DiabeticSense: A Non-Invasive, Multi-Sensor, IoT-Based Pre-Diagnostic System for Diabetes Detection Using Breath
Abstract
:1. Introduction
2. Background
3. Materials and Methods
3.1. Study Aim
3.2. Design
3.3. Details of Sensors Used
- TGS 826: The sensing element of TGS 826 is a metal–oxide–semiconductor that has low conductivity in clean air. In the presence of a detectable gas, the sensor’s conductivity increases depending on the gas concentration in the air. A simple electrical circuit can convert the change in conductivity to an output signal corresponding to the gas concentration. The TGS 826 has sensitivity to VOCs such as iso-butane, ethanol, ammonia, and hydrogen gas. The sensor can detect concentrations as low as 30 ppm in the air and is ideally suited to critical safety-related applications such as the detection of ammonia leaks in refrigeration systems and ammonia detection in the agricultural field [25].
- TGS 2610: TGS 2610 is a semiconductor-type gas sensor that combines very high sensitivity to Liquefied petroleum (LP) gas with low power consumption and long life. Due to the miniaturization of its sensing chip, TGS 2610 requires a heater current of only 56 mA and the device is housed in a standard TO-5 package. The TGS 2610 is available in two different models with different external housings but identical sensitivity to LP gas. Both models can satisfy the requirements of performance standards such as UL1484 and EN50194. TGS 2610-C00 possesses a small size and quick gas response, making it suitable for gas leakage checkers. TGS 2610-D00 uses filter material in its housing, eliminating the influence of interference gasses such as alcohol, resulting in a highly selective response to LP gas. This feature makes the sensor ideal for residential gas leakage detectors, which require durability and resistance against interference gas [26]. TGS 2610 shows sensitivity to ethanol, hydrogen, methane, iso-butane, and propane gas.
- TGS 822: The sensing element of TGS 822 Figaro gas sensors is a tin dioxide (Sn) semiconductor with low conductivity in clean air. In the presence of a detectable gas, the sensor’s conductivity increases depending on the gas concentration in the air. A simple electrical circuit can convert the change in conductivity to an output signal corresponding to the gas concentration. The TGS 822 is highly sensitive to the vapors of organic solvents and other volatile vapors. It is also sensitive to combustible gasses such as carbon monoxide, making it an excellent general-purpose sensor. It is also available with a ceramic base highly resistant to severe environments as high as 200 °C (in TGS 823). The complete list of gases that TGS 822 is sensitive to includes methane, carbon monoxide, iso-butane, n-hexane, benzene, ethanol, and acetone [27].
- TGS 2602: The sensing element consists of a metal–oxide–semiconductor layer formed on the alumina substrate of a sensing chip together with an integrated heater. The TGS 2602 is highly sensitive to low concentrations of odorous gasses such as hydrogen, hydrogen sulfide, and ammonia generated from waste materials in office and home environments. The sensor is also susceptible to low concentrations of VOCs, such as toluene and ethanol emitted from wood finishing and construction products. Due to the miniaturization of the sensing chip, TGS 2602 requires a heater current of only 56 mA and the device is housed in a standard TO-5 package [28].
- TGS 2600: This sensor is highly sensitive to low concentrations of gaseous air contaminants in cigarette smoke, such as hydrogen, methane, and carbon monoxide, and also shows sensitivity to iso-butane and ethanol. In the presence of a detectable gas, the sensor’s conductivity increases depending on the gas concentration in the air. The sensor can detect hydrogen at a level of several ppm. Due to the miniaturization of the sensing chip, TGS 2600 requires a heater current of only 42 mA and the device is housed in a standard TO-5 package [29].
- TGS 2603: The sensing element consists of a metal–oxide–semiconductor layer formed on an alumina substrate of a sensing chip together with an integrated heater. In the presence of a detectable gas, the sensor’s conductivity increases depending on the gas concentration in the air. The TGS 2603 is highly sensitive to low concentrations of odorous gasses such as amine-series and sulfurous odor generated from waste materials or spoiled foods such as fish, such as methyl mercaptan and trimethyl amine, and is also sensitive to hydrogen sulfide, hydrogen, and ethanol. By utilising the change ratio of sensor resistance from the resistance in clean air as the relative response, human perception of air contaminants can be simulated and practical air quality control can be achieved [30].
- TGS 2620: The sensing element consists of a metal–oxide–semiconductor layer formed on an alumina substrate of a sensing chip together with an integrated heater. In the presence of a detectable gas, the sensor’s conductivity increases depending on the gas concentration in the air. The TGS 2620 is highly sensitive to the vapors of organic solvents and other volatile vapors, making it suitable for organic vapor detectors/alarms. The complete list of VOCs TGS 2620 senses includes methane, carbon monoxide, iso-butane, hydrogen, and ethanol. Due to the sensing chip’s miniaturization, TGS 2620 requires a heater current of only 42mA and the device is housed in a standard TO-5 package [31].
- MQ 138: The sensor measures the change in conductivity of a tin dioxide SnO semiconductor when exposed to VOCs. In clean air, SnO has low conductivity. However, when VOCs are present, they react with the SnO and increase its conductivity. The change in conductivity can be measured as a voltage change, which can then be used to determine the concentration of VOCs in the air. The MQ138 sensor is sensitive to various VOCs, including formaldehyde, benzene, toluene, and acetone. It has a working range of 1 to 100 ppm for benzene [32].
- DHT 22: DHT22 is a commonly used temperature and humidity sensor. The sensor has a dedicated NTC thermistor to measure temperature and an 8-bit microcontroller to output temperature and humidity values as serial data. The sensor can measure temperature from −40 °C to 80 °C and humidity from 0% to 100% with an accuracy of ±1 °C and ±1% [33].
