; Kern, S.E. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat. Genet. 1994, 8, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Schutte, M.; Hruban, R.H.; Geradts, J.; Maynard, R.; Hilgers, W.; Rabindran, S.K.; Moskaluk, C.A.; Hahn, S.; Schwarte-Waldhoff, I.; Schmiegel, W.; et al. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997, 15, 126–3130. [Google Scholar]
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 Mutations in Human Cancers: Origins, Consequences, and Clinical Use. Cold Spring Harb. Perspect. Biol. 2009, 2, a001008. [Google Scholar] [CrossRef] [PubMed]
- Goggins, M.; Hruban, R.H.; Kern, S.E. BRCA2 is inactivated late in the development of pancreatic intraepithelial neoplasia: Evidence and implications. Am. J. Pathol. 2000, 156, 1767–1771. [Google Scholar] [CrossRef]
- Loss of Expression of Dpc4 in Pancreatic Intraepithelial Neoplasia: Evidence That DPC4 Inactivation Occurs Late in Neoplastic Progression. Available online: https://pubmed.ncbi.nlm.nih.gov/10766191/ (accessed on 8 September 2022).
- Montgomery, E.; Goggins, M.; Zhou, S.; Argani, P.; Wilentz, R.E.; Kaushal, M.; Booker, S.; Romans, K.; Bhargava, P.; Hruban, R.H.; et al. Nuclear localization of Dpc4 (Madh4, Smad4) in colorectal carcinomas and relation to mismatch repair/transforming growth factor-β receptor defects. Am. J. Pathol. 2001, 158, 537–542. [Google Scholar] [CrossRef]
- Rooman, I.; De Medts, N.; Baeyens, L.; Lardon, J.; De Breuck, S.; Heimberg, H.; Bouwens, L. Expression of the Notch Signaling Pathway and Effect on Exocrine Cell Proliferation in Adult Rat Pancreas. Am. J. Pathol. 2006, 169, 1206–1214. [Google Scholar] [CrossRef]
- Sheng, T.; Li, C.; Zhang, X.; Chi, S.; He, N.; Chen, K.; McCormick, F.; Gatalica, Z.; Xie, J. Activation of the hedgehog pathway in advanced prostate cancer. Mol. Cancer 2004, 3, 29. [Google Scholar] [CrossRef]
- Al-Refaie, W.; Choi, E.; Tseng, J.; Tamm, E.; Lee, J.; Evans, D.; Pisters, P. Intraductal Papillary Mucinous Neoplasms of the Pancreas. Med. Princ. Pract. 2006, 15, 245–252. [Google Scholar] [CrossRef]
- Fischer, C.G.; Wood, L.D. From somatic mutation to early detection: Insights from molecular characterization of pancreatic cancer precursor lesions. J. Pathol. 2018, 246, 395–404. [Google Scholar] [CrossRef]
- Adsay, N.V.; Merati, K.; Andea, A.; Sarkar, F.; Hruban, R.H.; Wilentz, R.E.; Goggins, M.; Iocobuzio-Donahue, C.; Longnecker, D.S.; Klimstra, D.S. The Dichotomy in the Preinvasive Neoplasia to Invasive Carcinoma Sequence in the Pancreas: Differential Expression of MUC1 and MUC2 Supports the Existence of Two Separate Pathways of Carcinogenesis. Mod. Pathol. 2002, 15, 1087–1095. [Google Scholar] [CrossRef]
- Launonen, V. Mutations in the human LKB1/STK11 gene. Hum. Mutat. 2005, 26, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Hollstein, P.E.; Shaw, R.J. GNAS shifts metabolism in pancreatic cancer. Nature 2018, 20, 740–741. [Google Scholar] [CrossRef]
- Naveed; Naveed, S.; Qari, H.; Banday, T.; Altaf, A.; Para, M. Mucinous Cystic Neoplasms of Pancreas. Gastroenterol. Res. 2014, 7, 44–50. [Google Scholar] [CrossRef]
- Fujikura, K.; Akita, M.; Abe-Suzuki, S.; Itoh, T.; Zen, Y. Mucinous cystic neoplasms of the liver and pancreas: Relationship between KRAS driver mutations and disease progression. Histopathology 2017, 71, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Matthaei, H.; Schulick, R.D.; Hruban, R.H.; Maitra, A. Cystic precursors to invasive pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 141–150. [Google Scholar] [CrossRef]
- Prabhu, L.; Mundade, R.; Korc, M.; Loehrer, P.J.; Lu, T. Critical role of NF-κB in pancreatic cancer. Oncotarget 2014, 5, 10969. [Google Scholar] [CrossRef]
- Yang, S.; Wang, X.; Contino, G.; Liesa, M.; Sahin, E.; Ying, H.; Bause, A.; Li, Y.; Stommel, J.M.; Dell’Antonio, G.; et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011, 25, 717. [Google Scholar] [CrossRef]
- Ying, H.; Kimmelman, A.C.; Lyssiotis, C.A.; Hua, S.; Chu, G.C.; Fletcher-Sananikone, E.; Locasale, J.W.; Son, J.; Zhang, H.; Coloff, J.L.; et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 2012, 149, 656–670. [Google Scholar] [CrossRef]
- Li, X.; Jiang, Y.; Meisenhelder, J.; Yang, W.; Hawke, D.H.; Zheng, Y.; Xia, Y.; Aldape, K.; He, J.; Hunter, T.; et al. Mitochondria-Translocated PGK1 Functions as a Protein Kinase to Coordinate Glycolysis and the TCA Cycle in Tumorigenesis. Mol. Cell 2016, 61, 705–719. [Google Scholar] [CrossRef]
- Christenson, E.S.; Jaffee, E.; Azad, N.S. Current and emerging therapies for patients with advanced pancreatic ductal adenocarcinoma: A bright future. Lancet Oncol. 2020, 21, e135–e145. [Google Scholar] [CrossRef]
- Mizrahi, J.D.; Surana, R.; Valle, J.W.; Shroff, R.T. Pancreatic cancer. Lancet 2020, 395, 2008–2020. [Google Scholar] [CrossRef] [PubMed]
- Biankin, A.V.; Waddell, N.; Kassahn, K.S.; Gingras, M.-C.; Muthuswamy, L.B.; Johns, A.L.; Miller, D.K.; Wilson, P.J.; Patch, A.-M.; Wu, J.; et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012, 491, 399–405. [Google Scholar] [CrossRef]
- Hermann, P.C.; Sainz, B. Pancreatic cancer stem cells: A state or an entity? Semin. Cancer Biol. 2018, 53, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Heidt, D.G.; Dalerba, P.; Burant, C.F.; Zhang, L.; Adsay, V.; Wicha, M.; Clarke, M.F.; Simeone, D.M. Identification of pancreatic cancer stem cells. Cancer Res 2007, 67, 1030–1037. [Google Scholar] [CrossRef]
- Hutcheson, J.; Balaji, U.; Porembka, M.R.; Wachsmann, M.B.; McCue, P.A.; Knudsen, E.S.; Witkiewicz, A.K. Immunologic and metabolic features of pancreatic ductal adenocarcinoma define prognostic subtypes of disease. Clin. Cancer Res. 2016, 22, 3606–3617. [Google Scholar] [CrossRef]
