Phenotypic Clustering of Beta-Thalassemia Intermedia Patients Using Cardiovascular Magnetic Resonance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Population
2.2. Patient Follow-Up and Outcomes
2.3. Magnetic Resonance Imaging
2.4. Cluster Analysis
2.5. Data Pre-Processing
2.6. Evaluation of Clustering Tendency
2.7. Definition of Phenogroups
2.8. Supervised Random Forest
2.9. Statistical Analysis
3. Results
3.1. Patients’ Characteristics
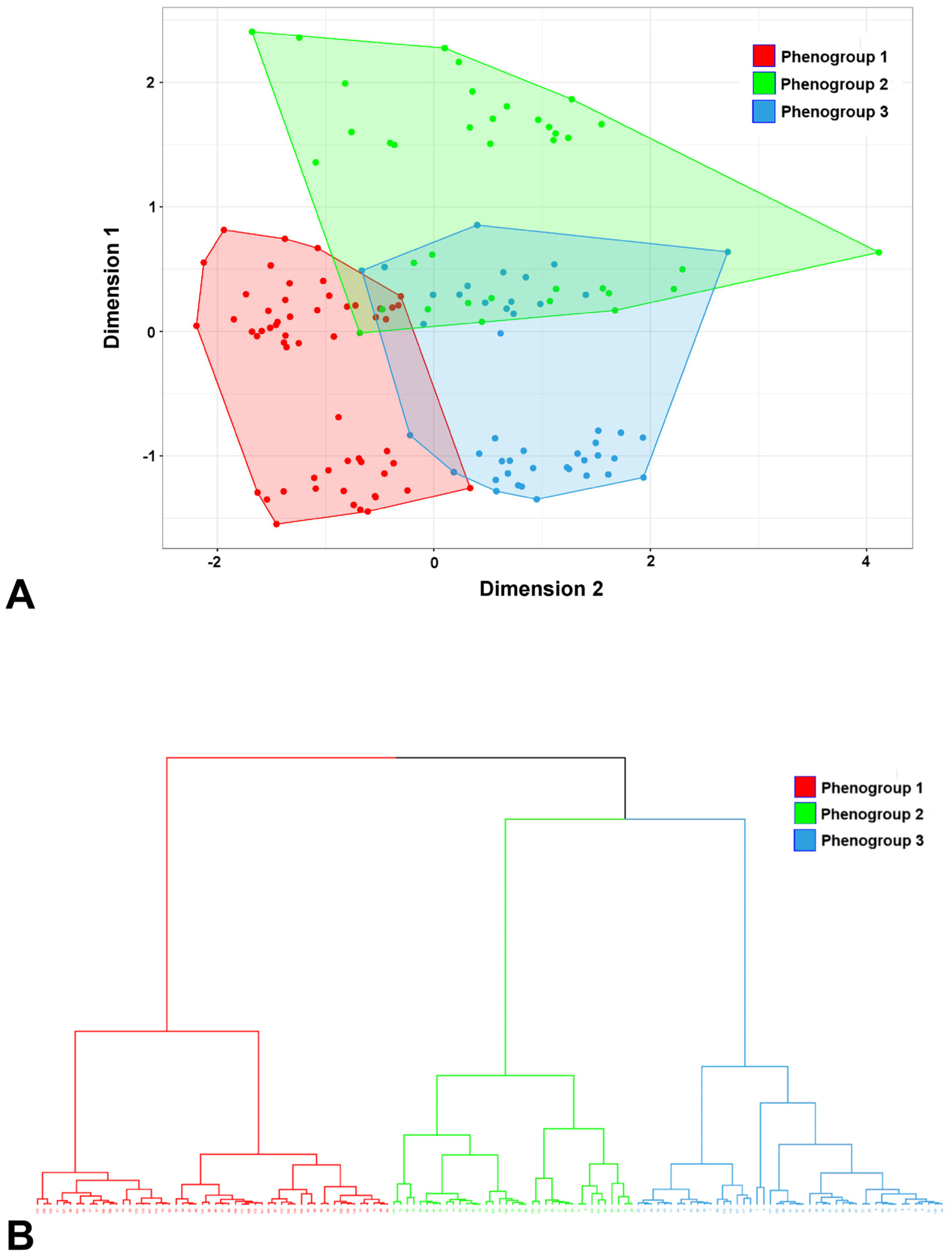
3.2. Clustering Results
3.3. Comparison among Phenogroups
3.4. Relevant Features for Clustering
3.5. Association of Phenogroups with Cardiovascular Complications
4. Discussion
Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weatherall, D.J. The thalassaemias. BMJ 1997, 314, 1675–1678. [Google Scholar] [CrossRef] [PubMed]
- Galanello, R.; Origa, R. Beta-thalassemia. Orphanet J. Rare Dis. 2010, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Rund, D.; Rachmilewitz, E. Beta-thalassemia. N. Engl. J. Med. 2005, 353, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Ben Salah, N.; Bou-Fakhredin, R.; Mellouli, F.; Taher, A.T. Revisiting beta thalassemia intermedia: Past, present, and future prospects. Hematology 2017, 22, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Weatherall, D.J. The definition and epidemiology of non-transfusion-dependent thalassemia. Blood Rev. 2012, 26 (Suppl. S1), S3–S6. [Google Scholar] [CrossRef] [PubMed]
- Taher, A.T.; Musallam, K.M.; El-Beshlawy, A.; Karimi, M.; Daar, S.; Belhoul, K.; Saned, M.S.; Graziadei, G.; Cappellini, M.D. Age-related complications in treatment-naive patients with thalassaemia intermedia. Br. J. Haematol. 2010, 150, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Taher, A.T.; Musallam, K.M.; Karimi, M.; El-Beshlawy, A.; Belhoul, K.; Daar, S.; Saned, M.S.; El-Chafic, A.H.; Fasulo, M.R.; Cappellini, M.D. Overview on practices in thalassemia intermedia management aiming for lowering complication rates across a region of endemicity: The OPTIMAL CARE study. Blood 2010, 115, 1886–1892. [Google Scholar] [CrossRef] [PubMed]
- Haddad, A.; Tyan, P.; Radwan, A.; Mallat, N.; Taher, A. beta-Thalassemia Intermedia: A Bird’s-Eye View. Turk. J. Haematol. 2014, 31, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Ginzburg, Y.; Rivella, S. beta-thalassemia: A model for elucidating the dynamic regulation of ineffective erythropoiesis and iron metabolism. Blood 2011, 118, 4321–4330. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.L.; Senecal, T.; Ghosh, M.C.; Ollivierre-Wilson, H.; Tu, T.; Rouault, T.A. Hepcidin regulates ferroportin expression and intracellular iron homeostasis of erythroblasts. Blood 2011, 118, 2868–2877. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, G. Role of hepcidin in the pathophysiology and diagnosis of anemia. Blood Res. 2013, 48, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Origa, R.; Barella, S.; Argiolas, G.M.; Bina, P.; Agus, A.; Galanello, R. No evidence of cardiac iron in 20 never- or minimally-transfused patients with thalassemia intermedia. Haematologica 2008, 93, 1095–1096. [Google Scholar] [CrossRef] [PubMed]
