Metabolomic Approach to Screening Homozygotes in Chinese Patients with Severe Familial Hypercholesterolemia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Sample and Clinical Data Collection
2.3. Sample Preparation for Metabolomic Analysis
2.4. Metabolomic Analysis and Data Processing
2.5. Statistical Analysis
3. Results
3.1. Subject Characteristics
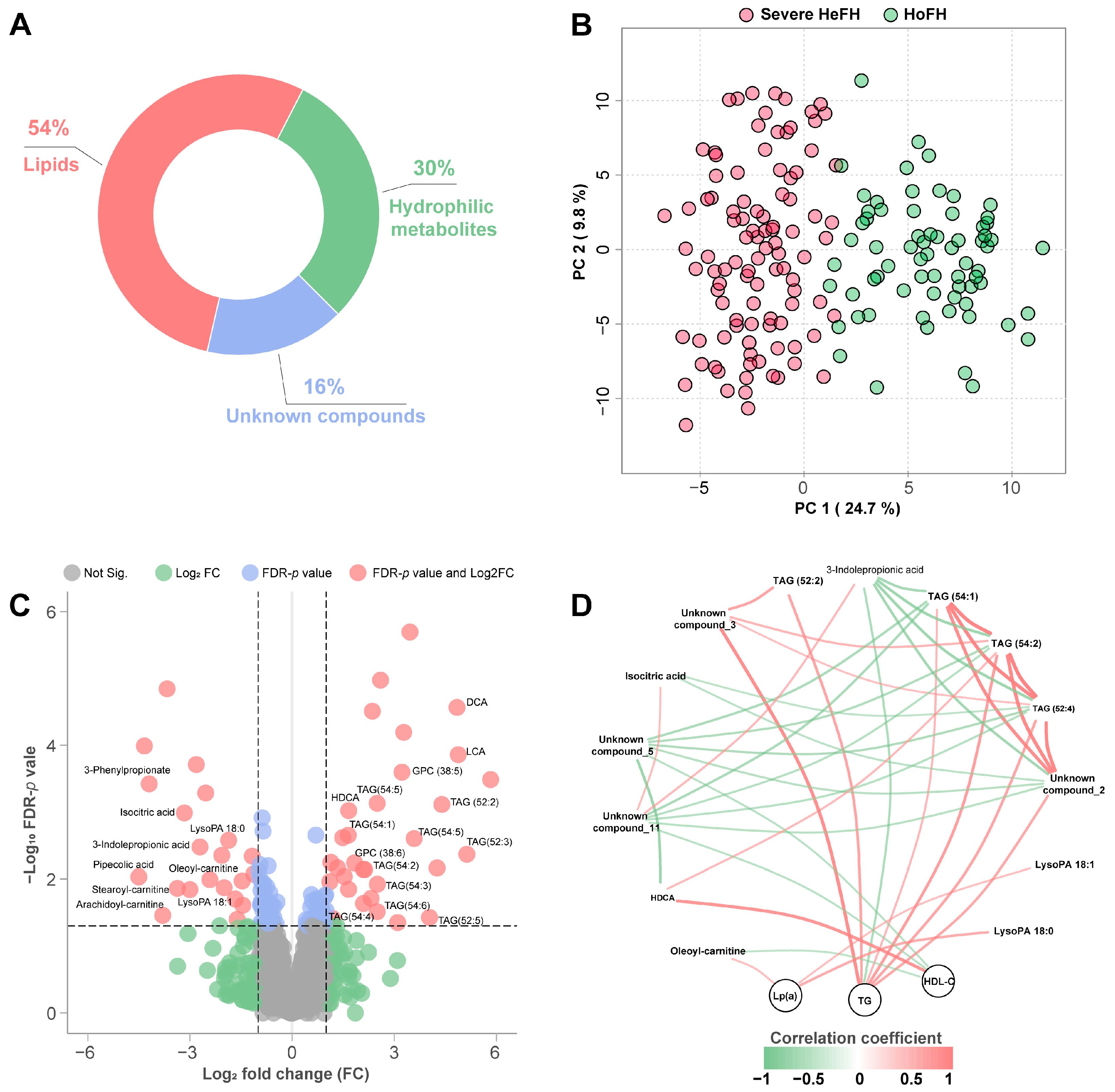
3.2. Sera Metabolome Profiles of HoFH and Severe HeFH
3.3. Specific Biosignatures for Differentiating HoFH from Severe HeFH
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Watts, G.F.; Gidding, S.S.; Mata, P.; Pang, J.; Sullivan, D.R.; Yamashita, S.; Raal, F.J.; Santos, R.D.; Ray, K.K. Familial hypercholesterolaemia: Evolving knowledge for designing adaptive models of care. Nat. Rev. Cardiol. 2020, 17, 360–377. [Google Scholar] [CrossRef] [PubMed]
- Argmann, C.A.; Houten, S.M.; Zhu, J.; Schadt, E.E. A next generation multiscale view of inborn errors of metabolism. Cell Metab. 2016, 23, 13–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reiner, Z. Management of patients with familial hypercholesterolaemia. Nat. Rev. Cardiol. 2015, 12, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Defesche, J.C.; Gidding, S.S.; Harada-Shiba, M.; Hegele, R.A.; Santos, R.D.; Wierzbicki, A.S. Familial hypercholesterolaemia. Nat. Rev. Dis. Primers 2017, 3, 17093. [Google Scholar] [CrossRef]
- Onorato, A.; Sturm, A.C. Heterozygous familial hypercholesterolemia. Circulation 2016, 133, e587–e589. [Google Scholar] [CrossRef] [Green Version]
- Beheshti, S.O.; Madsen, C.M.; Varbo, A.; Nordestgaard, B.G. Worldwide prevalence of familial hypercholesterolemia: Meta-analyses of 11 million subjects. J. Am. Coll. Cardiol. 2020, 75, 2553–2566. [Google Scholar] [CrossRef]
- Tromp, T.R.; Hartgers, M.L.; Hovingh, G.K.; Vallejo-Vaz, A.J.; Ray, K.K.; Soran, H.; Freiberger, T.; Bertolini, S.; Harada-Shiba, M.; Blom, D.J.; et al. Worldwide experience of homozygous familial hypercholesterolaemia: Retrospective cohort study. Lancet 2022, 399, 719–728. [Google Scholar] [CrossRef]
- Chen, P.; Chen, X.; Zhang, S. Current status of familial hypercholesterolemia in China: A need for patient FH registry systems. Front. Physiol. 2019, 10, 280. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Stoekenbroek, R.M.; Zhang, F.; Wang, Q.; Yu, W.; Yuan, H.; Cai, G.; Chen, Y.; Li, G.; Yang, Y.; et al. Homozygous familial hypercholesterolemia in China: Genetic and clinical characteristics from a real-world, multi-center, cohort study. J. Clin. Lipidol. 2022, 16, 306–314. [Google Scholar] [CrossRef]
- Blom, D.J.; Harada-Shiba, M.; Rubba, P.; Gaudet, D.; Kastelein, J.; Charng, M.J.; Pordy, R.; Donahue, S.; Ali, S.; Dong, Y.; et al. Efficacy and safety of alirocumab in adults with homozygous familial hypercholesterolemia: The ODYSSEY HoFH trial. J. Am. Coll. Cardiol. 2020, 76, 131–142. [Google Scholar] [CrossRef]