- Hermann, P.C.; Huber, S.L.; Herrler, T.; Aicher, A.; Ellwart, J.W.; Guba, M.; Bruns, C.J.; Heeschen, C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 2007, 1, 313–323. [Google Scholar] [CrossRef]
- Kim, M.P.; Fleming, J.B.; Wang, H.; Abbruzzese, J.L.; Choi, W.; Kopetz, S.; McConkey, D.J.; Evans, D.B.; Gallick, G.E. ALDH activity selectively defines an enhanced tumor-initiating cell population relative to cd133 expression in human pancreatic adenocarcinoma. PLoS ONE 2011, 6, e20636. [Google Scholar] [CrossRef] [PubMed]
- Gebauer, F.; Struck, L.; Tachezy, M.; Vashist, Y.; Wicklein, D.; Schumacher, U.; Izbicki, J.R.; Bockhorn, M. Serum EpCAM Expression in Pancreatic Cancer. Anticancer Res. 2014, 34, 4741–4746. [Google Scholar]
- Liu, M.; Hancock, S.E.; Sultani, G.; Wilkins, B.; Ding, E.; Osborne, B.; Quek, L.-E.; Turner, N. Snail-Overexpression Induces Epithelial-mesenchymal Transition and Metabolic Reprogramming in Human Pancreatic Ductal Adenocarcinoma and Non-tumorigenic Ductal Cells. J. Clin. Med. 2019, 8, 822. [Google Scholar] [CrossRef]
- Pelosi, E.; Castelli, G.; Testa, U. Pancreatic Cancer: Molecular Characterization, Clonal Evolution and Cancer Stem Cells. Biomedicines 2017, 5, 65. [Google Scholar] [CrossRef]
- Mueller, M.; Hermann, P.C.; Witthauer, J.; Rubio–Viqueira, B.; Leicht, S.F.; Huber, S.; Ellwart, J.W.; Mustafa, M.; Bartenstein, P.; D’Haese, J.G.; et al. Combined targeted treatment to eliminate tumorigenic cancer stem cells in human pancreatic cancer. Gastroenterology 2009, 137, 1102–1113. [Google Scholar] [CrossRef] [PubMed]
- Hermann, P.C.; Trabulo, S.M.; Sainz, B., Jr.; Balic, A.; Garcia, E.; Hahn, S.A.; Vandana, M.; Sahoo, S.K.; Tunici, P.; Bakker, A.; et al. Multimodal Treatment Eliminates Cancer Stem Cells and Leads to Long-Term Survival in Primary Human Pancreatic Cancer Tissue Xenografts. PLoS ONE 2013, 8, e66371. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, B.; Fong, C.; Luthra, A.; Smith, S.A.; DiNatale, R.G.; Nandakumar, S.; Walch, H.; Chatila, W.K.; Madupuri, R.; Kundra, R.; et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell 2022, 185, 563–575.e11. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Huang, C.; Cui Zhou, D.; Hu, Y.; Lih, T.M.; Savage, S.R.; Krug, K.; Clark, D.J.; Schnaubelt, M.; Chen, L.; et al. Proteogenomic characterization of pancreatic ductal adenocarcinoma. Cell 2021, 184, 5031–5052.e26. [Google Scholar] [CrossRef]
- Fraunhoffer, N.A.; Abuelafia, A.M.; Bigonnet, M.; Gayet, O.; Roques, J.; Nicolle, R.; Lomberk, G.; Urrutia, R.; Dusetti, N.; Iovanna, J. Multi-omics data integration and modeling unravels new mechanisms for pancreatic cancer and improves prognostic prediction. NPJ Precis. Oncol. 2022, 6, 57. [Google Scholar] [CrossRef]
- Kobayashi, T.; Honda, K. Trends in biomarker discoveries for the early detection and risk stratification of pancreatic cancer using omics studies. Expert Rev. Mol. Diagn. 2019, 19, 651–654. [Google Scholar] [CrossRef]
- Connor, A.A.; Gallinger, S. Pancreatic cancer evolution and heterogeneity: Integrating omics and clinical data. Nat. Rev. Cancer 2021, 22, 131–142. [Google Scholar] [CrossRef]
- Iacobuzio-Donahue, C.A.; Fu, B.; Yachida, S.; Luo, M.; Abe, H.; Henderson, C.M.; Vilardell, F.; Wang, Z.; Keller, J.W.; Banerjee, P.; et al. DPC4 Gene Status of the Primary Carcinoma Correlates With Patterns of Failure in Patients With Pancreatic Cancer. J. Clin. Oncol. 2009, 27, 1806–1813. [Google Scholar] [CrossRef]
- Yachida, S.; White, C.M.; Naito, Y.; Zhong, Y.; Brosnan, J.A.; Macgregor-Das, A.M.; Morgan, R.A.; Saunders, T.; Laheru, D.A.; Herman, J.M.; et al. Clinical significance of the genetic landscape of pancreatic cancer and implications for identification of potential long-term survivors. Clin. Cancer Res. 2012, 18, 6339–6347. [Google Scholar] [CrossRef]
- Haeno, H.; Gonen, M.; Davis, M.B.; Herman, J.M.; Iacobuzio-Donahue, C.A.; Michor, F. Computational modeling of pancreatic cancer reveals kinetics of metastasis suggesting optimum treatment strategies. Cell 2012, 148, 362–375. [Google Scholar] [CrossRef]
- Le Large, T.; Bijlsma, M.; Kazemier, G.; van Laarhoven, H.; Giovannetti, E.; Jimenez, C. Key biological processes driving metastatic spread of pancreatic cancer as identified by multi-omics studies. Semin. Cancer Biol. 2017, 44, 153–169. [Google Scholar] [CrossRef]
- Oldfield, L.E.; Connor, A.A.; Gallinger, S. Molecular Events in the Natural History of Pancreatic Cancer. Trends Cancer 2017, 3, 336–346. [Google Scholar] [CrossRef]
- Hegde, S.; Krisnawan, V.E.; Herzog, B.H.; Zuo, C.; Breden, M.A.; Knolhoff, B.L.; Hogg, G.D.; Tang, J.P.; Baer, J.M.; Mpoy, C.; et al. Dendritic cell paucity leads to dysfunctional immune surveillance in pancreatic cancer. Cancer Cell 2020, 37, 289–307.e9. [Google Scholar] [CrossRef]
- Chartier, C.; Raval, J.; Axelrod, F.; Bond, C.; Cain, J.; Dee-Hoskins, C.; Ma, S.; Fischer, M.M.; Shah, J.; Wei, J.; et al. Therapeutic targeting of tumor-derived r-spondin attenuates b-catenin signaling and tumorigenesis in multiple cancer types. Cancer Res. 2016, 76, 713–723. [Google Scholar] [CrossRef]
- Stratton, M.R. Exploring the genomes of cancer cells: Progress and promise. Science 2011, 331, 1553–1558. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Nakano, K.; Obchoei, S.; Setoguchi, K.; Matsumoto, M.; Yamamoto, T.; Obika, S.; Shimada, K.; Hiraoka, N. Small Nucleolar Noncoding RNA SNORA23, Up-Regulated in Human Pancreatic Ductal Adenocarcinoma, Regulates Expression of Spectrin Repeat-Containing Nuclear Envelope 2 to Promote Growth and Metastasis of Xenograft Tumors in Mice. Gastroenterology 2017, 153, 292–306.e2. [Google Scholar] [CrossRef]