- Roghi, A.; Cappellini, M.D.; Wood, J.C.; Musallam, K.M.; Patrizia, P.; Fasulo, M.R.; Cesaretti, C.; Taher, A.T. Absence of cardiac siderosis despite hepatic iron overload in Italian patients with thalassemia intermedia: An MRI T2* study. Ann. Hematol. 2010, 89, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Liguori, C.; Pitocco, F.; Di Giampietro, I.; De Vivo, A.E.; Schena, E.; Giurazza, F.; Sorrentino, F.; Zobel, B.B. Magnetic resonance comparison of left-right heart volumetric and functional parameters in thalassemia major and thalassemia intermedia patients. Biomed. Res. Int. 2015, 2015, 857642. [Google Scholar] [CrossRef] [PubMed]
- Taher, A.T.; Musallam, K.M.; Wood, J.C.; Cappellini, M.D. Magnetic resonance evaluation of hepatic and myocardial iron deposition in transfusion-independent thalassemia intermedia compared to regularly transfused thalassemia major patients. Am. J. Hematol. 2010, 85, 288–290. [Google Scholar] [CrossRef] [PubMed]
- Meloni, A.; Pistoia, L.; Gamberini, M.R.; Ricchi, P.; Cecinati, V.; Sorrentino, F.; Cuccia, L.; Allo, M.; Righi, R.; Fina, P.; et al. The Link of Pancreatic Iron with Glucose Metabolism and Cardiac Iron in Thalassemia Intermedia: A Large, Multicenter Observational Study. J. Clin. Med. 2021, 10, 5561. [Google Scholar] [CrossRef] [PubMed]
- Ricchi, P.; Meloni, A.; Pistoia, L.; Spasiano, A.; Spiga, A.; Allo, M.; Gamberini, M.R.; Lisi, R.; Campisi, S.; Peluso, A.; et al. The effect of desferrioxamine chelation versus no therapy in patients with non transfusion-dependent thalassaemia: A multicenter prospective comparison from the MIOT network. Ann. Hematol. 2018, 97, 1925–1932. [Google Scholar] [CrossRef] [PubMed]
- Varat, M.A.; Adolph, R.J.; Fowler, N.O. Cardiovascular effects of anemia. Am. Heart J. 1972, 83, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.M.; Levine, B.D.; Stembridge, M. A change of heart: Mechanisms of cardiac adaptation to acute and chronic hypoxia. J. Physiol. 2022, 600, 4089–4104. [Google Scholar] [CrossRef] [PubMed]
- Aessopos, A.; Berdoukas, V. Cardiac function and iron chelation in thalassemia major and intermedia: A review of the underlying pathophysiology and approach to chelation management. Mediterr. J. Hematol. Infect. Dis. 2009, 1, e2009002. [Google Scholar] [CrossRef] [PubMed]
- Pennell, D.J. Cardiovascular Magnetic Resonance. Circulation 2010, 121, 692–705. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Chen, Y.; Ferrari, V.A. Contemporary Application of Cardiovascular Magnetic Resonance Imaging. Annu. Rev. Med. 2020, 71, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.C. Diagnosis and management of transfusion iron overload: The role of imaging. Am. J. Hematol. 2007, 82, 1132–1135. [Google Scholar] [CrossRef] [PubMed]
- Pennell, D.J.; Udelson, J.E.; Arai, A.E.; Bozkurt, B.; Cohen, A.R.; Galanello, R.; Hoffman, T.M.; Kiernan, M.S.; Lerakis, S.; Piga, A.; et al. Cardiovascular function and treatment in beta-thalassemia major: A consensus statement from the American Heart Association. Circulation 2013, 128, 281–308. [Google Scholar] [CrossRef] [PubMed]
