Using Circulating Tumor DNA as a Novel Biomarker to Screen and Diagnose Colorectal Cancer: A Meta-Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Protocol and Registration
2.2. Search Strategy
2.3. Inclusion and Exclusion Criteria
2.4. Data Extraction
2.5. Quality Assessment
2.6. Statistical Analysis
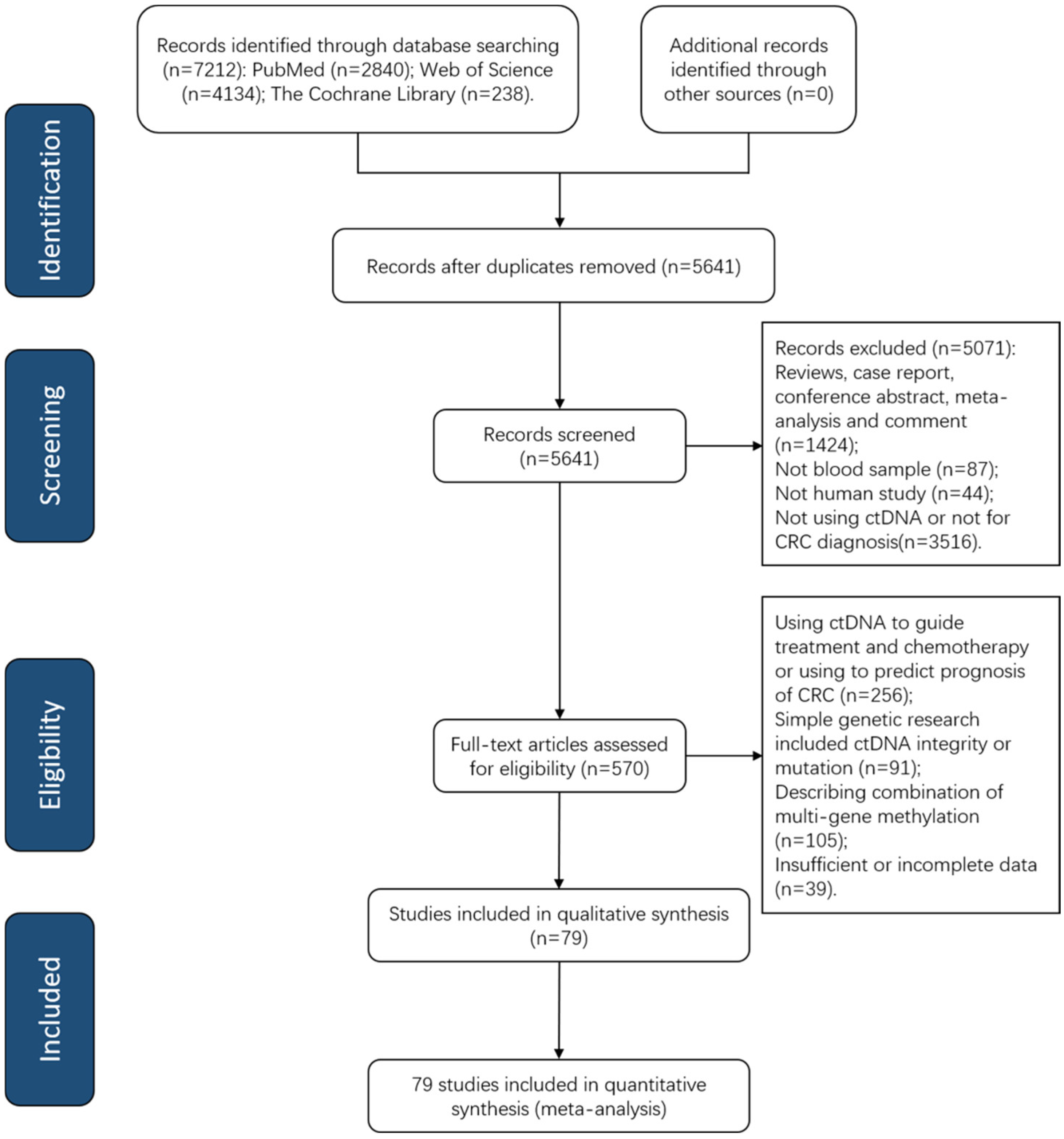
3. Results
3.1. Studies Characteristics
3.2. Quality Assessment
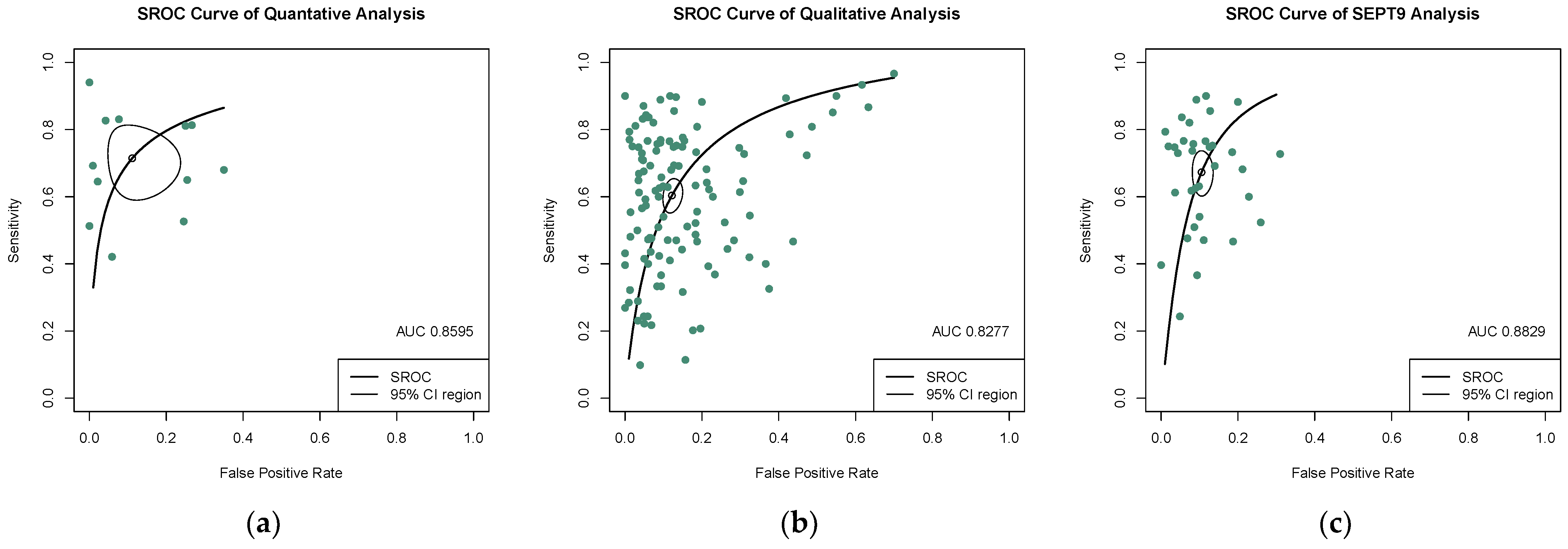
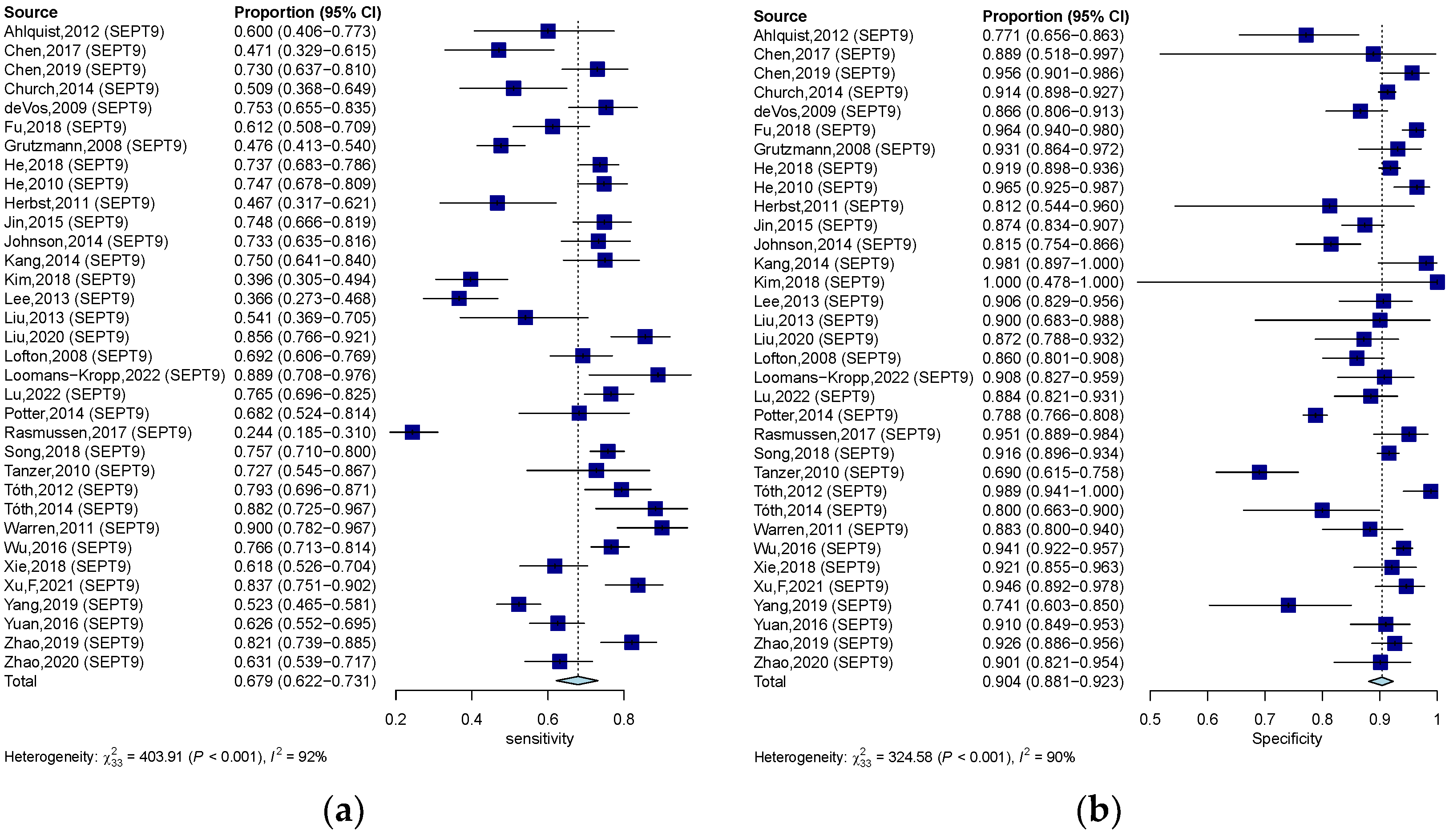
3.3. Diagnostic Value of Quantitative and Qualitative Analysis of ctDNA for CRC
3.4. Subgroup Analysis and Meta-Regression Analysis
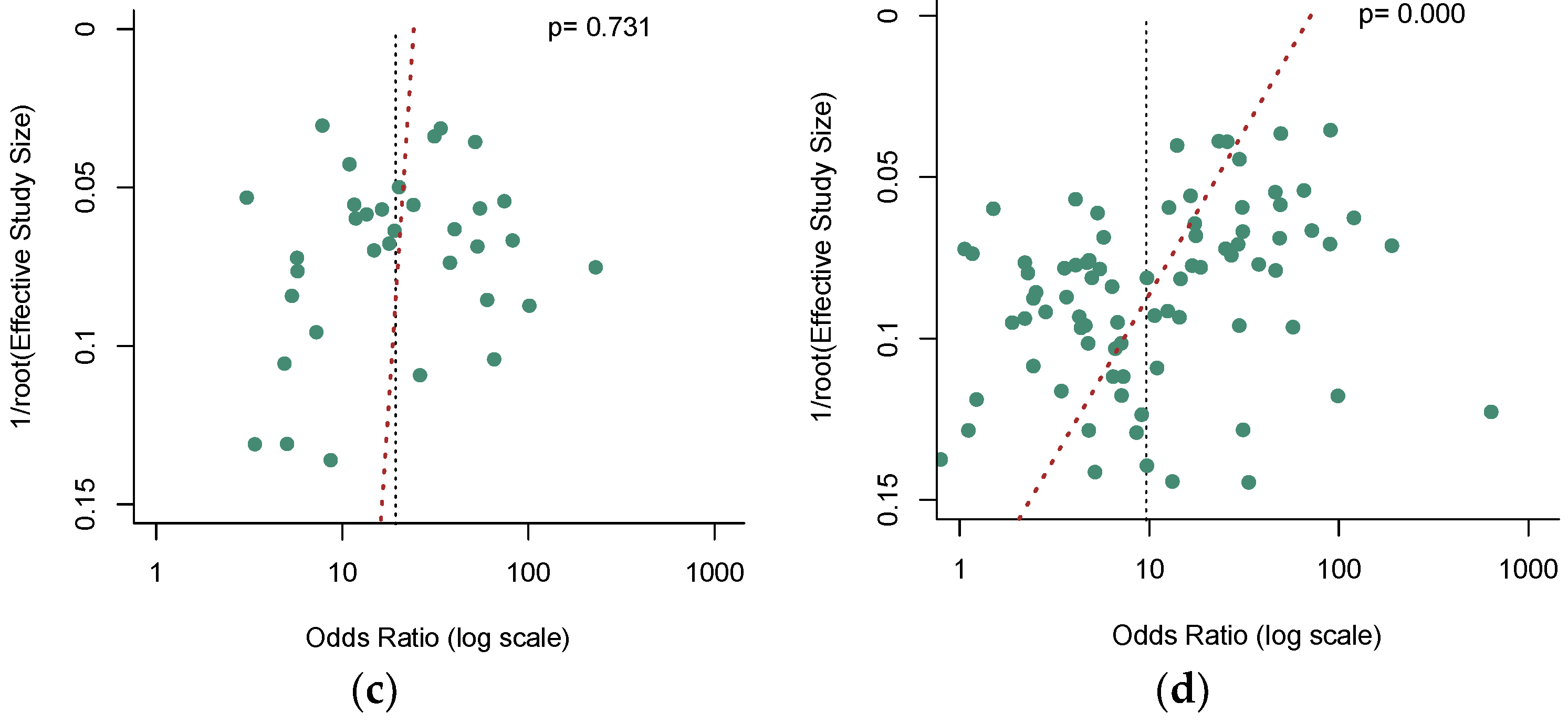
3.5. Publication Bias
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer Statistics for the Year 2020: An Overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Xu, P. Global Colorectal Cancer Burden in 2020 and Projections to 2040. Transl. Oncol. 2021, 14, 101174. [Google Scholar] [CrossRef] [PubMed]
- Allemani, C.; Matsuda, T.; Di Carlo, V.; Harewood, R.; Matz, M.; Nikšić, M.; Bonaventure, A.; Valkov, M.; Johnson, C.J.; Estève, J.; et al. Global Surveillance of Trends in Cancer Survival 2000-14 (CONCORD-3): Analysis of Individual Records for 37,513,025 Patients Diagnosed with One of 18 Cancers from 322 Population-Based Registries in 71 Countries. Lancet 2018, 391, 1023–1075. [Google Scholar] [CrossRef] [PubMed]
- Sawicki, T.; Ruszkowska, M.; Danielewicz, A.; Niedźwiedzka, E.; Arłukowicz, T.; Przybyłowicz, K.E. A Review of Colorectal Cancer in Terms of Epidemiology, Risk Factors, Development, Symptoms and Diagnosis. Cancers 2021, 13, 2025. [Google Scholar] [CrossRef] [PubMed]
- Ladabaum, U.; Dominitz, J.A.; Kahi, C.; Schoen, R.E. Strategies for Colorectal Cancer Screening. Gastroenterology 2020, 158, 418–432. [Google Scholar] [CrossRef]
- Ahlquist, D.A.; Sargent, D.J.; Loprinzi, C.L.; Levin, T.R.; Rex, D.K.; Ahnen, D.J.; Knigge, K.; Lance, M.P.; Burgart, L.J.; Hamilton, S.R.; et al. Stool DNA and Occult Blood Testing for Screen Detection of Colorectal Neoplasia. Ann. Intern. Med. 2008, 149, 441—450, W81. [Google Scholar] [CrossRef]
- Morikawa, T.; Kato, J.; Yamaji, Y.; Wada, R.; Mitsushima, T.; Shiratori, Y. A Comparison of the Immunochemical Fecal Occult Blood Test and Total Colonoscopy in the Asymptomatic Population. Gastroenterology 2005, 129, 422–428. [Google Scholar] [CrossRef]