3.4. Methodology
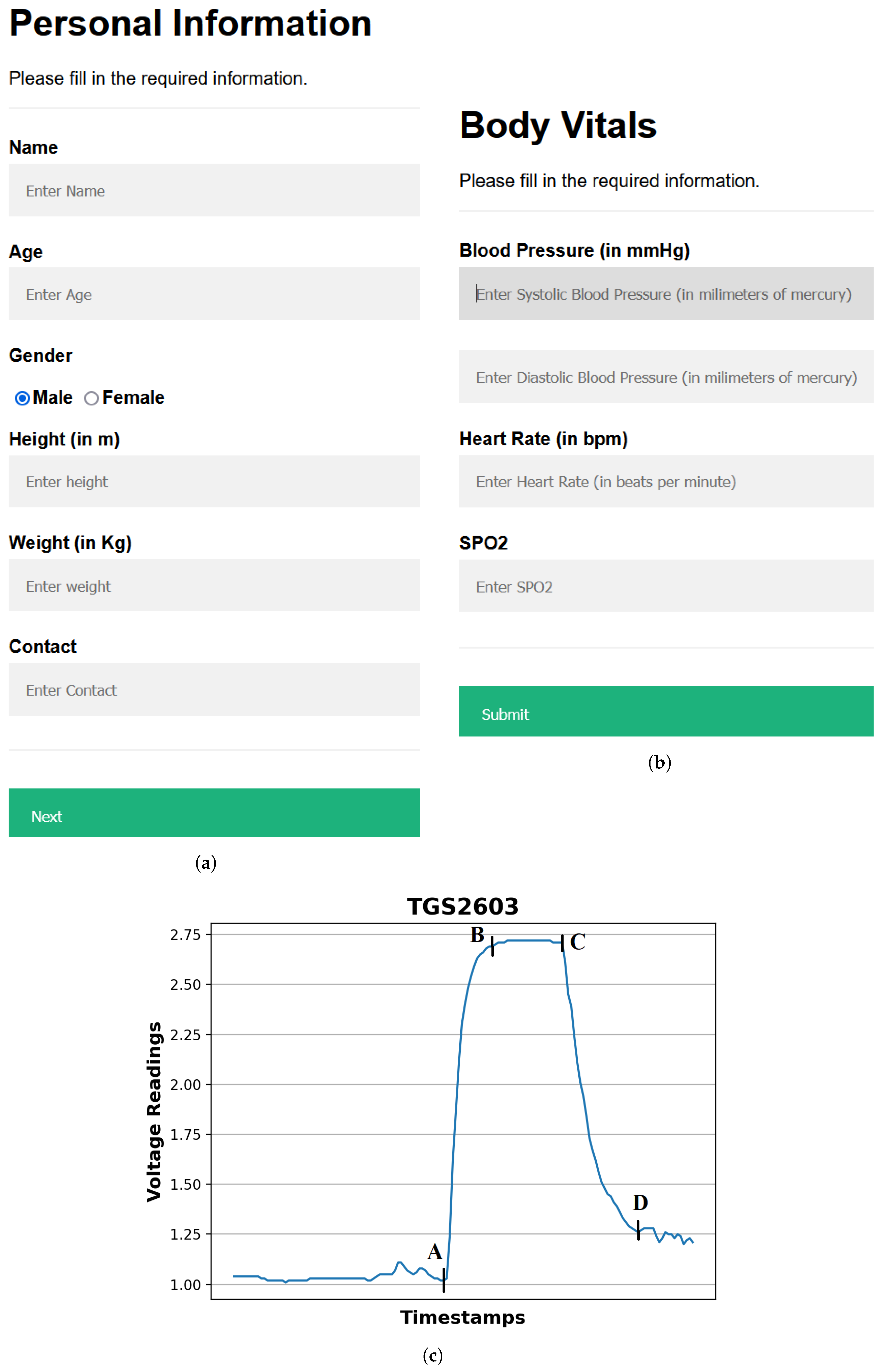
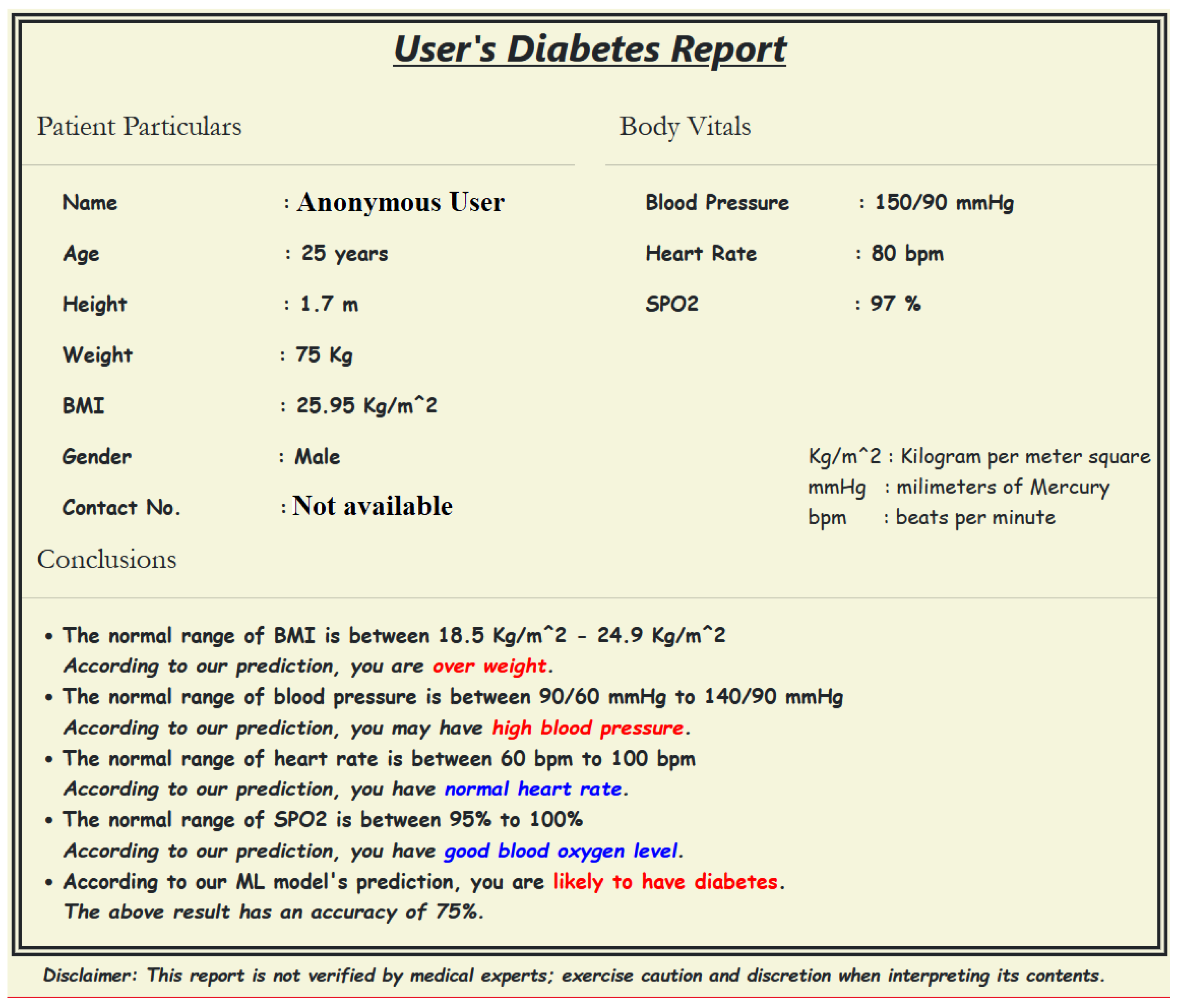
- Providing input details using a web-based interface: The procedure starts by entering the user’s demographic and body vitals information using a web-based interface (Figure 3a,b). The demographics include name, age, gender, height, and weight. To record the body vitals of a user, we make them sit in a stable position, rest for five minutes (to make their vitals stable if they have performed some physical activity), and then record their blood pressure, heart rate, and blood oxygen level using standard digital health devices available in the market. These measures can also be self-recorded by a user using digital health devices or a smartwatch and can be entered into the web-based interface. Note: the age recorded in the dataset is when they were detected with diabetes. Furthermore, since most of the T2DM cases occur at ages above 25 years, we have currently collected data from patients with an age of more than 25 years [34].
- Calibrating the sensors: To ensure accurate sensor readings, we calibrate the sensors to establish stable baselines by validating their readings under reference conditions using fresh air. This implies that the sensors were exposed to a known concentration of VOCs and their outputs were recorded. These data can then be used to create a calibration that can be used to correct the sensor readings for any variation. To obtain a stable baseline from the sensor output, the sensors were preheated with a microheater of the gas sensor. Once the sensor’s output stabilizes under fresh-air conditions, breath sample signatures are obtained from the sensor array (details of sensors’ calibration experiments are described in Section 3.5).
- Preheat the sensors: The sensors’ temperature increases to a relatively stable level during use, resulting in a change in the baseline response of the sensors. Therefore, the device was switched on for approximately 20 min until the baseline response shown on the host computer was stable.
- Regular weekly calibration of the sensors: In addition to the initial calibration, we also performed regular calibrations every two weeks to reduce the time drift. This is carried out by exposing the sensors to a set of 10 healthy or non-diabetic breath samples and 3 diabetic breath samples, and verifying that the sensor voltages’ range differs for diabetic participants in comparison to the non-diabetic participants. The results of the regular calibrations were used to update the calibration curve, which ensures that the sensor readings remain accurate over time.