- Meloni, A.; Maggio, A.; Positano, V.; Leto, F.; Angelini, A.; Putti, M.C.; Maresi, E.; Pucci, A.; Basso, C.; Perazzolo Marra, M.; et al. CMR for Myocardial Iron Overload Quantification: Calibration Curve from the MIOT Network. Eur. Radiol. 2020, 29, 2246–2252. [Google Scholar] [CrossRef] [PubMed]
- Mahrholdt, H.; Wagner, A.; Judd, R.M.; Sechtem, U.; Kim, R.J. Delayed enhancement cardiovascular magnetic resonance assessment of non-ischaemic cardiomyopathies. Eur. Heart J. 2005, 26, 1461–1474. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.; Barrett, C.J.; Crossman, D.J. Imaging tools for assessment of myocardial fibrosis in humans: The need for greater detail. Biophys. Rev. 2020, 12, 969–987. [Google Scholar] [CrossRef] [PubMed]
- Mahesh, B. Machine learning algorithms-a review. Int. J. Sci. Res. 2020, 9, 381–386. [Google Scholar]
- Pugliese, R.; Regondi, S.; Marini, R. Machine learning-based approach: Global trends, research directions, and regulatory standpoints. Data Sci. Manag. 2021, 4, 19–29. [Google Scholar] [CrossRef]
- Peng, J.; Jury, E.C.; Dönnes, P.; Ciurtin, C. Machine Learning Techniques for Personalised Medicine Approaches in Immune-Mediated Chronic Inflammatory Diseases: Applications and Challenges. Front. Pharmacol. 2021, 12, 720694. [Google Scholar] [CrossRef] [PubMed]
- Loftus, T.J.; Shickel, B.; Balch, J.A.; Tighe, P.J.; Abbott, K.L.; Fazzone, B.; Anderson, E.M.; Rozowsky, J.; Ozrazgat-Baslanti, T.; Ren, Y.; et al. Phenotype clustering in health care: A narrative review for clinicians. Front. Artif. Intell. 2022, 5, 842306. [Google Scholar] [CrossRef] [PubMed]
- Ezugwu, A.E.; Ikotun, A.M.; Oyelade, O.O.; Abualigah, L.; Agushaka, J.O.; Eke, C.I.; Akinyelu, A.A. A comprehensive survey of clustering algorithms: State-of-the-art machine learning applications, taxonomy, challenges, and future research prospects. Eng. Appl. Artif. Intell. 2022, 110, 104743. [Google Scholar] [CrossRef]
- Shah, S.J.; Katz, D.H.; Selvaraj, S.; Burke, M.A.; Yancy, C.W.; Gheorghiade, M.; Bonow, R.O.; Huang, C.C.; Deo, R.C. Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation 2015, 131, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Verdonschot, J.A.J.; Merlo, M.; Dominguez, F.; Wang, P.; Henkens, M.T.H.M.; Adriaens, M.E.; Hazebroek, M.R.; Masè, M.; Escobar, L.E.; Cobas-Paz, R.; et al. Phenotypic clustering of dilated cardiomyopathy patients highlights important pathophysiological differences. Eur. Heart J. 2020, 42, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Pezel, T.; Unterseeh, T.; Hovasse, T.; Asselin, A.; Lefèvre, T.; Chevalier, B.; Neylon, A.; Benamer, H.; Champagne, S.; Sanguineti, F.; et al. Phenotypic Clustering of Patients With Newly Diagnosed Coronary Artery Disease Using Cardiovascular Magnetic Resonance and Coronary Computed Tomography Angiography. Front. Cardiovasc. Med. 2021, 8, 760120. [Google Scholar] [CrossRef] [PubMed]
- Yamga, E.; Mullie, L.; Durand, M.; Cadrin-Chenevert, A.; Tang, A.; Montagnon, E.; Chartrand-Lefebvre, C.; Chassé, M. Interpretable clinical phenotypes among patients hospitalized with COVID-19 using cluster analysis. Front. Digit. Health 2023, 5, 1142822. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.-K.; Min, J.; Kim, H.W.; Ko, Y.; Oh, J.Y.; Jeong, Y.-J.; Kang, H.H.; Kang, J.Y.; Lee, S.-S.; Seo, M.; et al. Cluster analysis categorizes five phenotypes of pulmonary tuberculosis. Sci. Rep. 2022, 12, 10084. [Google Scholar] [CrossRef] [PubMed]