- Guo, X.; Zhou, Q.; Su, D.; Luo, Y.; Fu, Z.; Huang, L.; Li, Z.; Jiang, D.; Kong, Y.; Li, Z.; et al. Circular RNA circBFAR promotes the progression of pancreatic ductal adenocarcinoma via the miR-34b-5p/MET/Akt axis. Mol. Cancer 2020, 19, 83. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Cui, J.; Quan, M.; Xie, D.; Jia, Z.; Wei, D.; Wang, L.; Gao, Y.; Ma, Q.; Xie, K. The novel KLF4/MSI2 signaling pathway regulates growth and metastasis of pancreatic cancer. Clin. Cancer Res. 2017, 23, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Nishiwada, S.; Sho, M.; Banwait, J.K.; Yamamura, K.; Akahori, T.; Nakamura, K.; Baba, H.; Goel, A. A MicroRNA Signature Identifies Pancreatic Ductal Adenocarcinoma Patients at Risk for Lymph Node Metastases. Gastroenterology 2020, 159, 562–574. [Google Scholar] [CrossRef]
- Whittle, M.C.; Izeradjene, K.; Rani, P.G.; Feng, L.; Carlson, M.A.; DelGiorno, K.E.; Wood, L.D.; Goggins, M.; Hruban, R.H.; Chang, A.E.; et al. RUNX3 Controls a Metastatic Switch in Pancreatic Ductal Adenocarcinoma. Cell 2015, 161, 1345–1360. [Google Scholar] [CrossRef] [PubMed]
- Shindo, K.; Yu, J.; Suenaga, M.; Fesharakizadeh, S.; Cho, C.; Macgregor-Das, A.; Siddiqui, A.; Witmer, P.D.; Tamura, K.; Song, T.J.; et al. Deleterious Germline Mutations in Patients With Apparently Sporadic Pancreatic Adenocarcinoma. J. Clin. Oncol. 2017, 35, 3382–3390. [Google Scholar] [CrossRef] [PubMed]
- Liao, T.-T.; Yang, M.-H. Revisiting epithelial-mesenchymal transition in cancer metastasis: The connection between epithelial plasticity and stemness. Mol. Oncol. 2017, 11, 792–804. [Google Scholar] [CrossRef]
- Roe, J.-S.; Hwang, C.-I.; Somerville, T.D.; Milazzo, J.P.; Lee, E.J.; Da Silva, B.; Maiorino, L.; Tiriac, H.; Young, C.M.; Miyabayashi, K.; et al. Enhancer Reprogramming Promotes Pancreatic Cancer Metastasis. Cell 2017, 170, 875–888.e20. [Google Scholar] [CrossRef] [PubMed]
- Pennacchio, L.A.; Bickmore, W.; Dean, A.; Nobrega, M.A.; Bejerano, G. Enhancers: Five essential questions. Nat. Rev. Genet. 2013, 14, 288–295. [Google Scholar] [CrossRef]
- Rusek, A.M.; Abba, M.; Eljaszewicz, A.; Moniuszko, M.; Niklinski, J.; Allgayer, H. MicroRNA modulators of epigenetic regulation, the tumor microenvironment and the immune system in lung cancer. Mol. Cancer 2015, 14, 34. [Google Scholar] [CrossRef]
- Zhai, L.; Wu, R.; Han, W.; Zhang, Y.; Zhu, D. miR-127 enhances myogenic cell differentiation by targeting S1PR3. Cell Death Dis. 2017, 8, e2707. [Google Scholar] [CrossRef]
- Yu, Y.; Liu, L.; Ma, R.; Gong, H.; Xu, P.; Wang, C. MicroRNA-127 is aberrantly downregulated and acted as a functional tumor suppressor in human pancreatic cancer. Tumor Biol. 2016, 37, 14249–14257. [Google Scholar] [CrossRef]
- Dong, J.; Zhao, Y.-P.; Zhou, L.; Zhang, T.-P.; Chen, G. Bcl-2 upregulation induced by miR-21 via a direct interaction is associated with apoptosis and chemoresistance in MIA PaCa-2 pancreatic cancer cells. Arch. Med. Res. 2011, 42, 8–14. [Google Scholar] [CrossRef]
- Song, W.; Li, Q.; Wang, L.; Wang, L. Modulation of FoxO1 expression by miR-21 to promote growth of pancreatic ductal adenocarcinoma. Cell. Physiol. Biochem. 2015, 35, 184–190. [Google Scholar] [CrossRef]
- Nagao, Y.; Hisaoka, M.; Matsuyama, A.; Kanemitsu, S.; Hamada, T.; Fukuyama, T.; Nakano, R.; Uchiyama, A.; Kawamoto, M.; Yamaguchi, K.; et al. Association of microRNA-21 expression with its targets, PDCD4 and TIMP3, in pancreatic ductal adenocarcinoma. Mod. Pathol. 2012, 25, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Mark, J.; Park, J.K.; Lee, E.J.; Esau, C.; Schmittgen, T.D. Antisense inhibition of microRNA-21 or -221 arrests cell cycle, induces apoptosis, and sensitizesthe effects of gemcitabine in pancreatic adenocarcinoma. Pancreas 2009, 39, 190–199. [Google Scholar]
- Sarkar, S.; Dubaybo, H.; Ali, S.; Goncalves, P.; Kollepara, S.L.; Sethi, S.; APhilip, P.; Li, Y. Down-regulation of miR-221 inhibits proliferation of pancreatic cancer cells through up-regulation of PTEN, p27kip1, p57kip2, and PUMA. Am. J. Cancer Res. 2013, 3, 465–477. [Google Scholar] [PubMed]
- Xu, Q.; Li, P.; Chen, X.; Zong, L.; Jiang, Z.; Nan, L.; Lei, J.; Duan, W.; Zhang, D.; Li, X.; et al. miR-221/222 induces pancreatic cancer progression through the regulation of matrix metalloproteinases. Oncotarget 2015, 6, 14153–14164. [Google Scholar] [CrossRef]
- Su, A.; He, S.; Tian, B.; Hu, W.; Zhang, Z. MicroRNA-221 mediates the effects of PDGF-BB on migration, proliferation, and the epithelial-mesenchymal transition in pancreatic cancer cells. PLoS ONE 2013, 8, e71309. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, Y.; Yang, Y.; Liu, J.; Song, Y.; Cao, Y.; Chen, X.; Yang, W.; Wang, F.; Gao, J.; et al. MicroRNA-222 Controls Human Pancreatic Cancer Cell Line Capan-2 Proliferation by P57 Targeting. J. Cancer 2015, 6, 1230–1235. [Google Scholar] [CrossRef] [PubMed]
- Ohuchida, K.; Mizumoto, K.; Lin, C.; Yamaguchi, H.; Ohtsuka, T.; Sato, N.; Toma, H.; Nakamura, M.; Nagai, E.; Hashizume, M.; et al. MicroRNA-10a is overexpressed in human pancreatic cancer and involved in its invasiveness partially via suppression of the HOXA1 gene. Ann. Surg. Oncol. 2012, 19, 2394–2402. [Google Scholar] [CrossRef]
- Wang, P.; Zheng, D.; Qi, H.; Gao, Q. Thioredoxin-interacting protein is a favored target of miR-125b, promoting metastasis and progression of pancreatic cancer via the HIF1α pathway. J. Biochem. Mol. Toxicol. 2021, 35, e22782. [Google Scholar] [CrossRef]
- Long, L.-M.; Zhan, J.-K.; Wang, H.-Q.; Li, S.; Chen, Y.-Y.; Liu, Y.-S. The Clinical Significance of miR-34a in Pancreatic Ductal Carcinoma and Associated Molecular and Cellular Mechanisms. Pathobiology 2016, 84, 38–48. [Google Scholar] [CrossRef]
- Xie, F.; Li, C.; Zhang, X.; Peng, W.; Wen, T. MiR-143-3p suppresses tumorigenesis in pancreatic ductal adenocarcinoma by targeting KRAS. Biomed. Pharmacother. 2019, 119, 109424. [Google Scholar] [CrossRef]