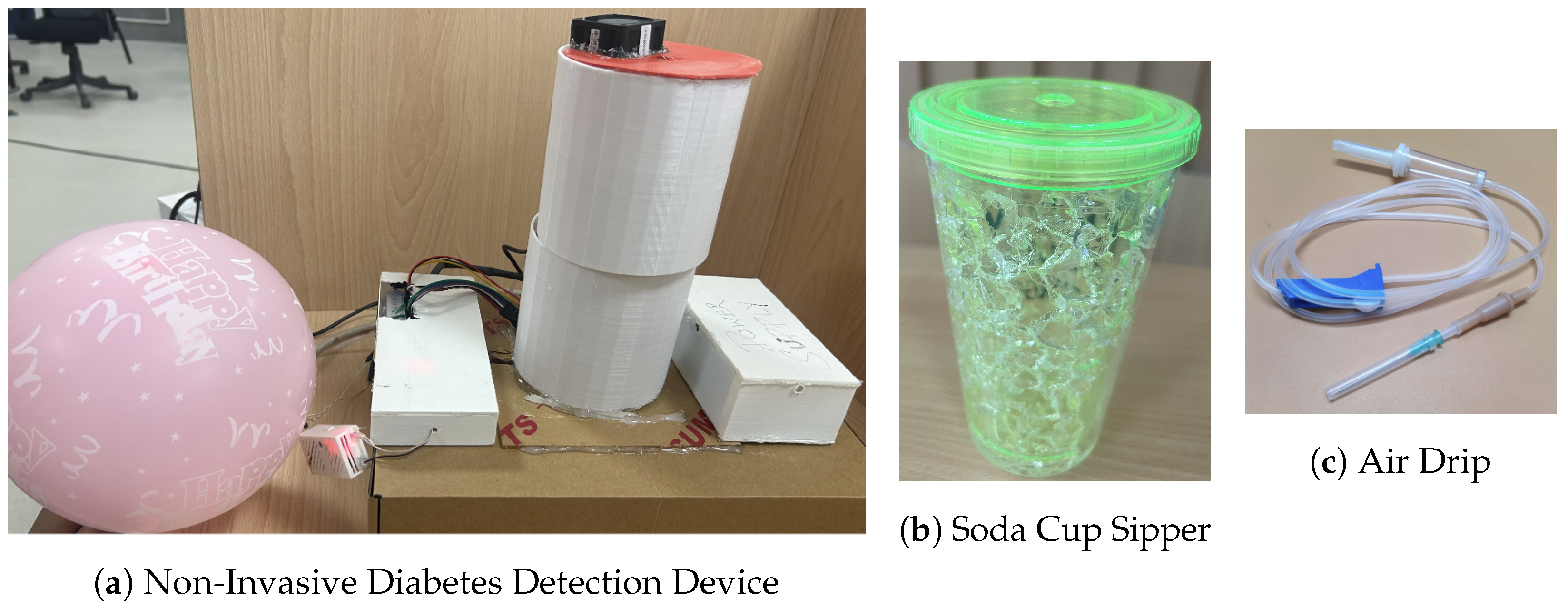
- Collecting and infusing the breath sample into the device: A (fasting sugar) breath sample collected in a balloon was infused into the sensor-based setup using a drip pipe mounted on top of the soda cup cap. We placed silica gel packets in a device to absorb moisture (or water) present in breath samples. The drip pipe’s one end was attached to the mouth of the inflated balloon, housing the breath sample. The other end was connected to a soda cup cap containing the embedded sensors.
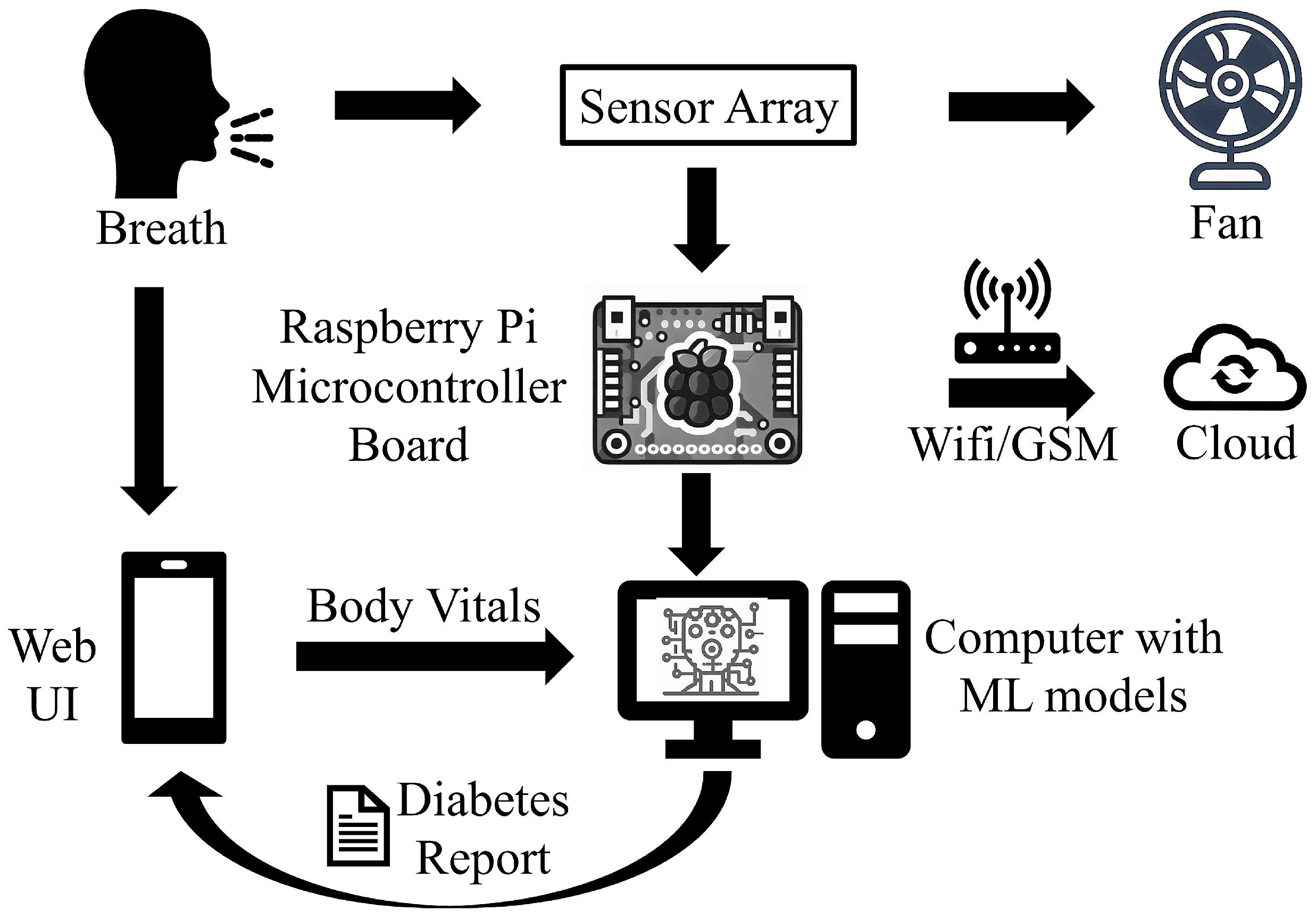
- Processing the breath sample and recording the data: Upon interaction with the VOCs present in the breath sample, the sensors showed deflection from their baseline (as showing in Figure 3c). The recorded deflection data conveyed through the MQTT protocol are directed into an InfluxDB time series database. The Grafana visualization dashboard facilitates the visualization of real-time sensor responses. The experimental setup comprised a Raspberry Pi hosting the MQTT server, Grafana, Node-RED, and Influx DB running as a Docker container. The sensor voltage readings acted as those of characteristics as they depended on the concentration of VOCs in the breath sample.