- Robinson, G.A.; Peng, J.; Dönnes, P.; Coelewij, L.; Naja, M.; Radziszewska, A.; Wincup, C.; Peckham, H.; Isenberg, D.A.; Ioannou, Y.; et al. Disease-associated and patient-specific immune cell signatures in juvenile-onset systemic lupus erythematosus: Patient stratification using a machine-learning approach. Lancet Rheumatol. 2020, 2, e485–e496. [Google Scholar] [CrossRef] [PubMed]
- Figgett, W.A.; Monaghan, K.; Ng, M.; Alhamdoosh, M.; Maraskovsky, E.; Wilson, N.J.; Hoi, A.Y.; Morand, E.F.; Mackay, F. Machine learning applied to whole-blood RNA-sequencing data uncovers distinct subsets of patients with systemic lupus erythematosus. Clin. Transl. Immunol. 2019, 8, e01093. [Google Scholar] [CrossRef] [PubMed]
- Positano, V.; Salani, B.; Pepe, A.; Santarelli, M.F.; De Marchi, D.; Ramazzotti, A.; Favilli, B.; Cracolici, E.; Midiri, M.; Cianciulli, P.; et al. Improved T2* assessment in liver iron overload by magnetic resonance imaging. Magn. Reson. Imaging 2009, 27, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Meloni, A.; Positano, V.; Pepe, A.; Rossi, G.; Dell’Amico, M.; Salvatori, C.; Keilberg, P.; Filosa, A.; Sallustio, G.; Midiri, M.; et al. Preferential patterns of myocardial iron overload by multislice multiecho T*2 CMR in thalassemia major patients. Magn. Reson. Med. 2010, 64, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Meloni, A.; Ramazzotti, A.; Positano, V.; Salvatori, C.; Mangione, M.; Marcheschi, P.; Favilli, B.; De Marchi, D.; Prato, S.; Pepe, A.; et al. Evaluation of a web-based network for reproducible T2* MRI assessment of iron overload in thalassemia. Int. J. Med. Inform. 2009, 78, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Jessup, M.; Abraham, W.T.; Casey, D.E.; Feldman, A.M.; Francis, G.S.; Ganiats, T.G.; Konstam, M.A.; Mancini, D.M.; Rahko, P.S.; Silver, M.A.; et al. 2009 focused update: ACCF/AHA Guidelines for the Diagnosis and Management of Heart Failure in Adults: A report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines: Developed in collaboration with the International Society for Heart and Lung Transplantation. Circulation 2009, 119, 1977–2016. [Google Scholar] [PubMed]
- Buxton, A.E.; Calkins, H.; Callans, D.J.; DiMarco, J.P.; Fisher, J.D.; Greene, H.L.; Haines, D.E.; Hayes, D.L.; Heidenreich, P.A.; Miller, J.M.; et al. ACC/AHA/HRS 2006 key data elements and definitions for electrophysiological studies and procedures: A report of the American College of Cardiology/American Heart Association Task Force on Clinical Data Standards (ACC/AHA/HRS Writing Committee to Develop Data Standards on Electrophysiology). Circulation 2006, 114, 2534–2570. [Google Scholar] [PubMed]