- Raal, F.J.; Rosenson, R.S.; Reeskamp, L.F.; Hovingh, G.K.; Kastelein, J.; Rubba, P.; Ali, S.; Banerjee, P.; Chan, K.C.; Gipe, D.A.; et al. Evinacumab for homozygous familial hypercholesterolemia. N. Engl. J. Med. 2020, 383, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Cuchel, M.; Bruckert, E.; Ginsberg, H.N.; Raal, F.J.; Santos, R.D.; Hegele, R.A.; Kuivenhoven, J.A.; Nordestgaard, B.G.; Descamps, O.S.; Steinhagen-Thiessen, E.; et al. Homozygous familial hypercholesterolaemia: New insights and guidance for clinicians to improve detection and clinical management. A position paper from the consensus panel on familial hypercholesterolaemia of the European atherosclerosis society. Eur. Heart J. 2014, 35, 2146–2157. [Google Scholar] [CrossRef] [PubMed]
- Sturm, A.C.; Truty, R.; Callis, T.E.; Aguilar, S.; Esplin, E.D.; Garcia, S.; Haverfield, E.V.; Morales, A.; Nussbaum, R.L.; Rojahn, S.; et al. Limited-variant screening vs comprehensive genetic testing for familial hypercholesterolemia diagnosis. JAMA Cardiol. 2021, 6, 902–909. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Li, F.; Li, L.; Wang, Y.; Li, J.; Yang, Y.; Jiang, L.; Wang, L.; Qin, Y. Low-density lipoprotein receptor genotypes modify the sera metabolome of patients with homozygous familial hypercholesterolemia. iScience 2022, 25, 105334. [Google Scholar] [CrossRef]
- Brandts, J.; Ray, K.K. Familial hypercholesterolemia: JACC focus seminar 4/4. J. Am. Coll. Cardiol. 2021, 78, 1831–1843. [Google Scholar] [CrossRef]
- Luirink, I.K.; Braamskamp, M.; Wiegman, A.; Hartgers, M.L.; Sjouke, B.; Defesche, J.C.; Hovingh, G.K. The clinical and molecular diversity of homozygous familial hypercholesterolemia in children: Results from the Genetics of clinical homozygous hypercholesterolemia (GoTCHA) study. J. Clin. Lipidol. 2019, 13, 272–278. [Google Scholar] [CrossRef]
- Liu, N.; Xiao, J.; Gijavanekar, C.; Pappan, K.L.; Glinton, K.E.; Shayota, B.J.; Kennedy, A.D.; Sun, Q.; Sutton, V.R.; Elsea, S.H. Comparison of untargeted metabolomic profiling vs traditional metabolic screening to identify inborn errors of metabolism. JAMA Netw. Open 2021, 4, e2114155. [Google Scholar] [CrossRef]
- Ismail, I.T.; Showalter, M.R.; Fiehn, O. Inborn errors of metabolism in the era of untargeted metabolomics and lipidomics. Metabolites 2019, 9, 242. [Google Scholar] [CrossRef] [Green Version]
- Haijes, H.A.; van der Ham, M.; Prinsen, H.; Broeks, M.H.; van Hasselt, P.M.; de Sain-van, D.V.M.; Verhoeven-Duif, N.M.; Jans, J. Untargeted metabolomics for metabolic diagnostic screening with automated data interpretation using a knowledge-based algorithm. Int. J. Mol. Sci. 2020, 21, 979. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Jiang, L.; Zhang, H.; Cheng, S.; Wen, W.; Xu, L.; Zhang, F.; Yang, Y.; Wang, L.; Chen, J. Integrated analysis of microRNA and mRNA expression profiles in homozygous familial hypercholesterolemia patients and validation of atherosclerosis associated critical regulatory network. Genomics 2021, 113, 2572–2582. [Google Scholar] [CrossRef]