- Getting the setup ready for the following sample: After a reaction time of two seconds, we removed the cup’s cap and mounted it with a fan assembly to expel the breath sample present in the device. Once the voltage readings were restored to their baselines, we stopped recording these for the present sample. At this stage, the setup is ready to process the next breath sample.
3.5. Sensor Calibration Experiments
3.6. Clinical Trial Study and Experimental Setting
3.7. Preprocessing the Sensor Voltages
3.8. Feature Extraction
3.9. ML Model Development
- Gradient Boosting (G-Boost): G-Boost creates a stage-wise model and generalises the model by allowing for the optimization of an arbitrary differentiable loss function. Gradient boosting combines weak learners into a single strong learner in an iterative fashion. As each weak learner is added, a new model is fitted to provide a more accurate estimate of the response variable [52,53].
- Decision Tree (DT): A DT is developed by recursively splitting data based on feature values to develop subsets that are as pure as feasible, which means that each subset mainly comprises instances of a single class [54].
- K-Nearest Neighbours (KNNs): KNNs do not make any underlying assumptions about data distribution. Given some prior data (training data), KNNs classify coordinates identified by an attribute [54].
- Ridge: Ridge regression enhances regular linear regression by slightly changing its cost function, which results in less overfit models [55].
- Lasso: Lasso is a regression analysis method that performs both variable selection and regularization to enhance the prediction accuracy and interpretability of the resulting statistical model. For lasso, the coefficient estimates do not need to be unique if covariates are collinear. Lasso’s ability to perform subset selection relies on the form of the constraint and has a variety of interpretations, including in terms of geometry, Bayesian statistics, and convex analysis [56,57].
- Elastic Net (ENet): ENet combines the two most popular regularized variants of linear regression: ridge and lasso. Ridge utilises an L2 penalty, and lasso uses an L1 penalty. ENet uses both the L2 and the L1 penalty [58].
- Logistic Regression: It is used for predicting the categorical dependent variable using a given set of independent variables. Logistic regression predicts the output of a categorical dependent variable. Therefore the outcome must be a categorical or discrete value. It can be either yes or no, 0 or 1, true or false, etc., but instead of giving the exact value as 0 and 1, it gives the probabilistic values between 0 and 1 [59].
- Support Vector Machines (SVMs): SVMs operate by determining the appropriate hyperplane for separating various classes in the data space. The hyperplane is chosen to maximize the margin, which is the distance between the hyperplane and the nearest data points of each class, also known as support vectors [60].
- eXtreme Gradient Boosting (XG-Boost): XG-Boost is an open-source software library with a regularising gradient boosting framework for C++, Java, Python, R, Julia, Perl, and Scala. It works on Linux, Windows, and macOS. From the project description, it aims to provide a scalable, portable and distributed gradient boosting (GBM, GBRT, GBDT) library. It runs on a single machine, and the distributed processing frameworks Apache Hadoop, Apache Spark, Apache Flink, and Dask [12].
- Random Forest (RF): The RF algorithm generates multiple DTs during training by selecting random subsets of the original dataset and random subsets of characteristics for each tree. Each DT in the RF is developed using a technique known as recursive partitioning, which involves repeatedly splitting the data into subsets depending on the most discriminatory attributes, resulting in a tree-like structure [60].
3.10. Evaluation Metrics
3.11. Selecting the Best-Performing Model for Diabetes Prediction
3.12. Ethical Consideration
4. Results
5. Discussion
5.1. Performance Evaluation
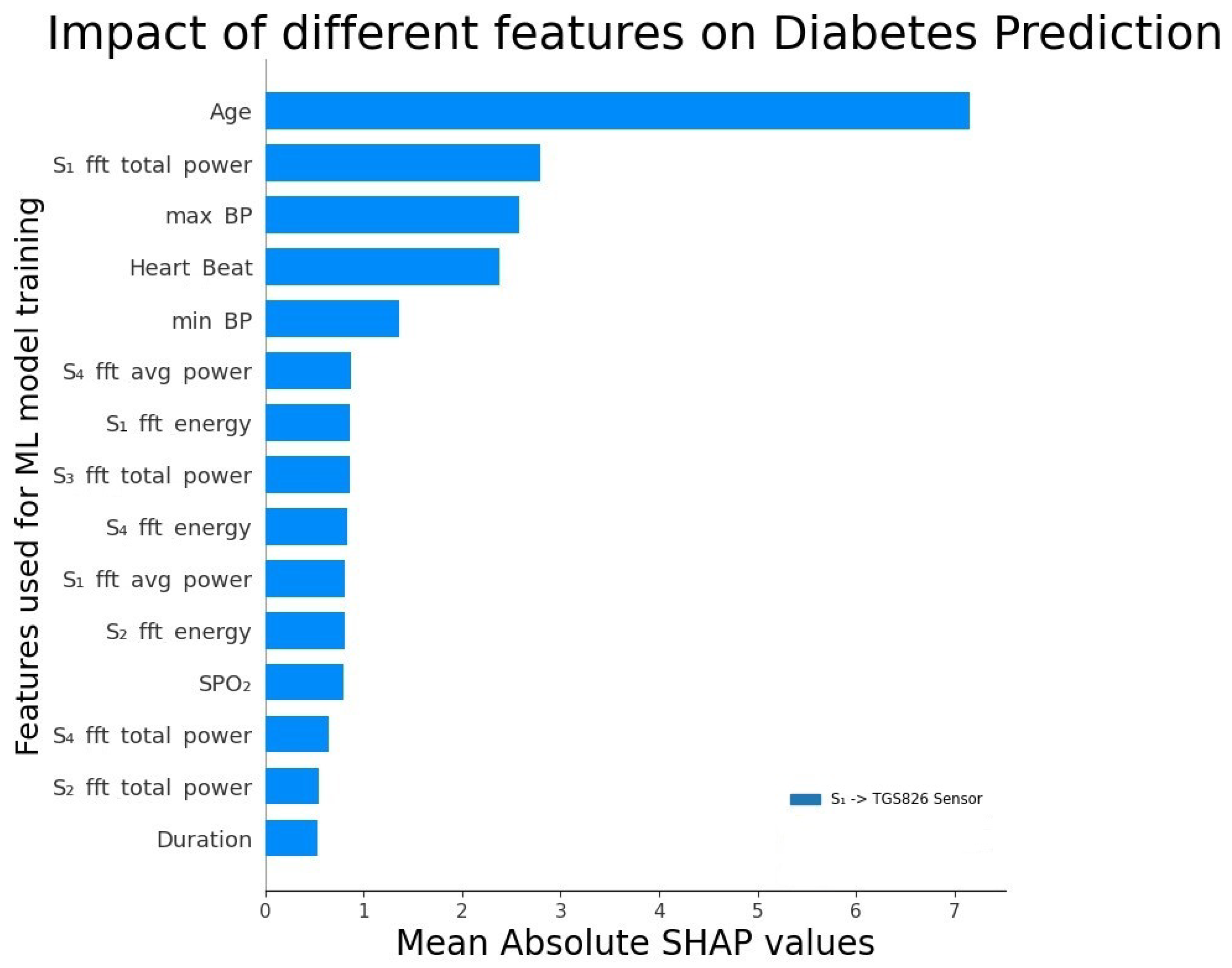
5.2. Feature Analysis
5.3. Performance Comparison with the Existing Works
6. Conclusions, Limitations, and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| T2DM | Type 2 diabetes mellitus |
| IoT | Internet of Things |
| IDF | International Diabetes Foundation |
| WHO | World Health Organization |
| VOC | Volatile Organic Compounds |
| MOS | Metal-Oxide-Semiconductor |
| ML | Machine Learning |
| BGL | Blood glucose level |
| E-Nose | Electronic Nose |
| DiabeticSense | Non-invasive multi-sensor diabetes detection device |
| ADC | Analog-to-digital converter |
| LP | Liquefied petroleum |
| ppm | parts per million |
| mL | milli-Litre |
| abs | absolute |
| max | maximum |
| min | minimum |
| mean | mean or average |
| stdDev | standard deviation |
| avg | average |
| G-Boost | Gradient Boosting |
| DT | Decision Tree |
| KNNs | K-Nearest Neighbours |
| ENet | Elastic Net |
| SVMs | Support Vector Machines |
| XG-Boost | eXtreme Gradient Boosting |
| RF | Random Forest |
| AUC | Area Under the Curve |
| MeanAcc | Mean Accuracy |
| SHAPs | Shapley additive explanations |
| BP | Blood Pressure |
| SPO | Oxygen level in blood |
| FFT | Fast Fourier Transform |
| SMOTE | Synthetic Minority Oversampling TEchnique |