- Cogliandro, T.; Derchi, G.; Mancuso, L.; Mayer, M.C.; Pannone, B.; Pepe, A.; Pili, M.; Bina, P.; Cianciulli, P.; De Sanctis, V.; et al. Guideline recommendations for heart complications in thalassemia major. J. Cardiovasc. Med. 2008, 9, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Positano, V.; Pepe, A.; Santarelli, M.F.; Scattini, B.; De Marchi, D.; Ramazzotti, A.; Forni, G.; Borgna-Pignatti, C.; Lai, M.E.; Midiri, M.; et al. Standardized T2* map of normal human heart in vivo to correct T2* segmental artefacts. NMR Biomed. 2007, 20, 578–590. [Google Scholar] [CrossRef] [PubMed]
- Meloni, A.; Luciani, A.; Positano, V.; De Marchi, D.; Valeri, G.; Restaino, G.; Cracolici, E.; Caruso, V.; Dell’amico, M.C.; Favilli, B.; et al. Single region of interest versus multislice T2* MRI approach for the quantification of hepatic iron overload. J. Magn. Reson. Imaging 2011, 33, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.C.; Enriquez, C.; Ghugre, N.; Tyzka, J.M.; Carson, S.; Nelson, M.D.; Coates, T.D. MRI R2 and R2* mapping accurately estimates hepatic iron concentration in transfusion-dependent thalassemia and sickle cell disease patients. Blood 2005, 106, 1460–1465. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, M.D.; Weissman, N.J.; Dilsizian, V.; Jacobs, A.K.; Kaul, S.; Laskey, W.K.; Pennell, D.J.; Rumberger, J.A.; Ryan, T.; Verani, M.S. Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart. A statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Circulation 2002, 105, 539–542. [Google Scholar] [PubMed]
- Meloni, A.; Righi, R.; Missere, M.; Renne, S.; Schicchi, N.; Gamberini, M.R.; Cuccia, L.; Lisi, R.; Spasiano, A.; Roberti, M.G.; et al. Biventricular Reference Values by Body Surface Area, Age, and Gender in a Large Cohort of Well-Treated Thalassemia Major Patients Without Heart Damage Using a Multiparametric CMR Approach. J. Magn. Reson. Imaging 2021, 53, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Meloni, A.; Favilli, B.; Positano, V.; Cianciulli, P.; Filosa, A.; Quarta, A.; D’Ascola, D.; Restaino, G.; Lombardi, M.; Pepe, A. Safety of cardiovascular magnetic resonance gadolinium chelates contrast agents in patients with hemoglobinopaties. Haematologica 2009, 94, 1625–1627. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, B.; Skellam, J.G. A New Method for determining the Type of Distribution of Plant Individuals. Ann. Bot. 1954, 18, 213–227. [Google Scholar] [CrossRef]
- Abdi, H.; Valentin, D. Multiple Factor Analysis (MFA). In Encyclopedia of Measurement and Statistics; Sage Publications: Thousand Oaks, CA, USA, 2007. [Google Scholar]
- Jolliffe, I.T. Principal Component Analysis; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar]
- Josse, J. Principal Component Methods—Hierarchical Clustering—Partitional Clustering: Why Would We Need to Choose for Visualizing Data? Agrocampus Ouest: Rennes, France, 2010. [Google Scholar]
- Rigatti, S.J. Random Forest. J. Insur. Med. 2017, 47, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Parmar, A.; Katariya, R.; Patel, V. A Review on Random Forest: An Ensemble Classifier. In Proceedings of the International Conference on Intelligent Data Communication Technologies and Internet of Things (ICICI) 2018, Coimbatore, India, 7–8 August 2018; Springer International Publishing: Cham, Switzerland, 2019; pp. 758–763. [Google Scholar]