- Song, L.; Jia, J.; Peng, X.; Xiao, W.; Li, Y. The Performance of the SEPT9 Gene Methylation Assay and a Comparison with Other CRC Screening Tests: A Meta-Analysis. Sci. Rep. 2017, 7, 3032. [Google Scholar] [CrossRef]
- Adler, A.; Geiger, S.; Keil, A.; Bias, H.; Schatz, P.; deVos, T.; Dhein, J.; Zimmermann, M.; Tauber, R.; Wiedenmann, B. Improving Compliance to Colorectal Cancer Screening Using Blood and Stool Based Tests in Patients Refusing Screening Colonoscopy in Germany. BMC Gastroenterol. 2014, 14, 183. [Google Scholar] [CrossRef]
- Niu, F.; Wen, J.; Fu, X.; Li, C.; Zhao, R.; Wu, S.; Yu, H.; Liu, X.; Zhao, X.; Liu, S.; et al. Stool DNA Test of Methylated Syndecan-2 for the Early Detection of Colorectal Neoplasia. Cancer Epidemiol. Biomark. Prev. 2017, 26, 1411–1419. [Google Scholar] [CrossRef]
- Meissner, H.I.; Breen, N.; Klabunde, C.N.; Vernon, S.W. Patterns of Colorectal Cancer Screening Uptake among Men and Women in the United States. Cancer Epidemiol. Biomark. Prev. 2006, 15, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Leon, S.; Shapiro, B.; Sklaroff, D.; Yaros, M. Free DNA in the Serum of Cancer Patients and the Effect of Therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar] [PubMed]
- Han, D.S.C.; Lo, Y.M.D. The Nexus of CfDNA and Nuclease Biology. Trends Genet. 2021, 37, 758–770. [Google Scholar] [CrossRef] [PubMed]
- Lui, Y.Y.; Chik, K.-W.; Chiu, R.W.; Ho, C.-Y.; Lam, C.W.; Lo, Y.D. Predominant Hematopoietic Origin of Cell-Free DNA in Plasma and Serum after Sex-Mismatched Bone Marrow Transplantation. Clin. Chem. 2002, 48, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.W.; Chan, K.A.; Sun, H.; Jiang, P.; Su, X.; Chen, E.Z.; Lun, F.M.; Hung, E.C.; Lee, V.; Wong, J. Nonhematopoietically Derived DNA Is Shorter than Hematopoietically Derived DNA in Plasma: A Transplantation Model. Clin. Chem. 2012, 58, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Hasenleithner, S.O.; Speicher, M.R. A Clinician’s Handbook for Using CtDNA throughout the Patient Journey. Mol. Cancer 2022, 21, 81. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A. Circulating Mutant DNA to Assess Tumor Dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Yi, X.; Ma, J.; Guan, Y.; Chen, R.; Yang, L.; Xia, X. The Feasibility of Using Mutation Detection in CtDNA to Assess Tumor Dynamics. Int. J. Cancer 2017, 140, 2642–2647. [Google Scholar] [CrossRef]
