Clinical Evaluation of Patients with Genetically Confirmed Familial Hypercholesterolemia
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Genetic Testing
2.3. LDLR Intron 16 Variant, Transcript Analysis
2.4. Statistical Analysis
3. Results
4. Discussion
5. Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Visseren, F.L.J.; Mach, F.; Smulders, Y.M.; Carballo, D.; Koskinas, K.C.; Bäck, M.; Benetos, A.; Biffi, A.; Boavida, J.-M.; Capodanno, D.; et al. 2021 ESC Guidelines on cardiovascular disease prevention in clinical practice. Eur. Heart J. 2021, 42, 3227–3337. [Google Scholar] [CrossRef] [PubMed]
- Ference, B.A.; Ginsberg, H.N.; Graham, I.; Ray, K.K.; Packard, C.J.; Bruckert, E.; Hegele, R.A.; Krauss, R.M.; Raal, F.J.; Schunkert, H.; et al. Low-density lipoproteins cause atherosclerotic cardiovascular disease. 1. Evidence from genetic, epidemiologic, and clinical studies. A consensus statement from the European Atherosclerosis Society Consensus Panel. Eur. Heart J. 2017, 38, 2459–2472. [Google Scholar] [CrossRef] [PubMed]
- Cholesterol Treatment Trialists’ (CTT) Collaborators; Mihaylova, B.; Emberson, J.; Blackwell, L.; Keech, A.; Simes, J.; Barnes, E.H.; Voysey, M.; Gray, A.; Collins, R.; et al. The effects of lowering LDL cholesterol with statin therapy in people at low risk of vascular disease: Meta-analysis of individual data from 27 randomised trials. Lancet 2012, 380, 581–590. [Google Scholar] [PubMed]
- Versmissen, J.; Oosterveer, D.M.; Yazdanpanah, M.; Defesche, J.C.; Basart, D.C.G.; Liem, A.H.; Heeringa, J.; Witteman, J.C.; Lansberg, P.J.; Kastelein, J.J.P.; et al. Efficacy of statins in familial hypercholesterolaemia: A long term cohort study. BMJ 2008, 337, a2423. [Google Scholar] [CrossRef]
- Neil, A.; Cooper, J.; Betteridge, J.; Capps, N.; McDowell, I.; Durrington, P.; Seed, M.; Humphries, S.E. Reductions in all-cause, cancer, and coronary mortality in statin-treated patients with heterozygous familial hypercholesterolaemia: A prospective registry study. Eur. Heart J. 2008, 29, 2625–2633. [Google Scholar] [CrossRef]
- de Ferranti, S.D.; Rodday, A.M.; Mendelson, M.M.; Wong, J.B.; Leslie, L.K.; Sheldrick, R.C. Prevalence of Familial Hypercholesterolemia in the 1999 to 2012 United States National Health and Nutrition Examination Surveys (NHANES). Circulation 2016, 133, 1067–1072. [Google Scholar] [CrossRef]
- Defesche, J.C.; Lansberg, P.J.; Umans-Eckenhausen, M.A.W.; Kastelein, J.J.P. Advanced method for the identification of patients with inherited hypercholesterolemia. Semin. Vasc. Med. 2004, 4, 59–65. [Google Scholar] [CrossRef]
- Williams, R.R.; Hunt, S.C.; Schumacher, M.C.; Hegele, R.A.; Leppert, M.F.; Ludwig, E.H.; Hopkins, P.N. Diagnosing heterozygous familial hypercholesterolemia using new practical criteria validated by molecular genetics. Am. J. Cardiol. 1993, 72, 171–176. [Google Scholar] [CrossRef]
- Scientific Steering Committee on behalf of the Simon Broome Register Group. Risk of fatal coronary heart disease in familial hypercholesterolaemia. BMJ 1991, 303, 893–896. [Google Scholar] [CrossRef]
- Sturm, A.C.; Knowles, J.W.; Gidding, S.S.; Ahmad, Z.S.; Ahmed, C.D.; Ballantyne, C.M.; Baum, S.J.; Bourbon, M.; Carrié, A.; Cuchel, M.; et al. Clinical Genetic Testing for Familial Hypercholesterolemia: JACC Scientific Expert Panel. J. Am. Coll. Cardiol. 2018, 72, 662–680. [Google Scholar] [CrossRef]