- Kent, O.A.; Chivukula, R.R.; Mullendore, M.; Wentzel, E.A.; Feldmann, G.; Lee, K.H.; Liu, S.; Leach, S.D.; Maitra, A.; Mendell, J.T. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev. 2010, 24, 2754–2759. [Google Scholar] [CrossRef]
- Gao, S.; Wang, P.; Hua, Y.; Xi, H.; Meng, Z.; Liu, T.; Chen, Z.; Liu, L. ROR functions as a ceRNA to regulate Nanog expression by sponging miR-145 and predicts poor prognosis in pancreatic cancer. Oncotarget 2016, 7, 1608–1618. [Google Scholar] [CrossRef]
- Khan, S.; Ebeling, M.C.; Zaman, M.S.; Sikander, M.; Yallapu, M.M.; Chauhan, N.; Yacoubian, A.M.; Behrman, S.W.; Zafar, N.; Kumar, D.; et al. MicroRNA-145 targets MUC13 and suppresses growth and invasion of pancreatic cancer. Oncotarget 2014, 5, 7599–7609. [Google Scholar] [CrossRef]
- Deng, S.; Zhu, S.; Wang, B.; Li, X.; Liu, Y.; Qin, Q.; Gong, Q.; Niu, Y.; Xiang, C.; Chen, J.; et al. Chronic pancreatitis and pancreatic cancer demonstrate active epithelial–mesenchymal transition profile, regulated by miR-217-SIRT1 pathway. Cancer Lett. 2014, 355, 184–191. [Google Scholar] [CrossRef]
- Zhao, W.-G.; Yu, S.-N.; Lu, Z.-H.; Ma, Y.-H.; Gu, Y.-M.; Chen, J. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinog. 2010, 31, 1726–1733. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Wang, B.; Liu, Y.; Zhang, J.-G.; Deng, S.-C.; Qin, Q.; Tian, K.; Li, X.; Zhu, S.; Niu, Y.; et al. miRNA-141, downregulated in pancreatic cancer, inhibits cell proliferation and invasion by directly targeting MAP4K4. Mol. Cancer Ther. 2013, 12, 2569–2580. [Google Scholar] [CrossRef]
- Zhu, Z.-M.; Xu, Y.-F.; Su, Q.-J.; Du, J.-D.; Tan, X.-L.; Tu, Y.-L.; Tan, J.-W.; Jiao, H.-B. Prognostic significance of microRNA-141 expression and its tumor suppressor function in human pancreatic ductal adenocarcinoma. Mol. Cell. Biochem. 2013, 388, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Li, Q.; Xu, D.; Wang, Q.; An, Y.; DU, Q.; Zhang, J.; Zhu, Y.; Miao, Y. hsa-miR-141 downregulates TM4SF1 to inhibit pancreatic cancer cell invasion and migration. Int. J. Oncol. 2013, 44, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Q.; Fang, Y.; Deng, X.; Chen, H.; Jin, J.; Lu, X.; Peng, C.; Li, H.; Shen, B. The Interplay Between miR-148a and DNMT1 Might be Exploited for Pancreatic Cancer Therapy. Cancer Investig. 2015, 33, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Li, M.; Zang, W.; Chen, X.; Wang, Y.; Li, P.; Du, Y.; Zhao, G.; Li, L. MiR-148a regulates the growth and apoptosis in pancreatic cancer by targeting CCKBR and Bcl-2. Tumor Biol. 2013, 35, 837–844. [Google Scholar] [CrossRef]
- Liffers, S.-T.; Munding, J.B.; Vogt, M.; Kuhlmann, J.D.; Verdoodt, B.; Nambiar, S.; Maghnouj, A.; Mirmohammadsadegh, A.; Hahn, S.A.; Tannapfel, A. MicroRNA-148a is down-regulated in human pancreatic ductal adenocarcinomas and regulates cell survival by targeting CDC25B. Lab. Investig. 2011, 91, 1472–1479. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Song, S.; He, S.; Zhu, X.; Zhang, Y.; Yi, B.; Zhang, B.; Qin, G.; Li, D. MicroRNA-375 targets PDK1 in pancreatic carcinoma and suppresses cell growth through the Akt signaling pathway. Int. J. Mol. Med. 2014, 33, 950–956. [Google Scholar] [CrossRef]
- Sun, W.; Lu, Y.; Hu, J.; Li, S.; Deng, S.; Li, M. MiR-29c inhibits cell growth, invasion, and migration of pancreatic cancer by targeting ITGB1. OncoTargets Ther. 2015, 9, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Li, J.; Chen, Z.; Li, X.; Zheng, S.; Yi, D.; Zhong, A.; Chen, J. miR-29c suppresses pancreatic cancer liver metastasis in an orthotopic implantation model in nude mice and affects survival in pancreatic cancer patients. Carcinog. 2015, 36, 676–684. [Google Scholar] [CrossRef]
- Zhao, G.; Zhang, J.-G.; Shi, Y.; Qin, Q.; Liu, Y.; Wang, B.; Tian, K.; Deng, S.-C.; Li, X.; Zhu, S.; et al. MiR-130b is a prognostic marker and inhibits cell proliferation and invasion in pancreatic cancer through targeting STAT3. PLoS ONE 2013, 8, e73803. [Google Scholar] [CrossRef]
- Radhakrishnan, P.; Mohr, A.M.; Grandgenett, P.M.; Steele, M.M.; Batra, S.K.; Hollingsworth, M.A. MicroRNA-200c modulates the expression of MUC4 and MUC16 by directly targeting their coding sequences in human pancreatic cancer. PLoS ONE 2013, 8, e73356. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Ohuchida, K.; Mizumoto, K.; Sato, N.; Kayashima, T.; Fujita, H.; Nakata, K.; Tanaka, M. MicroRNA, hsa-miR-200c, is an independent prognostic factor in pancreatic cancer and its upregulation inhibits pancreatic cancer invasion but increases cell proliferation. Mol. Cancer 2010, 9, 169. [Google Scholar] [CrossRef]
- Wang, S.; Chen, X.; Tang, M. MicroRNA-216a inhibits pancreatic cancer by directly targeting Janus kinase 2. Oncol. Rep. 2014, 32, 2824–2830. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, H.; Lin, S.; Ba, M.; Cui, S. MicroRNA-216a enhances the radiosensitivity of pancreatic cancer cells by inhibiting beclin-1-mediated autophagy. Oncol. Rep. 2015, 34, 1557–1564. [Google Scholar] [CrossRef]
- Zhang, X.-X.; Chen, H.; Li, H.-Y.; Chen, R.; He, L.; Yang, J.-L.; Xiao, L.-L.; Chen, J.-L. Long non-coding RNA small nucleolar RNA host gene 6 aggravates pancreatic cancer through upregulation of far upstream element binding protein 1 by sponging microRNA-26a-5p. Chin. Med. J. 2020, 133, 1211–1220. [Google Scholar] [CrossRef]
- Deng, J.; He, M.; Chen, L.; Chen, C.; Zheng, J.; Cai, Z. The Loss of miR-26a-Mediated Post-Transcriptional Regulation of Cyclin E2 in Pancreatic Cancer Cell Proliferation and Decreased Patient Survival. PLoS ONE 2013, 8, e76450. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Zhang, J.-G.; Liu, Y.; Qin, Q.; Wang, B.; Tian, K.; Liu, L.; Li, X.; Niu, Y.; Deng, S.-C.; et al. miR-148b functions as a tumor suppressor in pancreatic cancer by targeting AMPKα1. Mol. Cancer Ther. 2013, 12, 83–93. [Google Scholar] [CrossRef]