- Kawel-Boehm, N.; Maceira, A.; Valsangiacomo-Buechel, E.R.; Vogel-Claussen, J.; Turkbey, E.B.; Williams, R.; Plein, S.; Tee, M.; Eng, J.; Bluemke, D.A. Normal values for cardiovascular magnetic resonance in adults and children. J. Cardiovasc. Magn. Reson. 2015, 17, 29. [Google Scholar] [CrossRef] [PubMed]
- Petersen, S.E.; Aung, N.; Sanghvi, M.M.; Zemrak, F.; Fung, K.; Paiva, J.M.; Francis, J.M.; Khanji, M.Y.; Lukaschuk, E.; Lee, A.M.; et al. Reference ranges for cardiac structure and function using cardiovascular magnetic resonance (CMR) in Caucasians from the UK Biobank population cohort. J. Cardiovasc. Magn. Reson. 2017, 19, 18. [Google Scholar] [CrossRef] [PubMed]
- Meloni, A.; Detterich, J.; Berdoukas, V.; Pepe, A.; Lombardi, M.; Coates, T.D.; Wood, J.C. Comparison of biventricular dimensions and function between pediatric sickle-cell disease and thalassemia major patients without cardiac iron. Am. J. Hematol. 2013, 88, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Chung, A.K.; Das, S.R.; Leonard, D.; Peshock, R.M.; Kazi, F.; Abdullah, S.M.; Canham, R.M.; Levine, B.D.; Drazner, M.H. Women Have Higher Left Ventricular Ejection Fractions Than Men Independent of Differences in Left Ventricular Volume. Circulation 2006, 113, 1597–1604. [Google Scholar] [CrossRef] [PubMed]
- Yeon, S.B.; Salton, C.J.; Gona, P.; Chuang, M.L.; Blease, S.J.; Han, Y.; Tsao, C.W.; Danias, P.G.; Levy, D.; O’Donnell, C.J.; et al. Impact of age, sex, and indexation method on MR left ventricular reference values in the Framingham Heart Study offspring cohort. J. Magn. Reson. Imaging 2015, 41, 1038–1045. [Google Scholar] [CrossRef] [PubMed]
- Kerkhof, P.L.M.; Handly, N. Insights from physiology applied to interpretation of supranormal ejection fraction in women. Eur. Heart J. Cardiovasc. Imaging 2020, 21, 375–377. [Google Scholar] [CrossRef] [PubMed]
- Assomull, R.G.; Prasad, S.K.; Lyne, J.; Smith, G.; Burman, E.D.; Khan, M.; Sheppard, M.N.; Poole-Wilson, P.A.; Pennell, D.J. Cardiovascular magnetic resonance, fibrosis, and prognosis in dilated cardiomyopathy. J. Am. Coll. Cardiol. 2006, 48, 1977–1985. [Google Scholar] [CrossRef] [PubMed]
- Bruder, O.; Wagner, A.; Jensen, C.J.; Schneider, S.; Ong, P.; Kispert, E.-M.; Nassenstein, K.; Schlosser, T.; Sabin, G.V.; Sechtem, U.; et al. Myocardial Scar Visualized by Cardiovascular Magnetic Resonance Imaging Predicts Major Adverse Events in Patients With Hypertrophic Cardiomyopathy. J. Am. Coll. Cardiol. 2010, 56, 875–887. [Google Scholar] [CrossRef] [PubMed]
- Roes, S.D.; Kelle, S.; Kaandorp, T.A.; Kokocinski, T.; Poldermans, D.; Lamb, H.J.; Boersma, E.; van der Wall, E.E.; Fleck, E.; de Roos, A.; et al. Comparison of myocardial infarct size assessed with contrast-enhanced magnetic resonance imaging and left ventricular function and volumes to predict mortality in patients with healed myocardial infarction. Am. J. Cardiol. 2007, 100, 930–936. [Google Scholar] [CrossRef] [PubMed]