| MSE | Mean Square Error |
| sqrt | Square root |
| T | Temperature |
| H | Humidity |
| °C | degree Celsius |
References
- DeFronzo, R.A.; Ferrannini, E.; Groop, L.; Henry, R.R.; Herman, W.H.; Holst, J.J.; Hu, F.B.; Kahn, C.R.; Raz, I.; Shulman, G.I.; et al. Type 2 diabetes mellitus. Nat. Rev. Dis. Prim. 2015, 1, 15019. [Google Scholar] [CrossRef]
- Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B.B.; Stein, C.; Basit, A.; Chan, J.C.; Mbanya, J.C.; et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022, 183, 109119. [Google Scholar] [CrossRef]
- Roglic, G. WHO Global report on diabetes: A summary. Int. J. Noncommun. Dis. 2016, 1, 3–8. [Google Scholar] [CrossRef]
- Kou, L.; Zhang, D.; Liu, D. A novel medical e-nose signal analysis system. Sensors 2017, 17, 402. [Google Scholar] [CrossRef]
- Risby, T.H.; Solga, S. Current status of clinical breath analysis. Appl. Phys. B 2006, 85, 421–426. [Google Scholar] [CrossRef]
- Phillips, M.; Altorki, N.; Austin, J.H.; Cameron, R.B.; Cataneo, R.N.; Greenberg, J.; Kloss, R.; Maxfield, R.A.; Munawar, M.I.; Pass, H.I.; et al. Prediction of lung cancer using volatile biomarkers in breath. Cancer Biomarkers 2007, 3, 95–109. [Google Scholar] [CrossRef]
- Dent, A.G.; Sutedja, T.G.; Zimmerman, P.V. Exhaled breath analysis for lung cancer. J. Thorac. Dis. 2013, 5, S540. [Google Scholar]
- Marcondes-Braga, F.G.; Batista, G.L.; Bacal, F.; Gutz, I. Exhaled breath analysis in heart failure. Curr. Heart Fail. Rep. 2016, 13, 166–171. [Google Scholar] [CrossRef]
- Dixit, K.; Fardindoost, S.; Ravishankara, A.; Tasnim, N.; Hoorfar, M. Exhaled breath analysis for diabetes diagnosis and monitoring: Relevance, challenges and possibilities. Biosensors 2021, 11, 476. [Google Scholar] [CrossRef]
- Hoenes, J.; Müller, P.; Surridge, N. The technology behind glucose meters: Test strips. Diabetes Technol. Ther. 2008, 10, S-10. [Google Scholar]
- Canadian Agency for Drugs and Technologies in Health (CADTH). Systematic review of use of blood glucose test strips for the management of diabetes mellitus. CADTH Technol. Overviews 2010, 1, e0101.
- Paleczek, A.; Grochala, D.; Rydosz, A. Artificial breath classification using XG-Boost algorithm for diabetes detection. Sensors 2021, 21, 4187. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Zhang, D.; Zhang, L.; Lu, G. Non-invasive blood glucose monitoring for diabetics by means of breath signal analysis. Sens. Actuators B Chem. 2012, 173, 106–113. [Google Scholar] [CrossRef]
- Anderson, J.C. Measuring breath acetone for monitoring fat loss. Obesity 2015, 23, 2327–2334. [Google Scholar] [CrossRef]
- Sun, M.; Chen, Z.; Gong, Z.; Zhao, X.; Jiang, C.; Yuan, Y.; Wang, Z.; Li, Y.; Wang, C. Determination of breath acetone in 149 Type 2 diabetic patients using a ringdown breath-acetone analyzer. Anal. Bioanal. Chem. 2015, 407, 1641–1650. [Google Scholar] [CrossRef]
- Righettoni, M.; Schmid, A.; Amann, A.; Pratsinis, S.E. Correlations between blood glucose and breath components from portable gas sensors and PTR-TOF-MS. J. Breath Res. 2013, 7, 037110. [Google Scholar] [CrossRef]
- Pérez-Ros, P.; Navarro-Flores, E.; Julián-Rochina, I.; Martínez-Arnau, F.M.; Cauli, O. Changes in salivary amylase and glucose in diabetes: A scoping review. Diagnostics 2021, 11, 453. [Google Scholar] [CrossRef]
- Pamungkas, R.A.; Usman, A.M.; Chamroonsawasdi, K. A smartphone application of diabetes coaching intervention to prevent the onset of complications and to improve diabetes self-management: A randomized control trial. Diabetes Metab. Syndr. Clin. Res. Rev. 2022, 16, 102537. [Google Scholar] [CrossRef]
- Kirwan, M.; Vandelanotte, C.; Fenning, A.; Duncan, M.J. Diabetes self-management smartphone application for adults with type 1 diabetes: Randomized controlled trial. J. Med. Internet Res. 2013, 15, e235. [Google Scholar] [CrossRef]
- Španěl, P.; Smith, D. Progress in SIFT-MS: Breath analysis and other applications. Mass Spectrom. Rev. 2011, 30, 236–267. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, H.; Sun, W.; Lu, N.; Yan, M.; Wu, Y.; Hua, Z.; Fan, S. Development of a low-cost portable electronic nose for cigarette brands identification. Sensors 2020, 20, 4239. [Google Scholar] [CrossRef] [PubMed]
- Silva-Martinez, J.; Liu, X.; Zhou, D. Recent advances on linear low-dropout regulators. IEEE Trans. Circuits Syst. II Express Briefs 2020, 68, 568–573. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhu, J.; Shen, X.; Lin, C.; Zhang, Y.; Liang, Y.; Cao, B.; Li, J.; Liu, X.; Rao, W.; et al. Chinese diabetes datasets for data-driven machine learning. Sci. Data 2023, 10, 35. [Google Scholar] [CrossRef]
- Chicharro-Luna, E.; Pomares-Gómez, F.J.; Ortega-Ávila, A.B.; Marchena-Rodríguez, A.; Blanquer-Gregori, J.F.J.; Navarro-Flores, E. Predictive model to identify the risk of losing protective sensibility of the foot in patients with diabetes mellitus. Int. Wound J. 2020, 17, 220–227. [Google Scholar] [CrossRef]
- TGS 826-A00; FIGARO—Ammonia (NH3) Gas Sensor. Figaro Inc.: Osaka, Japan, 2023. Available online: https://www.soselectronic.com/en/products/figaro/tgs-826-a00-53106 (accessed on 22 August 2023).
- TGS2610-C00; [LP Gas Sensor]. Figaro Inc.: Osaka, Japan, 2023. Available online: https://www.figarosensor.com/product/entry/tgs2610-c00.html (accessed on 22 August 2023).