- Gao, L.; Yang, Y.; Xu, H.; Liu, R.; Li, D.; Hong, H.; Qin, M.; Wang, Y. miR-335 functions as a tumor suppressor in pancreatic cancer by targeting OCT4. Tumor Biol. 2014, 35, 8309–8318. [Google Scholar] [CrossRef]
- Hamada, S.; Masamune, A.; Miura, S.; Satoh, K.; Shimosegawa, T. MiR-365 induces gemcitabine resistance in pancreatic cancer cells by targeting the adaptor protein SHC1 and pro-apoptotic regulator BAX. Cell. Signal. 2014, 26, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhu, C.-F.; Ma, M.-Z.; Chen, G.; Song, M.; Zeng, Z.-L.; Lu, W.-H.; Yang, J.; Wen, S.; Chiao, P.J.; et al. Micro-RNA-155 is induced by K-Ras oncogenic signal and promotes ROS stress in pancreatic cancer. Oncotarget 2015, 6, 21148–21158. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Chen, J.; Wang, J.; Amos, C.; Killary, A.M.; Sen, S.; Wei, C.; Frazier, M.L. Putative tumor suppressor gene SEL1L was downregulated by aberrantly upregulated hsa-mir-155 in human pancreatic ductal adenocarcinoma. Mol. Carcinog. 2013, 53, 711–721. [Google Scholar] [CrossRef]
- Liu, W.-J.; Zhao, Y.-P.; Zhang, T.-P.; Zhou, L.; Cui, Q.-C.; Zhou, W.-X.; You, L.; Chen, G.; Shu, H. MLH1 as a direct target of MiR-155 and a potential predictor of favorable prognosis in pancreatic cancer. J. Gastrointest. Surg. 2013, 17, 1399–1405. [Google Scholar] [CrossRef]
- Huang, C.; Li, H.; Wu, W.; Jiang, T.; Qiu, Z. Regulation of miR-155 affects pancreatic cancer cell invasiveness and migration by modulating the STAT3 signaling pathway through SOCS1. Oncol. Rep. 2013, 30, 1223–1230. [Google Scholar] [CrossRef]
- Gironella, M.; Seux, M.; Xie, M.-J.; Cano, C.; Tomasini, R.; Gommeaux, J.; Garcia, S.; Nowak, J.; Yeung, M.L.; Jeang, K.-T.; et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc. Natl. Acad. Sci. USA 2007, 104, 16170–16175. [Google Scholar] [CrossRef]
- Liu, N.; Sun, Y.-Y.; Zhang, X.-W.; Chen, S.; Wang, Y.; Zhang, Z.-X.; Song, S.-W.; Qiu, G.-B.; Fu, W.-N. Oncogenic miR-23a in Pancreatic Ductal Adenocarcinogenesis Via Inhibiting APAF1. Dig. Dis. Sci. 2015, 60, 2000–2008. [Google Scholar] [CrossRef]
- Listing, H.; Mardin, W.A.; Wohlfromm, S.; Mees, S.T.; Haier, J. MiR-23a/-24-induced gene silencing results in mesothelial cell integration of pancreatic cancer. Br. J. Cancer 2014, 112, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Ou, Y.; Wu, K.; Chen, Y.; Sun, W. miR-143 inhibits the metastasis of pancreatic cancer and an associated signaling pathway. Tumor Biol. 2012, 33, 1863–1870. [Google Scholar] [CrossRef]
- Li, Y.; VandenBoom, T.; Wang, Z.; Kong, D.; Ali, S.; Philip, P.A.; Sarkar, F.H. Abstract 5703: Up-regulation of miR-146a contributes to the inhibition of invasion of pancreatic cancer cells. Cancer Res 2010, 70 (Suppl. 8), 5703. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.K.; Bhardwaj, A.; Singh, S.; Arora, S.; Bin Wang, B.; Grizzle, W.E.; Singh, A.P. MicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cells. Carcinogenesis 2011, 32, 1832–1839. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xu, D.; Wang, Q.; Zheng, D.; Jiang, X.; Xu, L. LPS induced miR-181a promotes pancreatic cancer cell migration via targeting PTEN and MAP2K4. Dig. Dis. Sci. 2014, 59, 1452–1460. [Google Scholar] [CrossRef]
- Zhang, P.; Guo, Z.; Hu, R.; He, X.; Jiao, X.; Zhu, X. Interaction between microRNA-181a and TNFAIP1 regulates pancreatic cancer proliferation and migration. Tumor Biol. 2015, 36, 9693–9701. [Google Scholar] [CrossRef]
- Zhang, X.J.; Ye, H.; Zeng, C.W.; He, B.; Zhang, H.; Chen, Y.Q. Dysregulation of miR-15a and miR-214 in human pancreatic cancer. J. Hematol. Oncol. 2010, 3, 46. [Google Scholar] [CrossRef]
- Zhang, W.-L.; Zhang, J.-H.; Wu, X.-Z.; Yan, T.; Lv, W. miR-15b promotes epithelial-mesenchymal transition by inhibiting SMURF2 in pancreatic cancer. Int. J. Oncol. 2015, 47, 1043–1053. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, J.; Zhang, L.; Zhu, Z.; Fan, J.; Chen, L.; Zhuang, L.; Luo, J.; Chen, H.; Liu, L.; et al. MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology 2013, 145, 1133–1143.e12. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, H.; Wang, X.; Zhou, L.; Li, H.; Deng, T.; Qu, Y.; Duan, J.; Bai, M.; Ge, S.; et al. The miR-24-Bim pathway promotes tumor growth and angiogenesis in pancreatic carcinoma. Oncotarget 2015, 6, 43831–43842. [Google Scholar] [CrossRef]
- He, G.; Zhang, L.; Li, Q.; Yang, L. miR-92a/DUSP10/JNK signalling axis promotes human pancreatic cancer cells proliferation. Biomed. Pharmacother. 2014, 68, 25–30. [Google Scholar] [CrossRef]
- Cai, B.; An, Y.; Lv, N.; Chen, J.; Tu, M.; Sun, J.; Wu, P.; Wei, J.; Jiang, K.; Miao, Y. miRNA-181b increases the sensitivity of pancreatic ductal adenocarcinoma cells to gemcitabine in vitro and in nude mice by targeting BCL-2. Oncol. Rep. 2013, 29, 1769–1776. [Google Scholar] [CrossRef]
- Takiuchi, D.; Eguchi, H.; Nagano, H.; Iwagami, Y.; Tomimaru, Y.; Wada, H.; Kawamoto, K.; Kobayashi, S.; Marubashi, S.; Tanemura, M.; et al. Involvement of microRNA-181b in the gemcitabine resistance of pancreatic cancer cells. Pancreatology 2013, 13, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Tang, J.; Zhuang, X.; Zhuang, Y.; Cheng, W.; Chen, W.; Yao, H.; Zhang, S. MiR-196a Promotes Pancreatic Cancer Progression by Targeting Nuclear Factor Kappa-B-Inhibitor Alpha. PLoS ONE 2014, 9, e87897. [Google Scholar] [CrossRef]
- Liu, M.B.; Du, Y.; Gao, J.; Liu, J.; Kong, X.; Gong, Y.B.; Li, Z.; Wu, H.B.; Chen, H.B. Aberrant expression miR-196a is associated with abnormal apoptosis, invasion, and proliferation of pancreatic cancer cells. Pancreas 2013, 42, 1169–1181. [Google Scholar] [CrossRef]