- Bing, R.; Dweck, M.R. Myocardial fibrosis: Why image, how to image and clinical implications. Heart 2019, 105, 1832–1840. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.C.; Weiss, R.G.; Thiemann, D.R.; Kitagawa, K.; Schmidt, A.; Dalal, D.; Lai, S.; Bluemke, D.A.; Gerstenblith, G.; Marbán, E.; et al. Late gadolinium enhancement by cardiovascular magnetic resonance heralds an adverse prognosis in nonischemic cardiomyopathy. J. Am. Coll. Cardiol. 2008, 51, 2414–2421. [Google Scholar] [CrossRef] [PubMed]
- Pepe, A.; Meloni, A.; Rossi, G.; Midiri, M.; Missere, M.; Valeri, G.; Sorrentino, F.; D’Ascola, D.G.; Spasiano, A.; Filosa, A.; et al. Prediction of cardiac complications for thalassemia major in the widespread cardiac magnetic resonance era: A prospective multicentre study by a multi-parametric approach. Eur. Heart J. Cardiovasc. Imaging 2018, 19, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Meloni, A.; Montanaro, D.; De Marchi, D.; Resta, M.; Keilberg, P.; Pistoia, L.; Positano, V.; Spasiano, A.; Casini, T.; De Bari, C.C.; et al. Absence of T1 Hyperintensity in the Brain of High-risk Patients After Multiple Administrations of High-dose Gadobutrol for Cardiac Magnetic Resonance. Clin. Neuroradiol. 2020, 31, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.C.; Ghugre, N. Magnetic resonance imaging assessment of excess iron in thalassemia, sickle cell disease and other iron overload diseases. Hemoglobin 2008, 32, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Meloni, A.; Martini, N.; Positano, V.; De Luca, A.; Pistoia, L.; Sbragi, S.; Spasiano, A.; Casini, T.; Bitti, P.P.; Allò, M.; et al. Myocardial iron overload by cardiovascular magnetic resonance native segmental T1 mapping: A sensitive approach that correlates with cardiac complications. J. Cardiovasc. Magn. Reson. 2021, 23, 70. [Google Scholar] [CrossRef] [PubMed]
- Vitrano, A.; Musallam, K.M.; Meloni, A.; Addario Pollina, S.; Karimi, M.; El-Beshlawy, A.; Hajipour, M.; Di Marco, V.; Ansari, S.H.; Filosa, A.; et al. Random Forest Clustering Identifies Three Subgroups of β-Thalassemia with Distinct Clinical Severity. Thalass. Rep. 2022, 12, 14–23. [Google Scholar]
- Bell, M.L.; Fiero, M.; Horton, N.J.; Hsu, C.-H. Handling missing data in RCTs; a review of the top medical journals. BMC Med. Res. Methodol. 2014, 14, 118. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, J.C.; Gluud, C.; Wetterslev, J.; Winkel, P. When and how should multiple imputation be used for handling missing data in randomised clinical trials—A practical guide with flowcharts. BMC Med. Res. Methodol. 2017, 17, 162. [Google Scholar] [CrossRef] [PubMed]



| All β-TI Patients (N = 138) | |
|---|---|
| Females, N (%) | 72 (52.2) |
| Age (years) | 38.1 ± 11.4 |
| Regular transfusions, N (%) | 73 (52.9) |
| Serum hemoglobin (g/dL) | 9.1 ± 1.1 |
| Serum ferritin (ng/L) | 813.8 ± 911.9 |
| MRI LIC (mg/g/dw) | 7.9 ± 7.7 |
| Global heart T2* (ms) | 35.9 ± 8.4 |
| LV EDVI (mL/m2) | 97.1 ± 20.6 |
| LV mass index (g/m2) | 63.6 ± 12.9 |
| LV EF (%) | 62.8 ± 5.9 |
| RV EDVI (mL/m2) | 90.0 ± 22.2 |
| RV mass index (g/m2) | 22.7 ± 6.2 |
| RV EF (%) | 65.3 ± 6.4 |
| Replacement myocardial fibrosis, N(%) | 37 (26.8) |
| LA area index (cm2/m2) | 14.2 ± 2.6 |
| RA area index (cm2/m2) | 13.1 ± 2.5 |
| Dimension | Variance/ Cumulative Variance (%) | Variables with Contribution > 5% |
|---|---|---|