- TGS 822; FIGARO—Organic Solvent Vapor Sensor. Figaro Inc.: Osaka, Japan, 2023. Available online: https://www.soselectronic.com/en/products/figaro/tgs-822-7719 (accessed on 22 August 2023).
- TGS 2602; Air Quality/VOC Sensor. Figaro Inc.: Osaka, Japan, 2023. Available online: https://www.figarosensor.com/product/entry/tgs2602.html (accessed on 22 August 2023).
- TGS 2600; Air Quality Sensor. Figaro Inc.: Osaka, Japan, 2023. Available online: https://www.figarosensor.com/product/entry/tgs2600.html (accessed on 22 August 2023).
- TGS 2603; Odorous Gas Sensor. Figaro Inc.: Osaka, Japan, 2023. Available online: https://www.figarosensor.com/product/entry/tgs2603.html (accessed on 22 August 2023).
- TGS 2620; For the Detection of Solvent Vapors. Figaro Inc.: Osaka, Japan, 2023. Available online: https://www.figarosensor.com/product/entry/tgs2620.html (accessed on 22 August 2023).
- MQ138; VOC Gas Sensor. Zhengzhou Winsen Electronics Technology Co., Ltd.: Zhengzhou, China, 2023. Available online: https://www.winsen-sensor.com/sensors/voc-sensor/mq138.html (accessed on 22 August 2023).
- DHT22/AM2302; Digital Temperature Humidity Sensor. Aosong Electronics Co., Ltd.: Guangzhou, China, 2023. Available online: https://www.kuongshun-ks.com/uno/uno-sensor/dht22-am2302-digital-temperature-humidity-sensor.html (accessed on 22 August 2023).
- Owen, K.; Shepherd, M.; Stride, A.; Ellard, S.; Hattersley, A.T. Heterogeneity in young adult-onset diabetes: Aetiology alters clinical characteristics. Diabet. Med. 2002, 19, 758–761. [Google Scholar] [CrossRef] [PubMed]
- numpy.absolute—SciPy v1.11.2 Manual. Available online: https://numpy.org/doc/stable/reference/generated/numpy.absolute.html (accessed on 22 August 2023).
- numpy.max—SciPy v1.11.2 Manual. Available online: https://numpy.org/doc/stable/reference/generated/numpy.max.html (accessed on 22 August 2023).
- numpy.min—SciPy v1.11.2 Manual. Available online: https://numpy.org/doc/stable/reference/generated/numpy.min.html (accessed on 22 August 2023).
- numpy.mean—SciPy v1.11.2 Manual. Available online: https://numpy.org/doc/stable/reference/generated/numpy.mean.html (accessed on 22 August 2023).
- numpy.std—SciPy v1.11.2 Manual. Available online: https://numpy.org/doc/stable/reference/generated/numpy.std.html (accessed on 22 August 2023).
- numpy.gradient—SciPy v1.11.2 Manual. Available online: https://numpy.org/doc/stable/reference/generated/numpy.gradient.html (accessed on 22 August 2023).
- Integration (scipy.integrate)—SciPy v1.11.2 Manual. Available online: https://docs.scipy.org/doc/scipy/tutorial/integrate.html (accessed on 22 August 2023).
- Hierlemann, A.; Gutierrez-Osuna, R. Higher-order chemical sensing. Chem. Rev. 2008, 108, 563–613. [Google Scholar] [CrossRef] [PubMed]
- Fourier Transforms (scipy.fft)—SciPy v1.11.2. Available online: https://docs.scipy.org/doc/scipy/tutorial/fft.html#fast-fourier-transforms (accessed on 22 August 2023).
- Discrete Fourier Transform (numpy.fft)—NumPy v1.25 Manual. Available online: https://numpy.org/doc/stable/reference/routines.fft.html (accessed on 22 August 2023).
- Wasilewski, F. Wavelet Transforms in Python; 2023. Available online: https://pywavelets.readthedocs.io/en/latest (accessed on 22 August 2023).
- scipy.signal.find_peaks—SciPy v1.11.2 Manual. Available online: https://docs.scipy.org/doc/scipy/reference/generated/scipy.signal.find_peaks.html (accessed on 22 August 2023).
- scipy.stats.skew—SciPy v1.11.2 Manual. Available online: https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.skew.html (accessed on 22 August 2023).
- scipy.stats.kurtosis—SciPy v1.11.2 Manual. Available online: https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.kurtosis.html (accessed on 22 August 2023).
- scipy.stats.entropy—SciPy v1.11.2 Manual. Available online: https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.entropy.html (accessed on 22 August 2023).
- statsmodels.tsa.ar_model.AutoReg—Statsmodels 0.15.0 (+49) Stable Release Manual. Available online: https://www.statsmodels.org/dev/generated/statsmodels.tsa.ar_model.AutoReg.html (accessed on 22 August 2023).
- librosa.stft—Librosa 0.10.1 Documentation. Available online: https://librosa.org/doc/main/generated/librosa.stft.html (accessed on 22 August 2023).
- Friedman, J.H. Stochastic gradient boosting. Comput. Stat. Data Anal. 2002, 38, 367–378. [Google Scholar] [CrossRef]
- Ahamed, B.S.; Arya, M.S.; Sangeetha, S.; Auxilia Osvin, N.V. Diabetes Mellitus Disease Prediction and Type Classification Involving Predictive Modeling Using Machine Learning Techniques and Classifiers. Appl. Comput. Intell. Soft Comput. 2022, 2022, 7899364. [Google Scholar] [CrossRef]
- Sharaff, A.; Gupta, H. Extra-tree classifier with metaheuristics approach for email classification. In Advances in Computer Communication and Computational Sciences, Proceedings of IC4S 2018; Springer: Berlin/Heidelberg, Germany, 2019; pp. 189–197. [Google Scholar]
- Gupta, D.; Choudhury, A.; Gupta, U.; Singh, P.; Prasad, M. Computational approach to clinical diagnosis of diabetes disease: A comparative study. Multimed. Tools Appl. 2021, 80, 30091–30116. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B Stat. Methodol. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Wang, X.; Zhai, M.; Ren, Z.; Ren, H.; Li, M.; Quan, D.; Chen, L.; Qiu, L. Exploratory study on classification of diabetes mellitus through a combined Random Forest Classifier. BMC Med. Inform. Decis. Mak. 2021, 21, 105. [Google Scholar] [CrossRef] [PubMed]
- Jayanthi, N.; Babu, B.V.; Rao, N.S. Survey on clinical prediction models for diabetes prediction. J. Big Data 2017, 4, 26. [Google Scholar] [CrossRef]
- Mujumdar, A.; Vaidehi, V. Diabetes prediction using machine learning algorithms. Procedia Comput. Sci. 2019, 165, 292–299. [Google Scholar] [CrossRef]
- Olisah, C.C.; Smith, L.; Smith, M. Diabetes mellitus prediction and diagnosis from a data preprocessing and machine learning perspective. Comput. Methods Programs Biomed. 2022, 220, 106773. [Google Scholar] [CrossRef] [PubMed]
| Sensor Model | VOCs Sensitivity | Sensitivity Range (in ppm) 1 |
|---|---|---|
| TGS 826 | iso-butane, ethanol, ammonia, and hydrogen | 30–5000 |