- Heitzer, E.; Ulz, P.; Geigl, J.B. Circulating Tumor DNA as a Liquid Biopsy for Cancer. Clin. Chem. 2015, 61, 112–123. [Google Scholar] [CrossRef]
- Roy, D.; Tiirikainen, M. Diagnostic Power of DNA Methylation Classifiers for Early Detection of Cancer. Trends Cancer 2020, 6, 78–81. [Google Scholar] [CrossRef]
- Gilson, P.; Merlin, J.-L.; Harlé, A. Detection of Microsatellite Instability: State of the Art and Future Applications in Circulating Tumour DNA (CtDNA). Cancers 2021, 13, 1491. [Google Scholar] [CrossRef] [PubMed]
- Symonds, E.L.; Pedersen, S.K.; Yeo, B.; Al Naji, H.; Byrne, S.E.; Roy, A.; Young, G.P. Assessment of Tumor Burden and Response to Therapy in Patients with Colorectal Cancer Using a Quantitative CtDNA Test for Methylated BCAT1/IKZF1. Mol. Oncol. 2022, 16, 2031–2041. [Google Scholar] [CrossRef] [PubMed]
- Anker, P.; Lefort, F.; Vasioukhin, V.; Lyautey, J.; Lederrey, C.; Chen, X.Q.; Stroun, M.; Mulcahy, H.E.; Farthing, M. K-Ras Mutations Are Found in DNA Extracted from the Plasma of Patients with Colorectal Cancer. Gastroenterology 1997, 112, 1114–1120. [Google Scholar] [CrossRef] [PubMed]
- Vymetalkova, V.; Cervena, K.; Bartu, L.; Vodicka, P. Circulating Cell-Free DNA and Colorectal Cancer: A Systematic Review. Int. J. Mol. Sci. 2018, 19, 3356. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. BMJ 2009, 339, b2535. [Google Scholar] [CrossRef]
- Stroup, D.F.; Berlin, J.A.; Morton, S.C.; Olkin, I.; Williamson, G.D.; Rennie, D.; Moher, D.; Becker, B.J.; Sipe, T.A.; Thacker, S.B. Meta-Analysis of Observational Studies in Epidemiology: A Proposal for Reporting. Meta-Analysis Of Observational Studies in Epidemiology (MOOSE) Group. JAMA 2000, 283, 2008–2012. [Google Scholar] [CrossRef]
- Whiting, P.F.; Rutjes, A.W.S.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.G.; Sterne, J.A.C.; Bossuyt, P.M.M.; QUADAS-2 Group. QUADAS-2: A Revised Tool for the Quality Assessment of Diagnostic Accuracy Studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef]
- Schwarzer, G. Meta: An R Package for Meta-Analysis. R News 2007, 7, 40–45. [Google Scholar]
- Doebler, P.; Holling, H. Meta-Analysis of Diagnostic Accuracy with Mada. R Packag. 2015, 1, 15. [Google Scholar]
- Viechtbauer, W.; Viechtbauer, M.W. Package ‘Metafor’. The Comprehensive R Archive Network. Package ‘Metafor’. 2015. Available online: http://cran.r-project.org/web/packages/metafor/metafor.pdf (accessed on 20 November 2022).