- Ma, Y.; Yu, S.; Zhao, W.; Lu, Z.; Chen, J. miR-27a regulates the growth, colony formation and migration of pancreatic cancer cells by targeting Sprouty2. Cancer Lett. 2010, 298, 150–158. [Google Scholar] [CrossRef]
- Ma, J.; Fang, B.; Zeng, F.; Ma, C.; Pang, H.; Cheng, L.; Shi, Y.; Wang, H.; Yin, B.; Xia, J.; et al. Down-regulation of miR-223 reverses epithelial-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Oncotarget 2015, 6, 1740–1749. [Google Scholar] [CrossRef]
- Yong, S.; Yabin, Y.; Bing, Z.; Chuanrong, Z.; Dianhua, G.; Jianhuai, Z.; Weidong, Y.; Shuming, W.; Ling, L. Reciprocal regulation of DGCR5 and miR-320a affects the cellular malignant phenotype and 5-FU response in pancreatic ductal adenocarcinoma. Oncotarget 2017, 8, 90868–90878. [Google Scholar] [CrossRef]
- Laurila, E.M.; Sandström, S.; Rantanen, L.M.; Autio, R.; Kallioniemi, A. Both inhibition and enhanced expression of miR-31 lead to reduced migration and invasion of pancreatic cancer cells. Genes Chromosom. Cancer 2012, 51, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Baradaran, B.; Shahbazi, R.; Khordadmehr, M. Dysregulation of key microRNAs in pancreatic cancer development. Biomed. Pharmacother. 2018, 109, 1008–1015. [Google Scholar] [CrossRef] [PubMed]
- Nweke, E.E.; Brand, M. Downregulation of the let-7 family of microRNAs may promote insulin receptor/insulin-like growth factor signalling pathways in pancreatic ductal adenocarcinoma. Oncol. Lett. 2020, 20, 2613–2620. [Google Scholar] [CrossRef]
- Li, Z.; Li, X.; Yu, C.; Wang, M.; Peng, F.; Xiao, J.; Tian, R.; Jiang, J.; Sun, C. MicroRNA-100 regulates pancreatic cancer cells growth and sensitivity to chemotherapy through targeting FGFR3. Tumor Biol. 2014, 35, 11751–11759. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.; Zhong, X.; Jiang, X.; Leng, K.; Xu, Y.; Li, Z.; Huang, L.; Li, J.; Cui, Y. The role of long non-coding RNA AFAP1-AS1 in human malignant tumors. Pathol.-Res. Pract. 2018, 214, 1524–1531. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, D.; Hua, R.; Zhang, J.; Liu, W.; Huo, Y.; Cheng, Y.; Hong, J.; Sun, Y. Long non-coding RNAs expressed in pancreatic ductal adenocarcinoma and lncRNA BC008363 an independent prognostic factor in PDAC. Pancreatology 2014, 14, 385–390. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, H.; Chen, C. Long non-coding RNA PCED1B-AS1 promotes pancreatic ductal adenocarcinoma progression by regulating the miR-411-3p/HIF-1α axis. Oncol. Rep. 2021, 46, 134. [Google Scholar] [CrossRef] [PubMed]
- Gong, R.; Jiang, Y. Non-coding RNAs in Pancreatic Ductal Adenocarcinoma. Front. Oncol. 2020, 10, 309. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, L.; Shang, K.; Liu, F.; Che, J.; Li, H.; Cao, B. Long non-coding RNA H19, a novel therapeutic target for pancreatic cancer. Mol. Med. 2020, 26, 30. [Google Scholar] [CrossRef]
- Olivero, C.E.; Dimitrova, N. Identification and characterization of functional long noncoding RNAs in cancer. FASEB J. 2020, 34, 15630–15646. [Google Scholar] [CrossRef]
- Su, Y.; Gu, X.; Zheng, Q.; Zhu, L.; Lu, J.; Li, L. LncRNA PCGEM1 in Human Cancers: Functions, Mechanisms and Promising Clinical Utility. Front. Oncol. 2022, 12, 847745. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, L.; Mi, Y.; Tang, Y.; Reng, F.; Liu, B.; Zhang, Y.; Zheng, P. A ceRNA network and a potential regulatory axis in gastric cancer with different degrees of immune cell infiltration. Cancer Sci. 2020, 111, 4041–4050. [Google Scholar] [CrossRef]
- Bettin, N.; Oss Pegorar, C.; Cusanelli, E. The Emerging Roles of TERRA in Telomere Maintenance and Genome Stability. Cells 2019, 8, 246. [Google Scholar] [CrossRef]
- Feng, Q.; Chen, W.-D.; Wang, Y.-D. Gut Microbiota: An Integral Moderator in Health and Disease. Front. Microbiol. 2018, 9, 151. [Google Scholar] [CrossRef]
- Zhao, L.; Zhou, X.; Cai, W.; Shi, R.; Yang, G.; Yuan, L. Host intestinal epithelium derived mirnas shape the microbiota and its implication in cardiovascular diseases. J. Am. Coll. Cardiol. 2017, 69, 1075. [Google Scholar] [CrossRef]
- Gesualdo, M.; Rizzi, F.; Bonetto, S.; Rizza, S.; Cravero, F.; Saracco, G.M.; De Angelis, C.G. Pancreatic Diseases and Microbiota: A Literature Review and Future Perspectives. J. Clin. Med. 2020, 9, 3535. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, E.; Zhang, Y.; Zhang, L.; Montiel, M.; Zoltan, M.; Dong, W.; Quesada, P.; Sahin, I.; Chandra, V.; Lucas, A.S.; et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell 2019, 178, 795–806.e12. [Google Scholar] [CrossRef] [PubMed]
- Wong-Rolle, A.; Wei, H.K.; Zhao, C.; Jin, C. Unexpected guests in the tumor microenvironment: Microbiome in cancer. Protein Cell 2020, 12, 426–435. [Google Scholar] [CrossRef]
- Sammallahti, H.; Sarhadi, V.K.; Kokkola, A.; Ghanbari, R.; Rezasoltani, S.; Aghdaei, H.A.; Puolakkainen, P.; Knuutila, S. Oncogenomic Changes in Pancreatic Cancer and Their Detection in Stool. Biomolecules 2022, 12, 652. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Burns, M.B.; Subramanian, S.; Blekhman, R. Interaction between Host MicroRNAs and the Gut Microbiota in Colorectal Cancer. Msystems 2018, 3, e00205-17. [Google Scholar] [CrossRef]
- Sarhadi, V.; Lahti, L.; Saberi, F.; Youssef, O.; Kokkola, A.; Karla, T.; Tikkanen, M.; Rautelin, H.; Puolakkainen, P.; Salehi, R.; et al. Gut Microbiota and Host Gene Mutations in Colorectal Cancer Patients and Controls of Iranian and Finnish Origin. Anticancer Res. 2020, 40, 1325–1334. [Google Scholar] [CrossRef]
- Marin-Muller, C.; Li, D.; Bharadwaj, U.; Li, M.; Chen, C.; Hodges, S.E.; Fisher, W.E.; Mo, Q.; Hung, M.-C.; Yao, Q. A tumorigenic factor interactome connected through tumor suppressor MicroRNA-198 in human pancreatic cancer. Clin. Cancer Res. 2013, 19, 5901–5913. [Google Scholar] [CrossRef]