- Jiang, L.; Benito-Vicente, A.; Tang, L.; Etxebarria, A.; Cui, W.; Uribe, K.B.; Pan, X.D.; Ostolaza, H.; Yang, S.W.; Zhou, Y.J.; et al. Analysis of LDLR variants from homozygous FH patients carrying multiple mutations in the LDLR gene. Atherosclerosis 2017, 263, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Lu, Y.; Sun, J.; Chang, K.; Lu, M.; Fang, M.; Zeng, X.; Zhang, W.; Song, J.; Guo, X.; et al. Pharmacokinetics/pharmacometabolomics-pharmacodynamics reveals the synergistic mechanism of a multicomponent herbal formula, Baoyuan decoction against cardiac hypertrophy. Biomed. Pharmacother. 2021, 139, 111665. [Google Scholar] [CrossRef] [PubMed]
- Sjouke, B.; Kusters, D.M.; Kindt, I.; Besseling, J.; Defesche, J.C.; Sijbrands, E.J.; Roeters, V.L.J.; Stalenhoef, A.F.; Wiegman, A.; de Graaf, J.; et al. Homozygous autosomal dominant hypercholesterolaemia in the Netherlands: Prevalence, genotype-phenotype relationship, and clinical outcome. Eur. Heart J. 2015, 36, 560–565. [Google Scholar] [CrossRef] [Green Version]
- Xiang, R.; Fan, L.L.; Lin, M.J.; Li, J.J.; Shi, X.Y.; Jin, J.Y.; Liu, Y.X.; Chen, Y.Q.; Xia, K.; Zhao, S.P. The genetic spectrum of familial hypercholesterolemia in the central south region of China. Atherosclerosis 2017, 258, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Perez, D.I.L.; Charriere, S.; Vimont, A.; Alonso, R.; Muniz-Grijalvo, O.; Diaz-Diaz, J.L.; Zambon, D.; Moulin, P.; Bruckert, E.; et al. The added value of coronary calcium score in predicting cardiovascular events in familial hypercholesterolemia. JACC Cardiovasc. Imaging 2021, 14, 2414–2424. [Google Scholar] [CrossRef]
- Tomlinson, B.; Hu, M.; Chow, E. Current status of familial hypercholesterolemia in Chinese populations. Curr. Opin. Lipidol. 2019, 30, 94–100. [Google Scholar] [CrossRef]
- Ford, L.; Kennedy, A.D.; Goodman, K.D.; Pappan, K.L.; Evans, A.M.; Miller, L.; Wulff, J.E.; Wiggs, B.R.; Lennon, J.J.; Elsea, S.; et al. Precision of a clinical metabolomics profiling platform for use in the identification of inborn errors of metabolism. J. Appl. Lab. Med. 2020, 5, 342–356. [Google Scholar] [CrossRef]
- Tebani, A.; Abily-Donval, L.; Afonso, C.; Marret, S.; Bekri, S. Clinical Metabolomics: The new metabolic window for inborn errors of metabolism investigations in the post-genomic era. Int. J. Mol. Sci. 2016, 17, 1167. [Google Scholar] [CrossRef] [Green Version]
- Santos, R.D.; Gidding, S.S.; Hegele, R.A.; Cuchel, M.A.; Barter, P.J.; Watts, G.F.; Baum, S.J.; Catapano, A.L.; Chapman, M.J.; Defesche, J.C.; et al. Defining severe familial hypercholesterolaemia and the implications for clinical management: A consensus statement from the International Atherosclerosis Society Severe Familial Hypercholesterolemia Panel. Lancet Diabetes Endocrinol. 2016, 4, 850–861. [Google Scholar] [CrossRef]