- Glas, A.S.; Lijmer, J.G.; Prins, M.H.; Bonsel, G.J.; Bossuyt, P.M.M. The Diagnostic Odds Ratio: A Single Indicator of Test Performance. J. Clin. Epidemiol. 2003, 56, 1129–1135. [Google Scholar] [CrossRef]
- Arends, L.R.; Hamza, T.H.; van Houwelingen, J.C.; Heijenbrok-Kal, M.H.; Hunink, M.G.M.; Stijnen, T. Bivariate Random Effects Meta-Analysis of ROC Curves. Med. Decis. Mak. 2008, 28, 621–638. [Google Scholar] [CrossRef] [PubMed]
- Reitsma, J.B.; Glas, A.S.; Rutjes, A.W.S.; Scholten, R.J.P.M.; Bossuyt, P.M.; Zwinderman, A.H. Bivariate Analysis of Sensitivity and Specificity Produces Informative Summary Measures in Diagnostic Reviews. J. Clin. Epidemiol. 2005, 58, 982–990. [Google Scholar] [CrossRef] [PubMed]
- Mandrekar, J.N. Receiver Operating Characteristic Curve in Diagnostic Test Assessment. J. Thorac. Oncol. 2010, 5, 1315–1316. [Google Scholar] [CrossRef] [PubMed]
- Walter, S. Properties of the Summary Receiver Operating Characteristic (SROC) Curve for Diagnostic Test Data. Stat. Med. 2002, 21, 1237–1256. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H. Overview of the Process of Conducting Meta-Analyses of the Diagnostic Test Accuracy. J. Rheum. Dis. 2018, 25, 3–10. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring Inconsistency in Meta-Analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef]
- Thompson, S.G.; Higgins, J.P.T. How Should Meta-Regression Analyses Be Undertaken and Interpreted? Stat. Med. 2002, 21, 1559–1573. [Google Scholar] [CrossRef]
- Sterne, J.A.C.; Sutton, A.J.; Ioannidis, J.P.A.; Terrin, N.; Jones, D.R.; Lau, J.; Carpenter, J.; Rücker, G.; Harbord, R.M.; Schmid, C.H.; et al. Recommendations for Examining and Interpreting Funnel Plot Asymmetry in Meta-Analyses of Randomised Controlled Trials. BMJ 2011, 343, d4002. [Google Scholar] [CrossRef]
- Agah, S.; Akbari, A.; Talebi, A.; Masoudi, M.; Sarveazad, A.; Mirzaei, A.; Nazmi, F. Quantification of Plasma Cell-Free Circulating DNA at Different Stages of Colorectal Cancer. Cancer Investig. 2017, 35, 625–632. [Google Scholar] [CrossRef]
- Agostini, M.; Pucciarelli, S.; Enzo, M.V.; Del Bianco, P.; Briarava, M.; Bedin, C.; Maretto, I.; Friso, M.L.; Lonardi, S.; Mescoli, C.; et al. Circulating Cell-Free DNA: A Promising Marker of Pathologic Tumor Response in Rectal Cancer Patients Receiving Preoperative Chemoradiotherapy. Ann. Surg. Oncol. 2011, 18, 2461–2468. [Google Scholar] [CrossRef]
- Allegretti, M.; Cottone, G.; Carboni, F.; Cotroneo, E.; Casini, B.; Giordani, E.; Amoreo, C.A.; Buglioni, S.; Diodoro, M.; Pescarmona, E.; et al. Cross-Sectional Analysis of Circulating Tumor DNA in Primary Colorectal Cancer at Surgery and during Post-Surgery Follow-up by Liquid Biopsy. J. Exp. Clin. Cancer Res. 2020, 39, 12. [Google Scholar] [CrossRef] [PubMed]
- Czeiger, D.; Shaked, G.; Eini, H.; Vered, I.; Belochitski, O.; Avriel, A.; Ariad, S.; Douvdevani, A. Measurement of Circulating Cell-Free DNA Levels by a New Simple Fluorescent Test in Patients With Primary Colorectal Cancer. Am. J. Clin. Pathol. 2011, 135, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Danese, E.; Montagnana, M.; Minicozzi, A.M.; De Matteis, G.; Scudo, G.; Salvagno, G.L.; Cordiano, C.; Lippi, G.; Guidi, G.C. Real-Time Polymerase Chain Reaction Quantification of Free DNA in Serum of Patients with Polyps and Colorectal Cancers. Clin. Chem. Lab. Med. 2010, 48, 1665–1668. [Google Scholar] [CrossRef] [PubMed]
- El-Gayar, D.; El-Abd, N.; Hassan, N.; Ali, R. Increased Free Circulating DNA Integrity Index as a Serum Biomarker in Patients with Colorectal Carcinoma. Asian Pac. J. Cancer Prev. APJCP 2016, 17, 939–944. [Google Scholar] [CrossRef]
- Flamini, E.; Mercatali, L.; Nanni, O.; Calistri, D.; Nunziatini, R.; Zoli, W.; Rosetti, P.; Gardini, N.; Lattuneddu, A.; Verdecchia, G.M.; et al. Free DNA and Carcinoembryonic Antigen Serum Levels: An Important Combination for Diagnosis of Colorectal Cancer. Clin. Cancer Res. 2006, 12, 6985–6988. [Google Scholar] [CrossRef]
- Hao, T.B.; Shi, W.; Shen, X.J.; Qi, J.; Wu, X.H.; Wu, Y.; Tang, Y.Y.; Ju, S.Q. Circulating Cell-Free DNA in Serum as a Biomarker for Diagnosis and Prognostic Prediction of Colorectal Cancer. Br. J. Cancer 2014, 111, 1482–1489. [Google Scholar] [CrossRef]
- Junca, A.; Tachon, G.; Evrard, C.; Villalva, C.; Frouin, E.; Karayan-Tapon, L.; Tougeron, D. Detection of Colorectal Cancer and Advanced Adenoma by Liquid Biopsy (Decalib Study): The DdPCR Challenge. Cancers 2020, 12, 1482. [Google Scholar] [CrossRef]
- Lan, Y.T.; Chen, M.H.; Fang, W.L.; Hsieh, C.C.; Lin, C.H.; Jhang, F.Y.; Yang, S.H.; Lin, J.K.; Chen, W.S.; Jiang, J.K.; et al. Clinical Relevance of Cell-Free DNA in Gastrointestinal Tract Malignancy. Oncotarget 2017, 8, 3009–3017. [Google Scholar] [CrossRef]
- Qi, J.; Qian, C.; Shi, W.; Wu, X.H.; Jing, R.R.; Zhang, L.R.; Wang, Z.W.; Ju, S.Q. Alu-Based Cell-Free DNA: A Potential Complementary Biomarker for Diagnosis of Colorectal Cancer. Clin. Biochem. 2013, 46, 64–69. [Google Scholar] [CrossRef]
- Ahlquist, D.A.; Taylor, W.R.; Mahoney, D.W.; Zou, H.; Domanico, M.; Thibodeau, S.N.; Boardman, L.A.; Berger, B.M.; Lidgard, G.P. The Stool DNA Test Is More Accurate than the Plasma Septin 9 Test in Detecting Colorectal Neoplasia. Clin. Gastroenterol. Hepatol. 2012, 10, 272–277.e1. [Google Scholar] [CrossRef]
- Alizadeh Naini, M.; Kavousipour, S.; Hasanzarini, M.; Nasrollah, A.; Monabati, A.; Mokarram, P. O6-Methyguanine-DNA Methyl Transferase (MGMT) Promoter Methylation in Serum DNA of Iranian Patients with Colorectal Cancer. Asian Pac. J. Cancer Prev. APJCP 2018, 19, 1223–1227. [Google Scholar] [CrossRef]
- Amiot, A.; Mansour, H.; Baumgaertner, I.; Delchier, J.C.; Tournigand, C.; Furet, J.P.; Carrau, J.P.; Canoui-Poitrine, F.; Sobhani, I.; the CRC Group of Val De Marne. The Detection of the Methylated Wif-1 Gene Is More Accurate than a Fecal Occult Blood Test for Colorectal Cancer Screening. PLoS ONE 2014, 9, e99233. [Google Scholar] [CrossRef] [PubMed]
- Ansar, M.; Wang, C.-J.; Wang, Y.-H.; Shen, T.-H.; Hung, C.-S.; Chang, S.-C.; Lin, R.-K. SMAD3 Hypomethylation as a Biomarker for Early Prediction of Colorectal Cancer. Int. J. Mol. Sci. 2020, 21, 7395. [Google Scholar] [CrossRef]