- Humphries, S.E.; Norbury, G.; Leigh, S.; Hadfield, S.G.; Nair, D. What is the clinical utility of DNA testing in patients with familial hypercholesterolaemia? Curr. Opin. Lipidol. 2008, 19, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Watts, G.F.; Gidding, S.; Wierzbicki, A.S.; Toth, P.P.; Alonso, R.; Brown, W.V.; Bruckert, E.; Defesche, J.; Lin, K.K.; Livingston, M.; et al. Integrated guidance on the care of familial hypercholesterolaemia from the International FH Foundation. Int. J. Cardiol. 2014, 171, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Perez de Isla, L.; Alonso, R.; Watts, G.F.; Mata, N.; Saltijeral Cerezo, A.; Muñiz, O.; Fuentes, F.; Diaz-Diaz, J.L.; de Andrés, R.; Zambón, D.; et al. Attainment of LDL-Cholesterol Treatment Goals in Patients With Familial Hypercholesterolemia: 5-Year SAFEHEART Registry Follow-Up. J. Am. Coll. Cardiol. 2016, 67, 1278–1285. [Google Scholar] [CrossRef] [PubMed]
- Sjouke, B.; Kusters, D.M.; Kindt, I.; Besseling, J.; Defesche, J.C.; Sijbrands, E.J.G.; Roeters van Lennep, J.E.; Stalenhoef, A.F.H.; Wiegman, A.; de Graaf, J.; et al. Homozygous autosomal dominant hypercholesterolaemia in the Netherlands: Prevalence, genotype-phenotype relationship, and clinical outcome. Eur. Heart J. 2015, 36, 560–565. [Google Scholar] [CrossRef]
- Goldstein, J.L.; Schrott, H.G.; Hazzard, W.R.; Bierman, E.L.; Motulsky, A.G. Hyperlipidemia in coronary heart disease. II. Genetic analysis of lipid levels in 176 families and delineation of a new inherited disorder, combined hyperlipidemia. J. Clin. Investig. 1973, 52, 1544–1568. [Google Scholar] [CrossRef]
- Zimmerman, J.; Duprez, D.; Veach, P.M.; Zierhut, H.A. Barriers to the identification of familial hypercholesterolemia among primary care providers. J. Community Genet. 2019, 10, 229–236. [Google Scholar] [CrossRef]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucl. Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef]
- Gómez, J.; Reguero, J.R.; Morís, C.; Martín, M.; Alvarez, V.; Alonso, B.; Iglesias, S.; Coto, E. Mutation Analysis of the Main Hypertrophic Cardiomyopathy Genes Using Multiplex Amplification and Semiconductor Next-Generation Sequencing. Circ. J. 2014, 78, 2963–2971. [Google Scholar] [CrossRef] [PubMed]
- Gómez, J.; Lorca, R.; Reguero, J.R.; Morís, C.; Martín, M.; Tranche, S.; Alonso, B.; Iglesias, S.; Alvarez, V.; Díaz-Molina, B.; et al. Screening of the Filamin C Gene in a Large Cohort of Hypertrophic Cardiomyopathy Patients. Circ. Cardiovasc. Genet. 2017, 10, e001584. [Google Scholar] [CrossRef]
- Lorca, R.; Aparicio, A.; Cuesta-Llavona, E.; Pascual, I.; Junco, A.; Hevia, S.; Villazón, F.; Hernandez-Vaquero, D.; Rodríguez Reguero, J.J.; Moris, C.; et al. Familial Hypercholesterolemia in Premature Acute Coronary Syndrome. Insights from CholeSTEMI Registry. J. Clin. Med. 2020, 9, 3489. [Google Scholar] [CrossRef]
- Lorca, R.; Junco-Vicente, A.; Pérez-Pérez, A.; Pascual, I.; Persia-Paulino, Y.R.; González-Urbistondo, F.; Cuesta-Llavona, E.; Fernández-Barrio, B.C.; Morís, C.; Rubín, J.M.; et al. KCNH2 p.Gly262AlafsTer98: A New Threatening Variant Associated with Long QT Syndrome in a Spanish Cohort. Life 2022, 12, 556. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Dedoussis, G.V.Z.; Schmidt, H.; Genschel, J. LDL-receptor mutations in Europe. Hum. Mutat. 2004, 24, 443–459. [Google Scholar] [CrossRef] [PubMed]