- Shirazi, M.S.R.; Al-Alo, K.Z.K.; Al-Yasiri, M.H.; Lateef, Z.M.; Ghasemian, A. Microbiome Dysbiosis and Predominant Bacterial Species as Human Cancer Biomarkers. J. Gastrointest. Cancer 2019, 51, 725–728. [Google Scholar] [CrossRef]
- Allegra, A.; Musolino, C.; Tonacci, A.; Pioggia, G.; Gangemi, S. Interactions between the MicroRNAs and Microbiota in Cancer Development: Roles and Therapeutic Opportunities. Cancers 2020, 12, 805. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Guan, L.L. Noncoding RNAs: Regulatory Molecules of Host–Microbiome Crosstalk. Trends Microbiol. 2021, 29, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Tomkovich, S.; Gharaibeh, R.Z.; Dejea, C.M.; Pope, J.L.; Jiang, J.; Winglee, K.; Gauthier, J.; Newsome, R.C.; Yang, Y.; Fodor, A.A.; et al. Human Colon Mucosal Biofilms and Murine Host Communicate via Altered mRNA and microRNA Expression during Cancer. Msystems 2020, 5, e00451-19. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Ren, Y.; Sayed, M.; Hu, X.; Lei, C.; Kumar, A.; Hutchins, E.; Mu, J.; Deng, Z.; Luo, C.; et al. Plant-Derived Exosomal MicroRNAs Shape the Gut Microbiota. Cell Host Microbe 2018, 24, 637–652.e8. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, H.; Bernstein, C.; Payne, C.M.; Dvorak, K. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J. Gastroenterol. 2009, 15, 3329–3340. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef]

| Mutation | PanIN | IPMN | MCN |
|---|---|---|---|
| KRAS codon 12 | (+) in 90% of PanINs | (+) KRAS2 most often | (+) in chr.12p from 20% of the least malignant to 89% in most advanced stages |
| loss of p16 (CDK22A/INK4A) gene on chr. 9p21 | (+) in 30% in I, 55% in II, 71% in III | (−) | (−) |
| tp53 in chr. 17 (inactivation of MADH4/SMAD/DPC4) | (+) in PanIN II and III | (+) only in invasive component | (+) only in invasive component |
| BRCA2 (ch.19q) | (+) in PanIN II and III | (−) | (−) |
| mucins (MUC-1, MUC-2,MUC3) | MUC-1 | MUC-1, MUC-2, MUC-3 | (+) in different glycoforms |
| STK11/LKB1 | (−) | (+) in 30% of lesions | (−) |
| RNF43 | (−) | (+) in 15% of lesions | (+) |
| PI3KCA | (−) | (+) | (−) |
| GNAS | (−) | (+) in 4% of lesions | (−) |
| Mutation | IPMN | PanIn I | PanIn II | PanIn III | MCN | PDAC | Metastasis |
|---|---|---|---|---|---|---|---|
| KRAS | + | + | + | + | + | + | + |
| GNAS | + | − | − | − | − | + | − |
| Telomere disintegrity | − | + | + | + | − | − | − |
| MUC1, MUC2, MUC3 | − | + | + | − | − | − | − |
| CDK2 | − | + | + | + | + | + | − |
| BRCA2 | − | + | + | − | − | − | − |
| RFN43 | − | − | − | − | + | + | − |
| PI3KCA | − | − | + | − | − | − | − |
| STK11/LKB1SSTK11/LB1 | − | − | + | − | − | − | − |
| SMAD4 | + | − | − | + | + | + | + |
| CDKN2A | + | − | − | − | − | + | − |
| tp53 | − | − | − | − | + | + | + |
| FOXA1 | − | − | − | − | + | − | − |
| miRNA | Up/Down-Regulated in PDAC | Targeted Genes | Pro/Anticancer Outcome | Description |
|---|---|---|---|---|
| miR-107 | up | CDK6 [78] | pro | stops growth |
| miR-127 | down | BAG5 [79] | anti | inhibits cancer development |
| miR-21 | up | Bcl-2, FasL [80] | pro | decreases apoptosis, increases gemcitabine resistance |
| FoxO1 [81] | pro | increases tumor growth | ||
| PDCD4 [82] | pro | increases proliferation and reduces cell death | ||
| PTEN, RECK [83] | intensifies the progression of cell cycle, increases proliferation | |||
| miR-221 | up | p27kip1 [83] | pro | enhances the progression of cell cycle, promotes proliferation |
| PTEN, P27KIP1, P57KIP2, PUMA [84] | pro | increases proliferation | ||
| TIMP2 [85] | pro | increases proliferation and invasion, stops apoptosis | ||
| TRPS1 [86] | pro | Mediates EMT phenotype, migration, and growth | ||
| miR-222 | up | p57 [87] | pro | enhances proliferation |
| MMP2, MMP9 [85] | pro | enhances proliferation, invasion, stops apoptosis | ||
| miR-10 a | up | HOXA1 [88] | pro | enhances invasion |
| miR-125 b | up | TXNIP [89] | pro | enhances tumorigenesis and progression |
| miR-34 a | down | CD133, Notch1, Notch2, Notch4 receptors [90] | anti | inhibits cell survival, invasion, migration, increases cell apoptosis |
| miR-143 | down | KRAS [91] | anti | inhibits cell proliferation, migration, and invasion |
| miR-145 | up | KRAS, RREB1 [92] | anti | stops tumor growth |
| ROR [93] | anti | stops proliferation, invasion, and cell cycle | ||
| MUC13 [94] | anti | stops tumor growth and invasion | ||
| miR-217 | down | SIRT1 [95] | regulates (epithelial–mesenchymal transition) EMT process | |
| KRAS [96] | anti | stops cell growth and colony forming | ||
| miR-141 | down | MAP4K4 [97] | anti | stops proliferation, colony formation, invasion by inhibiting G1-phase, and apoptosis |
| YAP [98] | anti | stops proliferation, forming colonies, and apoptosis | ||
| TM4SF1 [99] | anti | stops invasion and migration | ||
| miR-148 a | down | DNMT1 [100] | anti | stops proliferation and metastasis |
| CCKBR, Bcl-2 [101] | anti | stops tumor growth and apoptosis | ||
| CDC25B [102] | anti | stops cell survival | ||
| miR-375 | down | PDK1 [103] | anti | stop cell growth and enhances apoptosis |
| miR-29 c | down | ITGB1 [104] | anti | stops cell growth, invasion, and migration |
| MMP2 [105] | anti | stops migration, invasion, metastasis (in mice model) | ||
| FRAT2, LRP6, FZD4, FZD5 [106] | anti | stops migration and stem-cell-like phenotype | ||
| miR-130 b | down | STAT3 [106] | anti/pro | stops invasion and encourages proliferation |
| miR-200 c | down | MUC4, MUC6 [107] | anti | targets directly MUC4 and 6 |
| E-cadherin [108] | anti/pro | stops invasion and enhances proliferation | ||
| miR-216 a | down | JAK2 [109] | anti | stop proliferation and enhances apoptosis |