- Ashoori, H.; Ghamarchehreh, M.E.; Tavallaei, M.; Ganji, S.M.; Hosseini, M.; Zolfaghari, M.; Ghamarchehreh, Z.; Vahidian, F. Evaluation of the Epigenetic Biomarker Bone Morphogenic Protein 3 for Colorectal Cancer Diagnosis. J. Clin. Diagn. Res. 2018, 12, GC07–GC09. [Google Scholar] [CrossRef]
- Bartak, B.K.; Kalmar, A.; Peterfia, B.; Patai, A.V.; Galamb, O.; Valcz, G.; Spisak, S.; Wichmann, B.; Nagy, Z.B.; Toth, K.; et al. Colorectal Adenoma and Cancer Detection Based on Altered Methylation Pattern of SFRP1, SFRP2, SDC2, and PRIMA1 in Plasma Samples. Epigenetics 2017, 12, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Bedin, C.; Enzo, M.V.; Del Bianco, P.; Pucciarelli, S.; Nitti, D.; Agostini, M. Diagnostic and Prognostic Role of Cell-Free DNA Testing for Colorectal Cancer Patients. Int. J. Cancer 2017, 140, 1888–1898. [Google Scholar] [CrossRef] [PubMed]
- Cassinotti, E.; Melson, J.; Liggett, T.; Melnikov, A.; Yi, Q.L.; Replogle, C.; Mobarhan, S.; Boni, L.; Segato, S.; Levenson, V. DNA Methylation Patterns in Blood of Patients with Colorectal Cancer and Adenomatous Colorectal Polyps. Int. J. Cancer 2012, 131, 1153–1157. [Google Scholar] [CrossRef]
- Chen, C.H.; Yan, S.L.; Yang, T.H.; Chen, S.F.; Yeh, Y.H.; Ou, J.J.; Lin, C.H.; Lee, Y.T.; Chen, C.H. The Relationship between the Methylated Septin-9 DNA Blood Test and Stool Occult Blood Test for Diagnosing Colorectal Cancer in Taiwanese People. J. Clin. Lab. Anal. 2017, 31, e22013. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Z.; Zhao, G.; Sun, C.; Ma, Y.; Zhang, L.; Zheng, M.; Li, H. Performance of a Novel Blood-Based Early Colorectal Cancer Screening Assay in Remaining Serum after the Blood Biochemical Test. Dis. Mrk. 2019, 2019, 5232780. [Google Scholar] [CrossRef]
- Cho, N.-Y.; Park, J.-W.; Wen, X.; Shin, Y.-J.; Kang, J.-K.; Song, S.-H.; Kim, H.-P.; Kim, T.-Y.; Bae, J.M.; Kang, G.H. Blood-Based Detection of Colorectal Cancer Using Cancer-Specific DNA Methylation Markers. Diagnostics 2021, 11, 51. [Google Scholar] [CrossRef]
- Church, T.R.; Wandell, M.; Lofton-Day, C.; Mongin, S.J.; Burger, M.; Payne, S.R.; Castanos-Velez, E.; Blumenstein, B.A.; Rosch, T.; Osborn, N.; et al. Prospective Evaluation of Methylated SEPT9 in Plasma for Detection of Asymptomatic Colorectal Cancer. Gut 2014, 63, 317–325. [Google Scholar] [CrossRef] [PubMed]
- deVos, T.; Tetzner, R.; Model, F.; Weiss, G.; Schuster, M.; Distler, J.; Steiger, K.V.; Grützmann, R.; Pilarsky, C.; Habermann, J.K.; et al. Circulating Methylated SEPT9 DNA in Plasma Is a Biomarker for Colorectal Cancer. Clin. Chem. 2009, 55, 1337–1346. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Yan, P.; Zhang, S.; Lu, Y.; Pan, L.; Tang, W.Q.; Chen, S.; Chen, S.F.; Zhang, A.Q.; Liu, W. Cell-Free Circulating Methylated SEPT9 for Noninvasive Diagnosis and Monitoring of Colorectal Cancer. Dis. Markers 2018, 2018, 6437104. [Google Scholar] [CrossRef] [PubMed]
- Grutzmann, R.; Molnar, B.; Pilarsky, C.; Habermann, J.K.; Schlag, P.M.; Saeger, H.D.; Miehlke, S.; Stolz, T.; Model, F.; Roblick, U.J.; et al. Sensitive Detection of Colorectal Cancer in Peripheral Blood by Septin 9 DNA Methylation Assay. PLoS ONE 2008, 3, e3759. [Google Scholar] [CrossRef]
- He, N.; Song, L.L.; Kang, Q.; Jin, P.; Cai, G.X.; Zhou, J.F.; Zhou, G.P.; Sheng, J.Q.; Cai, S.J.; Wang, J.M.; et al. The Pathological Features of Colorectal Cancer Determine the Detection Performance on Blood CtDNA. Technol. Cancer Res. Treat. 2018, 17, 1533033818791794. [Google Scholar] [CrossRef]
- He, Q.O.; Chen, H.Y.; Bai, E.Q.; Luo, Y.X.; Fu, R.J.; He, Y.S.; Jiang, J.; Wang, H.Q. Development of a Multiplex MethyLight Assay for the Detection of Multigene Methylation in Human Colorectal Cancer. Cancer Genet. Cytogenet. 2010, 202, 1–10. [Google Scholar] [CrossRef]
- Herbst, A.; Rahmig, K.; Stieber, P.; Philipp, A.; Jung, A.; Ofner, A.; Crispin, A.; Neumann, J.; Lamerz, R.; Kolligs, F.T. Methylation of NEUROG1 in Serum Is a Sensitive Marker for the Detection of Early Colorectal Cancer. Am. J. Gastroenterol. 2011, 106, 1110–1118. [Google Scholar] [CrossRef]
- Jensen, S.; Øgaard, N.; Ørntoft, M.W.; Rasmussen, M.H.; Bramsen, J.B.; Kristensen, H.; Mouritzen, P.; Madsen, M.R.; Madsen, A.H.; Sunesen, K.G.; et al. Novel DNA Methylation Biomarkers Show High Sensitivity and Specificity for Blood-Based Detection of Colorectal Cancer-a Clinical Biomarker Discovery and Validation Study. Clin. Epigenetics 2019, 11, 158. [Google Scholar] [CrossRef]
- Jin, P.; Kang, Q.; Wang, X.; Yang, L.; Yu, Y.; Li, N.; He, Y.Q.; Han, X.L.; Hang, J.; Zhang, J.; et al. Performance of a Second-Generation Methylated SEPT9 Test in Detecting Colorectal Neoplasm. J. Gastroenterol. Hepatol. 2015, 30, 830–833. [Google Scholar] [CrossRef]
- Johnson, D.A.; Barclay, R.L.; Mergener, K.; Weiss, G.; König, T.; Beck, J.; Potter, N.T. Plasma Septin9 versus Fecal Immunochemical Testing for Colorectal Cancer Screening: A Prospective Multicenter Study. PLoS ONE 2014, 9, e98238. [Google Scholar] [CrossRef]
- Kang, Q.; Jin, P.; Yang, L.; Wang, X.; An, H.; Liu, L.; Li, N.; Sheng, J. Significance of Septin9 gene methylation detection of plasma circulation DNA in colorectal cancer screening. Zhonghua Yi Xue Za Zhi 2014, 94, 3839–3841. [Google Scholar] [PubMed]
- Karam, R.A.; Zidan, H.E.; Abd Elrahman, T.M.; Badr, S.A.; Amer, S.A. Study of P16 Promoter Methylation in Egyptian Colorectal Cancer Patients. J. Cell. Biochem. 2019, 120, 8581–8587. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, J.K.; Hong, Y.J.; Moon, S.M.; Shin, U.S.; Kwon, H.; Shin, K.; Chang, Y.H. Detection of Methylated SEPT9 in Korean Colorectal Cancer Patients: Comparison with Previous Studies. Clin. Lab. 2018, 64, 1573–1579. [Google Scholar] [CrossRef]