- Zhang, X.; Coker, O.O.; Chu, E.S.; Fu, K.; Lau, H.; Wang, Y.X.; Chan, A.; Wei, H.; Yang, X.; Sung, J.; et al. Dietary cholesterol drives fatty liver-associated liver cancer by modulating gut microbiota and metabolites. Gut 2021, 70, 761–774. [Google Scholar] [CrossRef]
- Molinaro, A.; Wahlstrom, A.; Marschall, H.U. Role of bile acids in metabolic control. Trends Endocrinol. Metab. 2018, 29, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ding, W.X.; Li, T. Cholesterol and bile acid-mediated regulation of autophagy in fatty liver diseases and atherosclerosis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Pellicciari, R.; Pruzanski, M.; Auwerx, J.; Schoonjans, K. Targeting bile-acid signalling for metabolic diseases. Nat. Rev. Drug Discov. 2008, 7, 678–693. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Soffer, G.; Westerterp, M. Beyond Lipoprotein(a) plasma measurements: Lipoprotein(a) and inflammation. Pharmacol. Res. 2021, 169, 105689. [Google Scholar] [CrossRef]
- Zhou, Y.; Little, P.J.; Ta, H.T.; Xu, S.; Kamato, D. Lysophosphatidic acid and its receptors: Pharmacology and therapeutic potential in atherosclerosis and vascular disease. Pharmacol. Ther. 2019, 204, 107404. [Google Scholar] [CrossRef] [PubMed]

| HoFH (n = 69) | Severe HeFH (n = 101) | p Values | |
|---|---|---|---|
| Ages | 23.4 ± 15.2 | 27.1 ± 12.1 | 0.076 |
| Male sex, n (%) | 35, (50.72%) | 56, (55.45%) | 0.54 |
| Hypertension, n (%) | 2, (2.90%) | 7, (6.93%) | 0.25 |
| Diabetes mellitus, n (%) | 0, (0.0%) | 4, (3.96%) | 0.094 |
| Current smokers, n (%) | 1, (1.45%) | 8, (7.92%) | 0.064 |
| ASCVD history, n (%) | 5, (7.25%) | 2, (1.98%) | 0.089 |
| Non-LLT, n (%) | 6, (8.70%) | 11, (10.89%) | 0.64 |
| Statins alone, n (%) | 38, (55.07%) | 65, (64.36%) | 0.22 |
| Statins and ezetimibe, n (%) | 25, (36.23%) | 27, (26.37%) | 0.19 |
| LDL-C, mmol/L | 9.39 ± 1.83 | 8.90 ± 1.95 | 0.091 |
| TC, mmol/L | 11.88 ± 2.27 | 11.35 ± 2.21 | 0.14 |
| TG, mmol/L | 0.84 [0.60, 1.27] | 1.33 [0.87, 1.87] | 0.0041 |
| HDL-C, mmol/L | 1.04 ± 0.37 | 1.26 ± 0.29 | 0.014 |
| Non-HDL, mmol/L | 10.86 ± 2.27 | 10.09 ± 2.24 | 0.032 |
| LP(a), mg/dL | 41.9 [23.3, 56.5] | 15.6 [6.6, 19.6] | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, Z.; Du, Y.; Li, L.; Sun, H.; Hu, C.; Jiang, L.; Wang, L.; Qin, Y. Metabolomic Approach to Screening Homozygotes in Chinese Patients with Severe Familial Hypercholesterolemia. J. Clin. Med. 2023, 12, 483. https://doi.org/10.3390/jcm12020483
Du Z, Du Y, Li L, Sun H, Hu C, Jiang L, Wang L, Qin Y. Metabolomic Approach to Screening Homozygotes in Chinese Patients with Severe Familial Hypercholesterolemia. Journal of Clinical Medicine. 2023; 12(2):483. https://doi.org/10.3390/jcm12020483
Chicago/Turabian StyleDu, Zhiyong, Yunhui Du, Linyi Li, Haili Sun, Chaowei Hu, Long Jiang, Luya Wang, and Yanwen Qin. 2023. "Metabolomic Approach to Screening Homozygotes in Chinese Patients with Severe Familial Hypercholesterolemia" Journal of Clinical Medicine 12, no. 2: 483. https://doi.org/10.3390/jcm12020483