| TGS 2610 | ethanol, hydrogen, methane, iso-butane, and propane | 500–10,000 |
| TGS 822 | methane, carbon monoxide, iso-butane, n-hexane, benzene, ethanol, and acetone | 50–5000 |
| TGS 2602 | hydrogen, hydrogen sulfide, ammonia, ethanol, and toluene | 1–30 |
| TGS 2600 | methane, carbon monoxide, iso-butane, ethanol, and hydrogen | 1–100 |
| TGS 2603 | hydrogen sulfide, hydrogen, methyl mercaptan, trimethyl amine, and ethanol | 1–10 |
| TGS 2620 | methane, carbon monoxide, iso-butane, hydrogen, and ethanol | 50–5000 |
| MQ 138 | formaldehyde, benzene, toluene, and acetone | 5–500 |
| DHT 22 | Humidity (H) and Temperature (T) | H: 0–100 RH, T: −40–80 °C |
| Base Feature | Feature Used | Description |
|---|---|---|
| CurveMagnitude | abs(CurveMagnitude) [35] | The absolute value of curve magnitude values. |
| max(CurveMagnitude) [36] | The maximum of curve magnitude values. | |
| min(CurveMagnitude) [37] | The minimum of curve magnitude values. | |
| mean(CurveMagnitude) [38] | The mean or average of curve magnitude values. | |
| stdDev(CurveMagnitude) [39] | The median curve magnitude values. | |
| FirstDerivative [40] | max(FirstDerivative) | The maximum of first derivative of signal values. |
| min(FirstDerivative) | The minimum of first derivative of signal values. | |
| mean(FirstDerivative) | The mean of first derivative of signal values. | |
| abs(FirstDerivative) | The absolute value of the first derivative. | |
| stdDev(FirstDerivative) | The square root of the variance of the first derivative. | |
| SecondDerivative [40] | max(SecondDerivative) | The maximum of second derivative of signal values. |
| min(SecondDerivative) | The minimum of second derivative of signal values. | |
| mean(SecondDerivative) | The mean of second derivative of signal values. | |
| abs(SecondDerivative) | The absolute value of the second derivative. | |
| stdDev(SecondDerivative) | The square root of the variance of the second derivative. | |
| Slope and Integral of five intervals [41] | Slope of five intervals | The slope of the five intervals of the curve 1. |
| Integral of five intervals | The integral of the five intervals of the curve 1. | |
| Phase | It represents the integral of derivative over the magnitude values [42]. | |
| Fast Fourier Transform (fft) [43,44] | phase | The phase is calculated based on the fft of the sensors’ response. |
| powerSpectrum | The square of the absolute value of fft transform. | |
| spectralEntropy | It represents the entropy of power spectrum. | |
| Wavelet [45] | waveletCoeffs | Coefficients of wavelet transformation of the sensor’s response signal. |
| Peak [46] | height | The height of the peak. |
| width | The width of the peak. | |
| area | The trapezoidal area of the peak. | |
| Shape | skewness [47] | The measure of the asymmetry of a distribution, where a positive skew indicates a longer tail on the right side and a negative skew indicates a longer tail on the left side. |
| kurtosis [48] | The measure of the tailedness of a distribution; a positive value indicates fatter tails and a negative value indicates thinner tails. | |
| entropy [49] | The measure of the disorder or randomness of a shape; a higher entropy indicates a more disordered or random shape. | |
| Auto-Regressive (AR) [50] | coefficients | These represent the relationships between past and current values of the model. |
| predictionError | The difference between the actual observed value and the AR model’s predicted value. | |
| Short-time Fourier transform (STFT) [51] | dominantFrequency | The frequency component that has the highest magnitude of the signal. |
| avg(magnitude(STFTcoeffs)) | The average magnitude of the STFT coefficients, calculated by taking the mean of the magnitudes over all the time frames. | |
| Sum(magnitude(STFTcoeffs)) | The sum of the magnitudes of all the STFT coefficients. | |
| energy(STFT) | The overall power of the signal in the frequency domain. | |
| centroid(STFTcoeffs) | The weighted average of the frequencies in the STFT, where the weights are the magnitudes of the STFT coefficients. | |
| bandwidth(STFT) | The range of frequencies represented by a single STFT coefficient, determined by the window length. | |
| rolloff(STFT) | The frequency at which the magnitude of the STFT coefficients drops to −3dB, typically used as a measure of the sharpness of the transition between the passband and the stopband. |
| ML Classifiers | Parameter Name | Parameter Values |
|---|---|---|
| Decision Tree | criterion | (‘gini’, ‘entropy’, ‘log_loss’) |
| splitter | (‘best’, ‘random’) | |
| max depth | (2 to 10, step size of 1) | |
| min samples split | (2 to 10, step size of 1) | |
| Support Vector | C | (0.1 to 10, step size of 0.1) |
| kernel | (‘linear’, ‘poly’, ‘rbf’, ‘sigmoid’, ‘precomputed’) | |
| degree | (3 to 10, step size of 1) | |
| gamma | (‘scale’, ‘auto’, ‘float’) with (0.001 to 1, step size of 0.005) for ‘float’ | |
| Gradient Boost | learning rate | (0.01 to 10, step size of 0.01) |
| n estimators | (5 to 500, step size of 5) | |
| subsample | (0.01 to 1, step size of 0.01) | |
| criterion | (‘friedman mse’, ‘squared error’) | |
| min samples split | (2 to 10, step size of 1) | |
| max depth | (2 to 10, step size of 1) | |
| Random Forest | n estimators | (5 to 500, step size of 5) |
| criterion | (‘gini’, ‘entropy’, ‘log loss’) | |
| min samples split | (2 to 10, step size of 1) | |
| max depth | (2 to 10, step size of 1) | |
| max features | (‘sqrt’, ‘log2’) | |
| min samples leaf | (1 to 10, step size of 1) | |
| K-NNeighbors | n neighbors | (5 to 100, step size of 5) |
| weights | (‘uniform’, ‘distance’) | |
| algorithm | (‘auto’, ‘ball tree’, ‘kd tree’, ‘brute’) | |
| leaf size | (30 to 100, step size of 3) | |
| Elastic Net | alpha | (0.01 to 1, step size of 0.01) |
| l1 ratio | (0.01 to 1, step size of 0.01) | |
| fit intercept | (True, False) | |
| max iter | (1000 to 5000, step size of 100) | |
| selection | (‘cyclic’, ‘random’) | |
| Ridge | solver | (‘auto’, ‘svd’, ‘cholesky’, ‘lsqr’, ‘sparse_cg’, ‘sag’) |
| fit intercept | (True, False) | |
| max iter | (1000 to 5000, step size of 100) | |
| Lasso | alpha | (0.1 to 10, step size of 0.1) |