- Lee, H.S.; Hwang, S.M.; Kim, T.S.; Kim, D.W.; Park, D.J.; Kang, S.B.; Kim, H.H.; Park, K.U. Circulating Methylated Septin 9 Nucleic Acid in the Plasma of Patients with Gastrointestinal Cancer in the Stomach and Colon. Transl. Oncol. 2013, 6, 290-U245. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, Z.Z.; Zhao, G.D.; Ma, Y.; Chen, Y.; Xue, Q.; Zheng, M.X.; Fei, S.J. Performance of a MethyLight Assay for Methylated SFRP2 DNA Detection in Colorectal Cancer Tissue and Serum. Int. J. Biol. Mrk. 2019, 34, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.-H.; Xiao, J.; Ye, Z.-Y.; Wei, D.-L.; Zhai, X.-H.; Xu, R.-H.; Zeng, Z.-L.; Luo, H.-Y. Circulating Tumor DNA Methylation Marker MYO1-G for Diagnosis and Monitoring of Colorectal Cancer. Clin. Epigenetics 2021, 13, 232. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tham, C.K.; Ong, S.Y.; Ho, K.S.; Lim, J.F.; Chew, M.H.; Lim, C.K.; Zhao, Y.; Tang, C.L.; Eu, K.W. Serum Methylation Levels of TAC1. SEPT9 and EYA4 as Diagnostic Markers for Early Colorectal Cancers: A Pilot Study. Biomarkers 2013, 18, 399–405. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, G.D.; Miao, J.; Li, H.; Ma, Y.; Liu, X.Y.; Li, S.M.; Zhu, Y.; Xiong, S.M.; Zheng, M.X.; et al. Performance Comparison Between Plasma and Stool Methylated SEPT9 Tests for Detecting Colorectal Cancer. Front. Genet. 2020, 11, 324. [Google Scholar] [CrossRef]
- Liu, Z.; Tang, H.; Zhang, W.; Wang, J.; Wan, L.; Li, X.; Ji, Y.; Kong, N.; Zhang, Y.; Wang, J.; et al. Coupling of Serum CK20 and Hyper-Methylated CLIP4 as Promising Biomarker for Colorectal Cancer Diagnosis: From Bioinformatics Screening to Clinical Validation. Aging 2021, 13, 26161–26179. [Google Scholar] [CrossRef]
- Lofton-Day, C.; Model, F.; Devos, T.; Tetzner, R.; Distler, J.; Schuster, M.; Song, X.; Lesche, R.; Liebenberg, V.; Ebert, M.; et al. DNA Methylation Biomarkers for Blood-Based Colorectal Cancer Screening. Clin. Chem. 2008, 54, 414–423. [Google Scholar] [CrossRef]
- Loomans-Kropp, H.A.; Song, Y.; Gala, M.; Parikh, A.R.; Van Seventer, E.E.; Alvarez, R.; Hitchins, M.P.; Shoemaker, R.H.; Umar, A. Methylated Septin9 (m SEPT9): A Promising Blood-Based Biomarker for the Detection and Screening of Early-Onset Colorectal Cancer. Cancer Res. Commun. 2022, 2, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Zhang, Q.; Li, L.; Luo, X.; Liang, B.; Lu, Y.; Hu, B.; Jiang, H. Methylated Septin9 Has Moderate Diagnostic Value in Colorectal Cancer Detection in Chinese Population: A Multicenter Study. BMC Gastroenterol. 2022, 22, 232. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.Y.; Zhao, Q.; Wei, W.; Zheng, L.H.; Yi, S.H.; Li, G.; Wang, W.Q.; Sheng, H.; Pu, H.Y.; Mo, H.Y.; et al. Circulating Tumor DNA Methylation Profiles Enable Early Diagnosis, Prognosis Prediction, and Screening for Colorectal Cancer. Sci. Transl. Med. 2020, 12, eaax7533. [Google Scholar] [CrossRef] [PubMed]
- Oh, T.; Kim, N.; Moon, Y.; Kim, M.S.; Hoehn, B.D.; Park, C.H.; Kim, T.S.; Kim, N.K.; Chung, H.C.; An, S. Genome-Wide Identification and Validation of a Novel Methylation Biomarker, SDC2, for Blood-Based Detection of Colorectal Cancer. J. Mol. Diagn. 2013, 15, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Otero-Estevez, O.; Gallardo-Gomez, M.; de la Cadena, M.P.; Rodriguez-Berrocal, F.J.; Cubiella, J.; Ramirez, V.H.; Garcia-Nimo, L.; De Chiara, L. Value of Serum NEUROG1 Methylation for the Detection of Advanced Adenomas and Colorectal Cancer. Diagnostics 2020, 10, 437. [Google Scholar] [CrossRef]
- Pasha, H.F.; Radwan, M.I.; Yehia, A.M.; Toam, M.M. Circulating Methylated RUNX3 and SFRP1 Genes as a Noninvasive Panel for Early Detection of Colorectal Cancer. Eur. J. Gastroenterol. Hepatol. 2019, 31, 1342–1349. [Google Scholar] [CrossRef]
- Pedersen, S.K.; Baker, R.T.; McEvoy, A.; Murray, D.H.; Thomas, M.; Molloy, P.L.; Mitchell, S.; Lockett, T.; Young, G.P.; LaPointe, L.C. A Two-Gene Blood Test for Methylated DNA Sensitive for Colorectal Cancer. PLoS ONE 2015, 10, e125041. [Google Scholar] [CrossRef]
- Pedersen, S.K.; Symonds, E.L.; Baker, R.T.; Murray, D.H.; McEvoy, A.; Van Doorn, S.C.; Mundt, M.W.; Cole, S.R.; Gopalsamy, G.; Mangira, D.; et al. Evaluation of an Assay for Methylated BCAT1 and IKZF1 in Plasma for Detection of Colorectal Neoplasia. BMC Cancer 2015, 15, 654. [Google Scholar] [CrossRef]
- Potter, N.T.; Hurban, P.; White, M.N.; Whitlock, K.D.; Lofton-Day, C.E.; Tetzner, R.; Koenig, T.; Quigley, N.B.; Weiss, G. Validation of a Real-Time PCR-Based Qualitative Assay for the Detection of Methylated SEPT9 DNA in Human Plasma. Clin. Chem. 2014, 60, 1183–1191. [Google Scholar] [CrossRef]
- Rasmussen, S.L.; Krarup, H.B.; Sunesen, K.G.; Johansen, M.B.; Stender, M.T.; Pedersen, I.S.; Madsen, P.H.; Thorlacius-Ussing, O. Hypermethylated DNA, a Circulating Biomarker for Colorectal Cancer Detection. PLoS One 2017, 12, e180809. [Google Scholar] [CrossRef]
- Rezvani, N.; Alibakhshi, R.; Vaisi-Raygani, A.; Bashiri, F.; Saidijam, M. Detection of SPG20 Gene Promoter-Methylated DNA, as a Novel Epigenetic Biomarker, in Plasma for Colorectal Cancer Diagnosis Using the MethyLight Method. Oncol. Lett. 2017, 13, 3277–3284. [Google Scholar] [CrossRef] [PubMed]
- Rokni, P.; Shariatpanahi, A.M.; Sakhinia, E.; Kerachian, M.A. BMP3 Promoter Hypermethylation in Plasma-Derived Cell-Free DNA in Colorectal Cancer Patients. Genes Genom. 2018, 40, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, J.; Fujiya, M.; Okamoto, K.; Nata, T.; Inaba, Y.; Moriichi, K.; Tanabe, H.; Mizukami, Y.; Watari, J.; Ashida, T.; et al. Immunoprecipitation of Nucleosomal DNA Is a Novel Procedure to Improve the Sensitivity of Serum Screening for the P16 Hypermethylation Associated with Colon Cancer. Cancer Epidemiol. 2010, 34, 194–199. [Google Scholar] [CrossRef]
- Salehi, R.; Atapour, N.; Vatandoust, N.; Farahani, N.; Ahangari, F.; Salehi, A.R. Methylation Pattern of ALX4 Gene Promoter as a Potential Biomarker for Blood-Based Early Detection of Colorectal Cancer. Adv. Biomed. Res. 2015, 4, 252. [Google Scholar] [CrossRef]
- Song, L.; Wang, J.; Wang, H.; Chen, Y.; Jia, J.; Guo, S.; Liu, H.; Peng, X.; Xiao, W.; Gong, Y.; et al. The Quantitative Profiling of Blood MSEPT9 Determines the Detection Performance on Colorectal Tumors. Epigenomics 2018, 10, 1569–1583. [Google Scholar] [CrossRef] [PubMed]
- Suehiro, Y.; Hashimoto, S.; Higaki, S.; Fujii, I.; Suzuki, C.; Hoshida, T.; Matsumoto, T.; Yamaoka, Y.; Takami, T.; Sakaida, I.; et al. Blood Free-Circulating DNA Testing by Highly Sensitive Methylation Assay to Diagnose Colorectal Neoplasias. Oncotarget 2018, 9, 16974–16987. [Google Scholar] [CrossRef] [PubMed]