- Leigh, S.; Futema, M.; Whittall, R.; Taylor-Beadling, A.; Williams, M.; den Dunnen, J.T.; Humphries, S.E. The UCL low-density lipoprotein receptor gene variant database: Pathogenicity update. J. Med. Genet. 2017, 54, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Iacocca, M.A.; Hegele, R.A. Recent advances in genetic testing for familial hypercholesterolemia. Expert. Rev. Mol. Diagn. 2017, 17, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.; Wang, D.; Patel, K.; Whittall, R.; Wood, G.; Farrer, M.; Neely, R.D.G.; Fairgrieve, S.; Nair, D.; Barbir, M.; et al. Mutation detection rate and spectrum in familial hypercholesterolaemia patients in the UK pilot cascade project. Clin. Genet. 2010, 77, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Humphries, S.E.; Whittall, R.A.; Hubbart, C.S.; Maplebeck, S.; Cooper, J.A.; Soutar, A.K.; Naoumova, R.; Thompson, G.R.; Seed, M.; Durrington, P.N.; et al. Genetic causes of familial hypercholesterolaemia in patients in the UK: Relation to plasma lipid levels and coronary heart disease risk. J. Med. Genet. 2006, 43, 943–949. [Google Scholar] [CrossRef]
- Kassner, U.; Wühle-Demuth, M.; Missala, I.; Humphries, S.E.; Steinhagen-Thiessen, E.; Demuth, I. Clinical utility gene card for: Hyperlipoproteinemia, TYPE II. Eur. J. Hum. Genet. 2014, 22, 953. [Google Scholar] [CrossRef]
- Hopkins, P.N.; Defesche, J.; Fouchier, S.W.; Bruckert, E.; Luc, G.; Cariou, B.; Sjouke, B.; Leren, T.P.; Harada-Shiba, M.; Mabuchi, H.; et al. Characterization of Autosomal Dominant Hypercholesterolemia Caused by PCSK9 Gain of Function Mutations and Its Specific Treatment With Alirocumab, a PCSK9 Monoclonal Antibody. Circ. Cardiovasc. Genet. 2015, 8, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Naoumova, R.P.; Tosi, I.; Patel, D.; Neuwirth, C.; Horswell, S.D.; Marais, A.D.; van Heyningen, C.; Soutar, A.K. Severe hypercholesterolemia in four British families with the D374Y mutation in the PCSK9 gene: Long-term follow-up and treatment response. Arter. Thromb. Vasc. Biol. 2005, 25, 2654–2660. [Google Scholar] [CrossRef]
- Myant, N.B.; Forbes, S.A.; Day, I.N.; Gallagher, J. Estimation of the age of the ancestral arginine3500-->glutamine mutation in human apoB-100. Genomics 1997, 45, 78–87. [Google Scholar] [CrossRef]
- Andersen, L.H.; Miserez, A.R.; Ahmad, Z.; Andersen, R.L. Familial defective apolipoprotein B-100: A review. J. Clin. Lipidol. 2016, 10, 1297–1302. [Google Scholar] [CrossRef]
- Motazacker, M.M.; Pirruccello, J.; Huijgen, R.; Do, R.; Gabriel, S.; Peter, J.; Kuivenhoven, J.A.; Defesche, J.C.; Kastelein, J.J.P.; Hovingh, G.K.; et al. Advances in genetics show the need for extending screening strategies for autosomal dominant hypercholesterolaemia. Eur. Heart J. 2012, 33, 1360–1366. [Google Scholar] [CrossRef]
- Thomas, E.R.A.; Atanur, S.S.; Norsworthy, P.J.; Encheva, V.; Snijders, A.P.; Game, L.; Vandrovcova, J.; Siddiq, A.; Seed, M.; Soutar, A.K.; et al. Identification and biochemical analysis of a novel APOB mutation that causes autosomal dominant hypercholesterolemia. Mol. Genet. Genom. Med. 2013, 1, 155–161. [Google Scholar] [CrossRef]
- Pullinger, C.R.; Hennessy, L.K.; Chatterton, J.E.; Liu, W.; Love, J.A.; Mendel, C.M.; Frost, P.H.; Malloy, M.J.; Schumaker, V.N.; Kane, J.P. Familial ligand-defective apolipoprotein B. Identification of a new mutation that decreases LDL receptor binding affinity. J. Clin. Investig. 1995, 95, 1225–1234. [Google Scholar] [CrossRef]