| Beclin-1 [110] | anti | enhances radiosensitivity | ||
| miR-26 a | down | p53 [111] | anti | stops proliferation by phosphorylation of p53 |
| Cyclin E2 [112] | anti | stops proliferation | ||
| miR-148 b | down | AMPKalfa1 [113] | anti | stops cell cycle and cell growth |
| DNMT1 [114] | anti | influences methylation of tumor suppressor genes | ||
| miR-335 | down | OCT4 [114] | anti | stops progression and influences stem cell properties |
| miR-365 | down | SHC1-BAX [115] | pro | elicits gemcitabine resistance |
| miR-155 | up | Foxo3a, KRAS, ROS [116] | anti | decreases proliferation induced by ROS generation |
| SEL1L [117] | downregulates SEL1L | |||
| MLH1 [118] | downregulates MLH1 | |||
| SOCS1 [119] | enhances invasion and migration | |||
| up | TP53INP1 [120] | enhances tumor growth | ||
| miR-23 a | up | APAF1 [121] | pro | increases proliferation and decreases apoptosis |
| FZD5, HNF1B, TMEM92 [122] | pro | increases EMT-like cell transformation | ||
| miR-143 | up | ARHGEF1, ARHGEF2, KRAS [123] | anti | decreases migration, invasion, and metastasis to liver |
| miR-146 a | up | EGFR, IRAK1, MTA-2 [124] | anti | stops invasion |
| miR-150 | up | MUC4 [125] | anti | stops growth, clonogenicity, migration, invasion, enhances intercellular adhesion |
| miR-181 a | up | PTEN, MAP2K4 [126] | pro | enhances migration |
| TNFAIP1 [127] | pro | enhances proliferation and migration | ||
| miR-214 | up | ING4 [128] | pro | decreases sensitivity to gemcitabine |
| miR-15 b | up | SMURF2 [129] | pro | enhances EMT |
| miR-23 b | up | ATG12 [130] | pro/anti | regulates autophagy associated with radioresistance |
| miR-24 | up | Bim [131] | pro | increases cell growth |
| FZD5, HNF1B, TMEM92 [122] | pro | increases EMT-like cell share transformation | ||
| miR-92 a | up | DUSP10 [132] | pro | enhances proliferation |
| miR-181 b | up | BCL-2 [133] | anti | sensitizes particular cells to gemcitabine |
| CYLD [134] | anti | increases gemcitabine resistance of some cells | ||
| miR-196 a | up | NFKBIA [135] | pro | enhances proliferation and migration |
| ING5 [136] | pro | enhances proliferation, migration, and decreases apoptosis | ||
| miR-27 a | up | Sprouty2 [137] | pro | enhances growth, colony formation, and migration |
| miR-223 | up | FBw7 [138] | pro | secures EMT phenotype |
| miR-320 a | down | DGCR5 (lncRNA) [139] | anti | regulates proliferation, migration, and 5-FU resistance |
| miR-31 | up | APBB2 [140] | anti | reduces migration of cancer cells |
| miR-451 | up | CAB39 [141] | pro | enhances cell proliferation and lymphatic metastasis |
| miR-let7 | down | numerous genes in the insulin signaling pathway [142] | anti | inhibits tumor progression and increases therapy sensitivity |
| miR-100 | FGFR3 [143] | anti | inhibits proliferation and enhances sensitivity to cisplatin | |
| lncRNA | Up/Down-Regulated in PDAC | Targeted Genes | Pro/Anticancer Outcome | Description |
| AF339813 | up | NUF2, CDK1, CDK4/CDK6 [14] | pro | apoptosis, controls cell cycle |
| AFAPI1-AS1 | up | E-cadherin, N-adherin, Snail [14,144] | pro | regulates cell proliferation, migration, invasion |
| BC008363 | down | many protein-coding genes involved in tumor growth and drug resistance [14,145] | anti | diminishes tumor growth and rug resistance |
| CDKN2B-ASI | up | miRN-411-3p [146] | pro | regulates miRN-411-3p and HIF-1alfa (hypoxia-inducible factor) |
| ENST00000480739 | down | OS-9, HIF-1 [14] | pro | controls invasion and migration |
| GAS5 | up | miR-32-5p [147] | anti | regulates the cell cycle, involved in PI3K/Akt signaling pathway |
| GAS5 | down | CDK6 [14] | ani | stops cell proliferation |
| H19 | up | Let-7, HMGA2, miR-194, miR-138, miR-200 [14,148] | pro | mesenchymal–epithelial transition (MET), involved in development and progression |
| HOTAIR | up | PRC2, GDF15 [14] | pro | related to invasion, proliferation, progression of PC |
| HOTTIP | up | AURKA, WDR5, HOXA10, HOXB2, HOXA11, HOXA9, HOXA1, HOXA13 [14] | pro | cell cancer proliferation, stops cell apoptosis, enhances migration of cancer cells |
| MALAT-1 | up | Sox2, E-cadherin, N-adherin, vimentin, VEGF [14] | pro | regulates cell cycle, growth, migration, and invasion |
| PCGEM1 | up in most cancers | numerous miRNA [149,150] | pro | involved in cell invasion and migration, associated with cancer progression |
| PLACT1 | up | hnRNPA1 [151] | pro | regulates tumorigenesis through NF-kappa beta signaling pathway, involved in lung metastasis progression |
| TERRA | up | TRF2 [152] | anti | induces apoptosis, inhibits cell proliferation, invasion, metastasis, regulates cell cycle |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Izdebska, W.M.; Daniluk, J.; Niklinski, J. Microbiome and MicroRNA or Long Non-Coding RNA—Two Modern Approaches to Understanding Pancreatic Ductal Adenocarcinoma. J. Clin. Med. 2023, 12, 5643. https://doi.org/10.3390/jcm12175643
Izdebska WM, Daniluk J, Niklinski J. Microbiome and MicroRNA or Long Non-Coding RNA—Two Modern Approaches to Understanding Pancreatic Ductal Adenocarcinoma. Journal of Clinical Medicine. 2023; 12(17):5643. https://doi.org/10.3390/jcm12175643
Chicago/Turabian StyleIzdebska, Wiktoria Maria, Jaroslaw Daniluk, and Jacek Niklinski. 2023. "Microbiome and MicroRNA or Long Non-Coding RNA—Two Modern Approaches to Understanding Pancreatic Ductal Adenocarcinoma" Journal of Clinical Medicine 12, no. 17: 5643. https://doi.org/10.3390/jcm12175643
APA StyleIzdebska, W. M., Daniluk, J., & Niklinski, J. (2023). Microbiome and MicroRNA or Long Non-Coding RNA—Two Modern Approaches to Understanding Pancreatic Ductal Adenocarcinoma. Journal of Clinical Medicine, 12(17), 5643. https://doi.org/10.3390/jcm12175643