| 1 | 22.3/22.3 | Sex, LV EDVI, RV EDVI, LV mass index, RV mass index |
| 2 | 16.5/38.8 | Regular transfusions, replacement myocardial fibrosis |
| 3 | 13.2/52.0 | Replacement myocardial fibrosis, regular transfusions |
| 4 | 11.4/63.4 | Sex, RA area index, LA area index, RV EDVI, hemoglobin, LV EDVI, RV mass index |
| 5 | 7.9/71.3 | Ferritin, RV EF |
| 6 | 6.9/78.2 | Age, LV EF, RV EF, ferritin |
| 7 | 4.6/82.8 | Age, LA area index, RA area index, hemoglobin, RV EF, RV mass index |
| Phenogroup 1 (N = 56) | Phenogroup 2 (N = 37) | Phenogroup 3 (N = 45) | p-Value | Pairwise Comparisons | |
|---|---|---|---|---|---|
| Females, N (%) | 55 (98.2) | 17 (45.9) | 0 (0.0) | <0.0001 | 1 vs. 2: p < 0.0001 1 vs. 3: p < 0.0001 2 vs. 3: p < 0.0001 |
| Age (years) | 36.6 ± 12.3 | 39.8 ± 9.9 | 38.8 ± 11.5 | 0.380 | |
| Regular transfusions, N (%) | 34 (60.7) | 21 (56.8) | 18 (40.0) | 0.100 | |
| Serum hemoglobin (g/dL) | 8.9 ± 0.8 | 9.0 ± 0.9 | 9.3 ± 1.4 | 0.349 | |
| Serum ferritin (ng/L) | 860.1 ± 956.1 | 977.9 ± 1142.9 | 621.1 ± 560.7 | 0.351 | |
| MRI LIC (mg/g/dw) | 8.5 ± 8.1 | 8.7 ± 8.4 | 6.5 ± 6.4 | 0.228 | |
| Global heart T2* (ms) | 36.3 ± 8.5 | 34.6 ± 10.6 | 36.6 ± 6.2 | 0.931 | |
| LV EDVI (ml/m2) | 87.6 ± 15.1 | 101.2 ± 20.8 | 105.7 ± 21.8 | <0.0001 | 1 vs. 2: p = 0.003 1 vs. 3: p < 0.0001 |
| LV mass index (g/m2) | 56.7 ± 10.5 | 65.6 ± 12.2 | 70.6 ± 11.8 | <0.0001 | 1 vs. 2: p = 0.001 1 vs. 3: p < 0.0001 |
| LV EF (%) | 64.9 ± 6.7 | 61.7 ± 4.9 | 61.0 ± 4.8 | 0.005 | 1 vs. 2: p = 0.045 1 vs. 3: p = 0.009 |
| RV EDVI (mL/m2) | 80.3 ± 17.2 | 96.3 ± 23.4 | 97.0 ± 22.6 | <0.0001 | 1 vs. 2: p = 0.001 1 vs. 3: p < 0.0001 |
| RV mass index (g/m2) | 20.3 ± 3.8 | 24.7 ± 8.1 | 23.9 ± 6.0 | <0.0001 | 1 vs. 2: p = 0.009 1 vs. 3: p < 0.0001 |
| RV EF (%) | 66.3 ± 7.1 | 63.9 ± 5.5 | 65.1 ± 6.2 | 0.201 | |
| Replacement myocardial fibrosis, N (%) | 0 (0.0) | 37 (100.0) | 0 (0.0) | <0.0001 | 1 vs. 2: p < 0.0001 2 vs. 3: p < 0.0001 |
| LA area index (cm2/m2) | 14.0 ± 2.6 | 14.3 ± 3.1 | 14.4 ± 2.3 | 0.765 | |
| RA area index (cm2/m2) | 12.7 ± 2.5 | 12.7 ± 2.5 | 13.8 ± 2.4 | 0.053 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meloni, A.; Parravano, M.; Pistoia, L.; Cossu, A.; Grassedonio, E.; Renne, S.; Fina, P.; Spasiano, A.; Salvo, A.; Bagnato, S.; et al. Phenotypic Clustering of Beta-Thalassemia Intermedia Patients Using Cardiovascular Magnetic Resonance. J. Clin. Med. 2023, 12, 6706. https://doi.org/10.3390/jcm12216706
Meloni A, Parravano M, Pistoia L, Cossu A, Grassedonio E, Renne S, Fina P, Spasiano A, Salvo A, Bagnato S, et al. Phenotypic Clustering of Beta-Thalassemia Intermedia Patients Using Cardiovascular Magnetic Resonance. Journal of Clinical Medicine. 2023; 12(21):6706. https://doi.org/10.3390/jcm12216706
Chicago/Turabian StyleMeloni, Antonella, Michela Parravano, Laura Pistoia, Alberto Cossu, Emanuele Grassedonio, Stefania Renne, Priscilla Fina, Anna Spasiano, Alessandra Salvo, Sergio Bagnato, and et al. 2023. "Phenotypic Clustering of Beta-Thalassemia Intermedia Patients Using Cardiovascular Magnetic Resonance" Journal of Clinical Medicine 12, no. 21: 6706. https://doi.org/10.3390/jcm12216706