- Takane, K.; Midorikawa, Y.; Yagi, K.; Sakai, A.; Aburatani, H.; Takayama, T.; Kaneda, A. Aberrant Promoter Methylation of PPP1R3C and EFHD1 in Plasma of Colorectal Cancer Patients. Cancer Med. 2014, 3, 1235–1245. [Google Scholar] [CrossRef]
- Tang, D.; Liu, J.; Wang, D.R.; Yu, H.F.; Li, Y.K.; Zhang, J.Q. Diagnostic and Prognostic Value of the Methylation Status of Secreted Frizzled-Related Protein 2 in Colorectal Cancer. Clin. Investig. Med. 2011, 34, E88–E95. [Google Scholar] [CrossRef]
- Tanzer, M.; Balluff, B.; Distler, J.; Hale, K.; Leodolter, A.; Rocken, C.; Molnar, B.; Schmid, R.; Lofton-Day, C.; Schuster, T.; et al. Performance of Epigenetic Markers SEPT9 and ALX4 in Plasma for Detection of Colorectal Precancerous Lesions. PLoS ONE 2010, 5, e9061. [Google Scholar] [CrossRef]
- Tóth, K.; Sipos, F.; Kalmár, A.; Patai, A.V.; Wichmann, B.; Stoehr, R.; Golcher, H.; Schellerer, V.; Tulassay, Z.; Molnár, B. Detection of Methylated SEPT9 in Plasma Is a Reliable Screening Method for Both Left- and Right-Sided Colon Cancers. PLoS One 2012, 7, e46000. [Google Scholar] [CrossRef]
- Tóth, K.; Wasserkort, R.; Sipos, F.; Kalmár, A.; Wichmann, B.; Leiszter, K.; Valcz, G.; Juhász, M.; Miheller, P.; Patai, Á.V.; et al. Detection of Methylated Septin 9 in Tissue and Plasma of Colorectal Patients with Neoplasia and the Relationship to the Amount of Circulating Cell-Free DNA. PLoS One 2014, 9, e115415. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Yu, Z.H.; Liu, C.; Xu, L.Z.; Yu, W.; Lu, J.; Zhu, R.M.; Li, G.L.; Xia, X.Y.; Wei, X.W.; et al. Detection of RASSF1A Promoter Hypermethylation in Serum from Gastric and Colorectal Adenocarcinoma Patients. World J. Gastroenterol. 2008, 14, 3074–3080. [Google Scholar] [CrossRef] [PubMed]
- Warren, J.D.; Xiong, W.; Bunker, A.M.; Vaughn, C.P.; Furtado, L.V.; Roberts, W.L.; Fang, J.C.; Samowitz, W.S.; Heichman, K.A. Septin 9 Methylated DNA Is a Sensitive and Specific Blood Test for Colorectal Cancer. BMC Med. 2011, 9, 133. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Zhou, G.P.; Jin, P.; Zhu, J.Q.; Li, S.J.; Wu, Q.; Wang, G.Q.; Sheng, J.Q.; Wang, J.M.; Song, L.L.; et al. Detection of Colorectal Cancer Using a Simplified SEPT9 Gene Methylation Assay Is a Reliable Method for Opportunistic Screening. J. Mol. Diagn. 2016, 18, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.P.; Zou, J.H.; Tang, R.N.; Yao, Y.; You, C.Z. Detection and Clinical Significance of DLC1 Gene Methylation in Serum DNA from Colorectal Cancer Patients. Chin. J. Cancer Res. 2011, 23, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Jiang, X.Y.; Li, Q.; Sun, Z.J.; Quan, W.Q.; Duan, Y.P.; Li, D.; Chen, T.H. Diagnostic Value of Methylated Septin9 for Colorectal Cancer Detection. Front. Oncol. 2018, 8, 247. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Yu, S.; Han, J.; Zong, M.; Tan, Q.; Zeng, X.; Fan, L. Detection of Circulating Tumor DNA Methylation in Diagnosis of Colorectal Cancer. Clin. Transl. Gastroenterol. 2021, 12, e00386. [Google Scholar] [CrossRef]
- Xue, G.; Lu, C.J.; Pan, S.J.; Zhang, Y.L.; Miao, H.; Shan, S.; Zhu, X.T.; Zhang, Y. DNA Hypomethylation of CBS Promoter Induced by Folate Deficiency Is a Potential Noninvasive Circulating Biomarker for Colorectal Adenocarcinomas. Oncotarget 2017, 8, 51387–51401. [Google Scholar] [CrossRef]
- Yang, X.; Xu, Z.J.; Chen, X.; Zeng, S.S.; Qian, L.; Wei, J.; Peng, M.; Wang, X.; Liu, W.L.; Ma, H.Y.; et al. Clinical Value of Preoperative Methylated Septin 9 in Chinese Colorectal Cancer Patients. World J. Gastroenterol. 2019, 25, 2099–2109. [Google Scholar] [CrossRef]
- Young, G.P.; Symonds, E.L.; Nielsen, H.J.; Ferm, L.; Christensen, I.J.; Dekker, E.; van der Vlugt, M.; Mallant-Hent, R.C.; Boulter, N.; Yu, B.; et al. Evaluation of a Panel of Tumor-Specific Differentially-Methylated DNA Regions in IRF4, IKZF1 and BCAT1 for Blood-Based Detection of Colorectal Cancer. Clin. Epigenetics 2021, 13, 14. [Google Scholar] [CrossRef]
- Yuan, P.; Cheng, X.J.; Wu, X.J.; Li, L.; Zhang, L.H.; Li, Z.Y.; Xing, X.F.; Du, H.; Wang, X.H.; Hu, Y.; et al. OSMR and SEPT9: Promising Biomarkers for Detection of Colorectal Cancer Based on Blood-Based Tests. Transl. Cancer Res. 2016, 5, 131–139. [Google Scholar] [CrossRef]
- Zhan, J.; Li, X.; Yu, Z.; Yuan, Y.H.; Hou, J. Value of fecal and blood adenomatous polyposis coli gene and K-ras gene mutation detection in colorectal neoplasm screening. Nan Fang Yi Ke Da Xue Xue Bao J. South. Med. Univ. 2007, 27, 1018–1021. [Google Scholar]
- Zhang, X.; Song, Y.F.; Lu, H.N.; Wang, D.P.; Zhang, X.S.; Huang, S.L.; Sun, B.L.; Huang, Z.G. Combined Detection of Plasma GATA5 and SFRP2 Methylation Is a Valid Noninvasive Biomarker for Colorectal Cancer and Adenomas. World J. Gastroenterol. 2015, 21, 2629–2637. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.D.; Li, H.; Yang, Z.X.; Wang, Z.Z.; Xu, M.Q.; Xiong, S.M.; Li, S.M.; Wu, X.T.; Liu, X.Y.; Wang, Z.W.; et al. Multiplex Methylated DNA Testing in Plasma with High Sensitivity and Specificity for Colorectal Cancer Screening. Cancer Med. 2019, 8, 5619–5628. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.D.; Ma, Y.; Li, H.; Li, S.M.; Zhu, Y.; Liu, X.Y.; Xiong, S.M.; Liu, Y.; Miao, J.; Fei, S.J.; et al. A Novel Plasma Based Early Colorectal Cancer Screening Assay Base on Methylated SDC2 and SFRP2. Clin. Chim. Acta 2020, 503, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Zhang, Y.W.; Huang, X.; Chen, L.B. Analysis of the RUNX3 Gene Methylation in Serum DNA from Esophagus Squamous Cell Carcinoma, Gastric and Colorectal Adenocarcinoma Patients. Hepato-Gastroenterology 2011, 58, 2007–2011. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.Z.; Yu, B.M.; Wang, Z.W.; Sun, J.Y.; Cang, H.; Gao, F.; Li, D.H.; Zhao, R.; Feng, G.G.; Yi, J. Detection of Aberrant P16 Methylation in the Serum of Colorectal Cancer Patients. Clin. Cancer Res. 2002, 8, 188–191. [Google Scholar]
- Nagai, Y.; Sunami, E.; Yamamoto, Y.; Hata, K.; Okada, S.; Murono, K.; Yasuda, K.; Otani, K.; Nishikawa, T.; Tanaka, T.; et al. LINE-1 Hypomethylation Status of Circulating Cell-Free DNA in Plasma as a Biomarker for Colorectal Cancer. Oncotarget 2017, 8, 11906–11916. [Google Scholar] [CrossRef]