- Rabès, J.P.; Varret, M.; Devillers, M.; Aegerter, P.; Villéger, L.; Krempf, M.; Junien, C.; Boileau, C. R3531C mutation in the apolipoprotein B gene is not sufficient to cause hypercholesterolemia. Arter. Thromb. Vasc. Biol. 2000, 20, E76–E82. [Google Scholar] [CrossRef]
- Tybjaerg-Hansen, A.; Steffensen, R.; Meinertz, H.; Schnohr, P.; Nordestgaard, B.G. Association of mutations in the apolipoprotein B gene with hypercholesterolemia and the risk of ischemic heart disease. N. Engl. J. Med. 1998, 338, 1577–1584. [Google Scholar] [CrossRef]
- deGoma, E.M.; Ahmad, Z.S.; O’Brien, E.C.; Kindt, I.; Shrader, P.; Newman, C.B.; Pokharel, Y.; Baum, S.J.; Hemphill, L.C.; Hudgins, L.C.; et al. Treatment Gaps in Adults With Heterozygous Familial Hypercholesterolemia in the United States: Data from the CASCADE-FH Registry. Circ. Cardiovasc. Genet. 2016, 9, 240–249. [Google Scholar] [CrossRef]
- Iyen, B.; Qureshi, N.; Weng, S.; Roderick, P.; Kai, J.; Capps, N.; Durrington, P.N.; McDowell, I.F.; Soran, H.; Neil, A.; et al. Sex differences in cardiovascular morbidity associated with familial hypercholesterolaemia: A retrospective cohort study of the UK Simon Broome register linked to national hospital records. Atherosclerosis 2020, 315, 131–137. [Google Scholar] [CrossRef]
- Amor-Salamanca, A.; Castillo, S.; Gonzalez-Vioque, E.; Dominguez, F.; Quintana, L.; Lluís-Ganella, C.; Escudier, J.M.; Ortega, J.; Lara-Pezzi, E.; Alonso-Pulpon, L.; et al. Genetically Confirmed Familial Hypercholesterolemia in Patients with Acute Coronary Syndrome. J. Am. Coll. Cardiol. 2017, 70, 1732–1740. [Google Scholar] [CrossRef]
- Silva, P.R.S.; Jannes, C.E.; Oliveira, T.G.M.; Miname, M.H.; Rocha, V.Z.; Chacra, A.P.; Gurgel, M.H.C.; Montenegro, R.M.; Rodrigues Sobrinho, C.R.M.; Bello Moreira, A.S.; et al. Evaluation of clinical and laboratory parameters used in the identification of index cases for genetic screening of familial hypercholesterolemia in Brazil. Atherosclerosis 2017, 263, 257–262. [Google Scholar] [CrossRef]
- Haralambos, K.; Whatley, S.D.; Edwards, R.; Gingell, R.; Townsend, D.; Ashfield-Watt, P.; Lansberg, P.; Datta, D.B.N.; McDowell, I.F.W. Clinical experience of scoring criteria for Familial Hypercholesterolaemia (FH) genetic testing in Wales. Atherosclerosis 2015, 240, 190–196. [Google Scholar] [CrossRef]
- Khera, A.V.; Won, H.-H.; Peloso, G.M.; Lawson, K.S.; Bartz, T.M.; Deng, X.; van Leeuwen, E.M.; Natarajan, P.; Emdin, C.A.; Bick, A.G.; et al. Diagnostic Yield and Clinical Utility of Sequencing Familial Hypercholesterolemia Genes in Patients With Severe Hypercholesterolemia. J. Am. Coll. Cardiol. 2016, 67, 2578–2589. [Google Scholar] [CrossRef]
- Raal, F.J.; Pilcher, G.J.; Panz, V.R.; van Deventer, H.E.; Brice, B.C.; Blom, D.J.; Marais, A.D. Reduction in mortality in subjects with homozygous familial hypercholesterolemia associated with advances in lipid-lowering therapy. Circulation 2011, 124, 2202–2207. [Google Scholar] [CrossRef]
- Slack, J. Risks of ischaemic heart-disease in familial hyperlipoproteinaemic states. Lancet 1969, 2, 1380–1382. [Google Scholar] [CrossRef]
- Marks, D.; Thorogood, M.; Neil, H.A.W.; Humphries, S.E. A review on the diagnosis, natural history, and treatment of familial hypercholesterolaemia. Atherosclerosis 2003, 168, 1–14. [Google Scholar] [CrossRef]