| fit intercept | (True, False) | |
| copy X | (True, False) | |
| max iter | (1000 to 5000, step size of 100) | |
| selection | (‘cyclic’, ‘random’) | |
| Logistic Regression | penalty | (‘l1’, ‘l2’, ‘Elastic Net’, None) |
| dual | (True, False) | |
| C | (0.1 to 10, step size of 0.1) | |
| fit intercept | (True, False) | |
| solver | (‘lbfgs’, ‘liblinear’, ‘newton-cg’, ‘newton-cholesky’, ‘saga’, ‘sag’) | |
| max iter | (1000 to 5000, step size of 100) | |
| multi class | (‘auto’, ‘ovr’, ‘multinomial’) | |
| XG-Boost | max depth | (1 to 10, step size of 1) |
| alpha | (0.1 to 10, step size of 0.1) | |
| booster | (‘gbtree’, ‘gblinear’) | |
| eta | (0.01 to 1, step size of 0.01) | |
| min child weight | (1 to 10, step size of 1) |
| Feature | Description |
|---|---|
| Age | Age of the user |
| Gender | Gender of the user, i.e., male, female, or other |
| BP | User’s max and min BP values |
| SPO | Oxygen level in blood |
| Heart Rate | Heart rate of the patient |
| Fast Fourier Transform (fft) | phase |
| powerSpectrum | |
| spectralEntropy | |
| Phase | |
| FirstDerivative | max(FirstDerivative) |
| min(FirstDerivative) | |
| mean(FirstDerivative) | |
| abs(FirstDerivative) | |
| stdDev(FirstDerivative) | |
| SecondDerivative | max(SecondDerivative) |
| min(SecondDerivative) | |
| mean(SecondDerivative) | |
| abs(SecondDerivative) | |
| stdDev(SecondDerivative) | |
| Slope and Integral of five intervals | Slope of five intervals 1. |
| Integral of five intervals 1. |
| (a) Five-Fold Cross Validation Performance Metrics Scores of Various ML Classifiers | |||||
|---|---|---|---|---|---|
| ML Classifiers | BalancingTech | MeanAccuracy | MeanF1Score | MeanROC | MeanAcc 1 |
| Decision Tree | ADASYN | 0.847 | 0.843 | 0.844 | 0.845 |
| SMOTE | 0.796 | 0.793 | 0.801 | 0.797 | |
| UnBalanced | 0.713 | 0.662 | 0.666 | 0.68 | |
| Support Vector | ADASYN | 0.695 | 0.69 | 0.693 | 0.693 |
| SMOTE | 0.717 | 0.706 | 0.715 | 0.713 | |
| UnBalanced | 0.625 | 0.382 | 0.492 | 0.5 | |
| Gradient Boost | ADASYN | 0.866 | 0.865 | 0.868 | 0.866 |
| SMOTE | 0.816 | 0.812 | 0.819 | 0.816 | |
| UnBalanced | 0.763 | 0.741 | 0.774 | 0.759 | |
| Random Forest | ADASYN | 0.817 | 0.813 | 0.815 | 0.815 |
| SMOTE | 0.737 | 0.73 | 0.753 | 0.74 | |
| UnBalanced | 0.663 | 0.582 | 0.591 | 0.612 | |
| K-NNeighbors | ADASYN | 0.601 | 0.597 | 0.604 | 0.601 |
| SMOTE | 0.668 | 0.656 | 0.664 | 0.663 | |
| UnBalanced | 0.575 | 0.484 | 0.548 | 0.536 | |
| Elastic Net | ADASYN | 0.726 | 0.684 | 0.749 | 0.72 |
| SMOTE | 0.775 | 0.763 | 0.77 | 0.769 | |
| UnBalanced | 0.688 | 0.634 | 0.648 | 0.657 | |
| Ridge | ADASYN | 0.754 | 0.746 | 0.762 | 0.754 |
| SMOTE | 0.814 | 0.807 | 0.809 | 0.81 | |
| UnBalanced | 0.725 | 0.679 | 0.69 | 0.698 | |
| Lasso | ADASYN | 0.785 | 0.732 | 0.805 | 0.774 |
| SMOTE | 0.755 | 0.745 | 0.754 | 0.751 | |
| UnBalanced | 0.7 | 0.637 | 0.652 | 0.663 | |
| Logistic Regression | ADASYN | 0.683 | 0.669 | 0.693 | 0.687 |
| SMOTE | 0.824 | 0.816 | 0.817 | 0.819 | |
| UnBalanced | 0.713 | 0.66 | 0.668 | 0.68 | |
| XG-Boost | ADASYN | 0.867 | 0.857 | 0.864 | 0.86.3 |
| SMOTE | 0.815 | 0.808 | 0.81 | 0.811 | |
| UnBalanced | 0.688 | 0.626 | 0.646 | 0.653 | |
| (b) Test Performance Metrics Scores of various ML Classifiers | |||||
| ML Classifiers | Accuracy | F1 Score | ROC Area | MeanAcc 1 | |
| Decision Tree | 0.65 | 0.601 | 0.621 | 0.624 | |
| Support Vector | 0.55 | 0.54 | 0.54 | 0.543 | |
| Gradient Boost | 0.85 | 0.84 | 0.833 | 0.841 | |
| Random Forest | 0.55 | 0.436 | 0.51 | 0.499 | |
| K-NNeighbors | 0.45 | 0.449 | 0.449 | 0.449 | |
| Elastic Net | 0.5 | 0.479 | 0.485 | 0.488 | |
| Ridge | 0.6 | 0.56 | 0.576 | 0.579 | |
| Lasso | 0.45 | 0.437 | 0.439 | 0.442 | |
| Logistic Regression | 0.5 | 0.479 | 0.485 | 0.488 | |
| XG-Boost | 0.75 | 0.733 | 0.732 | 0.738 | |
| ML Classifiers | Hyper Tuned Parameter Values |
|---|---|
| Decision Tree | criterion: ‘entropy’, splitter: ‘best’, max depth: 5, min samples split: 2 |
| Support Vector | C: 10, kernel: ‘rbf’, degree: not relevant 1, gamma: ‘auto’ |
| Gradient Boost | learning rate: 1, n estimators: 100, subsample: 1, criterion: ‘friedman mse’, min samples split: 2, max depth: 3 |
| Random Forest | n estimators: 100, criterion: ‘entropy’, min samples split: 2, max depth: 9, max features: ‘sqrt’, min samples leaf: 1 |
| K-NNeighbors | n neighbors: 7, weights: ‘distance’, algorithm: ‘auto’, leaf size: 30 |
| Elastic Net | alpha: 0.1, l1 ratio: 0.5, fit intercept: ‘True’, max iter: 1000, selection: ‘cyclic’ |
| Ridge | solver: ‘auto’, fit intercept: ‘True’, max iter: 1000 |
| Lasso | alpha: 0.1, fit intercept: ‘True’, copy X: ‘True’, max iter: 1000, selection: ‘cyclic’ |
| Logistic Regression | penalty: ‘l2’, dual: ‘False’, C: 10, fit intercept: ‘True’, solver: ‘lbfgs’, max iter: 1000, multi class: ‘ovr’ |
| XG-Boost | max depth: 5, alpha: 0.1, booster: ‘gbtree’, eta: 0.3, min child weight: 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kapur, R.; Kumar, Y.; Sharma, S.; Rastogi, V.; Sharma, S.; Kanwar, V.; Sharma, T.; Bhavsar, A.; Dutt, V. DiabeticSense: A Non-Invasive, Multi-Sensor, IoT-Based Pre-Diagnostic System for Diabetes Detection Using Breath. J. Clin. Med. 2023, 12, 6439. https://doi.org/10.3390/jcm12206439
Kapur R, Kumar Y, Sharma S, Rastogi V, Sharma S, Kanwar V, Sharma T, Bhavsar A, Dutt V. DiabeticSense: A Non-Invasive, Multi-Sensor, IoT-Based Pre-Diagnostic System for Diabetes Detection Using Breath. Journal of Clinical Medicine. 2023; 12(20):6439. https://doi.org/10.3390/jcm12206439
Chicago/Turabian StyleKapur, Ritu, Yashwant Kumar, Swati Sharma, Vedant Rastogi, Shivani Sharma, Vikrant Kanwar, Tarun Sharma, Arnav Bhavsar, and Varun Dutt. 2023. "DiabeticSense: A Non-Invasive, Multi-Sensor, IoT-Based Pre-Diagnostic System for Diabetes Detection Using Breath" Journal of Clinical Medicine 12, no. 20: 6439. https://doi.org/10.3390/jcm12206439
APA StyleKapur, R., Kumar, Y., Sharma, S., Rastogi, V., Sharma, S., Kanwar, V., Sharma, T., Bhavsar, A., & Dutt, V. (2023). DiabeticSense: A Non-Invasive, Multi-Sensor, IoT-Based Pre-Diagnostic System for Diabetes Detection Using Breath. Journal of Clinical Medicine, 12(20), 6439. https://doi.org/10.3390/jcm12206439