- Bresalier, R.S.; Kopetz, S.; Brenner, D.E. Blood-Based Tests for Colorectal Cancer Screening: Do They Threaten the Survival of the FIT Test? Dig. Dis. Sci. 2015, 60, 664–671. [Google Scholar] [CrossRef]
- Ma, M.; Zhu, H.; Zhang, C.; Sun, X.; Gao, X.; Chen, G. “Liquid Biopsy”—CtDNA Detection with Great Potential and Challenges. Ann. Transl. Med. 2015, 3, 235. [Google Scholar] [CrossRef]
- Connolly, D.; Abdesselam, I.; Verdier-Pinard, P.; Montagna, C. Septin Roles in Tumorigenesis. Biol. Chem. 2011, 392, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Østevold, K.; Meléndez, A.V.; Lehmann, F.; Schmidt, G.; Aktories, K.; Schwan, C. Septin Remodeling Is Essential for the Formation of Cell Membrane Protrusions (Microtentacles) in Detached Tumor Cells. Oncotarget 2017, 8, 76686–76698. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, P.-M.; Liu, R.-B. Advance in Plasma SEPT9 Gene Methylation Assay for Colorectal Cancer Early Detection. World J. Gastrointest. Oncol. 2018, 10, 15–22. [Google Scholar] [CrossRef] [PubMed]




| Group | SEN (95% CI) | SPE (95% CI) | PLR (95% CI) | NLR (95% CI) | DOR (95% CI) | AUC |
|---|---|---|---|---|---|---|
| Quantitative analysis of ctDNA | 0.723 (0.623–0.803) | 0.920 (0.827–0.966) | 6.820 (3.335–12.800) | 0.326 (0.227–0.446) | 23.305 (9.378–57.906) | 0.860 |
| Qualitative analysis of ctDNA | 0.609 (0.563–0.650) | 0.891 (0.864–0.910) | 4.960 (4.190–5.860) | 0.453 (0.406–0.498) | 12.621 (10.004–15.922) | 0.828 |
| SEPT9 methylation assay | 0.679 (0.622–0.732) | 0.904 (0.881–0.923) | 6.420 (5.110–7.970) | 0.367 (0.307–0.430) | 20.551 (14.684–28.760) | 0.883 |
| Analysis | Group | Subgroup | SEN (95% CI) | SPE (95% CI) | DOR (95% CI) | AUC |
|---|---|---|---|---|---|---|
| Quantitative analysis | Control type | HC | 0.723 (0.626–0.803) | 0.927 (0.833–0.967) | 24.751 (9.704–63.129) | 0.861 |
| NCD | 0.647 (0.508–0.765) | 0.878 (0.563–0.976) | 13.444 (1.37–131.721) | 0.641 | ||
| Sample size | ≥100 | 0.771 (0.681–0.842) | 0.924 (0.815–0.971) | 32.219 (10.540–98.491) | 0.878 | |
| <100 | 0.545 (0.417–0.667) | 0.904 (0.604–0.983) | 6.753 (2.534–17.998) | 0.694 | ||
| Sample source | Plasma | 0.747 (0.584–0.861) | 0.908 (0.735–0.972) | 22.297 (4.819–103.165) | 0.868 | |
| Serum | 0.701 (0.587–0.795) | 0.929 (0.793–0.978) | 25.591 (7.815–83.805) | 0.841 | ||
| Assay method | RT-PCR | 0.782 (0.680–0.853) | 0.907 (0.767–0.967) | 28.510 (7.995–101.662) | 0.874 | |
| Other | 0.542 (0.445–0.635) | 0.942 (0.786–0.986) | 16.431 (4.418–61.105) | 0.663 | ||
| Qualitative analysis | Region | Asia | 0.623 (0.574–0.669) | 0.906 (0.886–0.922) | 15.968 (11.843–21.529) | 0.862 |
| Other | 0.594 (0.519–0.665) | 0.869 (0.823–0.904) | 9.672 (6.808–13.741) | 0.804 | ||
| Control type | HC | 0.601 (0.554–0.647) | 0.915 (0.895–0.932) | 15.1337 (11.413–20.067) | 0.844 | |
| NCD | 0.672 (0.617–0.723) | 0.809 (0.737–0.865) | 9.188 (6.525–12.934) | 0.774 | ||
| Sample size | >=100 | 0.598 (0.552–0.643) | 0.903 (0.884–0.919) | 13.760 (10.676–17.734) | 0.849 | |
| <100 | 0.662 (0.544–0.762) | 0.819 (0.694–0.900) | 7.445 (4.461–12.425) | 0.759 | ||
| Sample source | Plasma | 0.620 (0.570–0.667) | 0.884 (0.859–0.905) | 12.547 (9.745–16.155) | 0.831 | |
| Serum | 0.563 (0.477–0.645) | 0.920 (0.873–0.951) | 13.108 (7.187–23.906) | 0.817 | ||
| Assay method | MSP | 0.594 (0.551–0.636) | 0.901 (0.882–0.917) | 13.031 (10.216–16.623) | 0.838 | |
| Other | 0.781 (0.611–0.890) | 0.685 (0.482–0.836) | 8.531 (3.834–18.981) | 0.800 | ||
| Methylation gene location | SEPT-9 | 0.679 (0.622–0.732) | 0.904 (0.881–0.923) | 20.551 (14.684–28.760) | 0.883 | |
| Other | 0.577 (0.522–0.631) | 0.885 (0.854–0.910) | 10.297 (7.750–13.681) | 0.802 | ||
| SEPT9 methylation Assay | Region | Asia | 0.679 (0.616–0.737) | 0.922 (0.902–0.938) | 26.096 (17.252–39.471) | 0.904 |
| Other | 0.684 (0.572–0.777) | 0.874 (0.824–0.917) | 14.379 (8.808–23.472) | 0.867 | ||
| Control type | HC | 0.676 (0.609–0.737) | 0.927 (0.900–0.947) | 25.800 (15.869–41.945) | 0.887 | |
| NCD | 0.723 (0.650–0.783) | 0.859 (0.803–0.900) | 17.164 (11.237–26.218) | 0.854 | ||
| Sample size | ≥100 | 0.688 (0.629–0.741) | 0.908 (0.884–0.927) | 21.949 (15.393–31.298) | 0.888 | |
| <100 | 0.603 (0.403–0.773) | 0.832 (0.743–0.894) | 10.289 (3.742–28.293) | 0.847 | ||
| Sample source | Plasma | 0.693 (0.631–0.748) | 0.906 (0.883–0.925) | 22.488 (16.257–31.105) | 0.893 | |
| Serum | 0.575 (0.467–0.676) | 0.880(0.750–0.947) | 9.253 (2.297–37.269) | 0.735 |
| Analysis | Covariates | Coefficient | SE | p-Value |
|---|---|---|---|---|
| Quantitative analysis | Region | 0.229 | 1.107 | 0.836 |
| Control type | 0.607 | 1.268 | 0.632 | |
| Sample size | 1.467 | 1.333 | 0.271 | |
| Sample source | −0.544 | 1.057 | 0.606 | |
| Assay method | 0.038 | 1.163 | 0.974 | |
| Qualitative analysis | Region | 0.422 | 0.255 | 0.067 |
| Control type | 0.502 | 0.237 | 0.034 | |
| Sample size | 0.504 | 0.357 | 0.158 | |
| Sample source | −0.192 | 0.313 | 0.541 | |
| Assay method | −0.122 | 0.482 | 0.800 | |
| Methylation gene location | 0.661 | 0.255 | 0.010 | |
| SEPT9 | Region | 0.676 | 0.322 | 0.045 |
| Control type | 0.733 | 0.348 | 0.035 | |
| Sample size | 0.171 | 0.637 | 0.788 | |
| Sample source | 0.989 | 0.638 | 0.079 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Min, L.; Chen, J.; Yu, M.; Liu, D. Using Circulating Tumor DNA as a Novel Biomarker to Screen and Diagnose Colorectal Cancer: A Meta-Analysis. J. Clin. Med. 2023, 12, 408. https://doi.org/10.3390/jcm12020408
Min L, Chen J, Yu M, Liu D. Using Circulating Tumor DNA as a Novel Biomarker to Screen and Diagnose Colorectal Cancer: A Meta-Analysis. Journal of Clinical Medicine. 2023; 12(2):408. https://doi.org/10.3390/jcm12020408
Chicago/Turabian StyleMin, Liang, Jinghua Chen, Meihong Yu, and Deliang Liu. 2023. "Using Circulating Tumor DNA as a Novel Biomarker to Screen and Diagnose Colorectal Cancer: A Meta-Analysis" Journal of Clinical Medicine 12, no. 2: 408. https://doi.org/10.3390/jcm12020408
APA StyleMin, L., Chen, J., Yu, M., & Liu, D. (2023). Using Circulating Tumor DNA as a Novel Biomarker to Screen and Diagnose Colorectal Cancer: A Meta-Analysis. Journal of Clinical Medicine, 12(2), 408. https://doi.org/10.3390/jcm12020408