- Humphries, S.E.; Cooper, J.A.; Seed, M.; Capps, N.; Durrington, P.N.; Jones, B.; McDowell, I.F.W.; Soran, H.; Neil, H.a.W. Simon Broome Familial Hyperlipidaemia Register Group Coronary heart disease mortality in treated familial hypercholesterolaemia: Update of the UK Simon Broome FH register. Atherosclerosis 2018, 274, 41–46. [Google Scholar] [CrossRef]
- Zamora, A.; Masana, L.; Comas-Cufí, M.; Vila, À.; Plana, N.; García-Gil, M.; Alves-Cabratosa, L.; Marrugat, J.; Roman, I.; Ramos, R.; et al. Familial hypercholesterolemia in a European Mediterranean population-Prevalence and clinical data from 2.5 million primary care patients. J. Clin. Lipidol. 2017, 11, 1013–1022. [Google Scholar] [CrossRef]
- Amrock, S.M.; Duell, P.B.; Knickelbine, T.; Martin, S.S.; O’Brien, E.C.; Watson, K.E.; Mitri, J.; Kindt, I.; Shrader, P.; Baum, S.J.; et al. Health disparities among adult patients with a phenotypic diagnosis of familial hypercholesterolemia in the CASCADE-FHTM patient registry. Atherosclerosis 2017, 267, 19–26. [Google Scholar] [CrossRef]
- O’Keeffe, A.G.; Nazareth, I.; Petersen, I. Time trends in the prescription of statins for the primary prevention of cardiovascular disease in the United Kingdom: A cohort study using The Health Improvement Network primary care data. Clin. Epidemiol. 2016, 8, 123–132. [Google Scholar] [CrossRef]
- Maas, A.H.E.M.; Appelman, Y.E.A. Gender differences in coronary heart disease. Neth. Heart J. 2010, 18, 598–602. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Khambhati, J.; Varghese, T.; Stahl, E.P.; Kumar, S.; Sandesara, P.B.; Wenger, N.K.; Sperling, L.S. Comprehensive primary prevention of cardiovascular disease in women. Clin. Cardiol. 2017, 40, 832–838. [Google Scholar] [CrossRef] [PubMed]
- Gidding, S.S.; Champagne, M.A.; de Ferranti, S.D.; Defesche, J.; Ito, M.K.; Knowles, J.W.; McCrindle, B.; Raal, F.; Rader, D.; Santos, R.D.; et al. The Agenda for Familial Hypercholesterolemia: A Scientific Statement From the American Heart Association. Circulation 2015, 132, 2167–2192. [Google Scholar] [CrossRef]
- Goldberg, A.C.; Robinson, J.G.; Cromwell, W.C.; Ross, J.L.; Ziajka, P.E. Future issues, public policy, and public awareness of familial hypercholesterolemias: Recommendations from the National Lipid Association Expert Panel on Familial Hypercholesterolemia. J. Clin. Lipidol. 2011, 5, S46–S51. [Google Scholar] [CrossRef] [PubMed]
- Brett, T.; Qureshi, N.; Gidding, S.; Watts, G.F. Screening for familial hypercholesterolaemia in primary care: Time for general practice to play its part. Atherosclerosis 2018, 277, 399–406. [Google Scholar] [CrossRef]
- Wald, D.S.; Bestwick, J.P.; Morris, J.K.; Whyte, K.; Jenkins, L.; Wald, N.J. Child-Parent Familial Hypercholesterolemia Screening in Primary Care. N. Engl. J. Med. 2016, 375, 1628–1637. [Google Scholar] [CrossRef]
- Beheshti, S.O.; Madsen, C.M.; Varbo, A.; Nordestgaard, B.G. Worldwide Prevalence of Familial Hypercholesterolemia: Meta-Analyses of 11 Million Subjects. J. Am. Coll. Cardiol. 2020, 75, 2553–2566. [Google Scholar] [CrossRef]


| Gene | Transcript | Protein | cDNA | gnomAD v2.1 (European Non-Finnish) | Carriers | ACMG |
|---|---|---|---|---|---|---|
| LDLR | NM_000527 | (p.Glu31Ter) | c.91G>T | - | 1 | P |
| LDLR | NM_000527 | (p.Cys155Tyr) | c.464G>A | - | 1 | P |
| LDLR | NM_000527 | (p.Glu174Ter) | c.520G>T | - | 1 | P |
| LDLR | NM_000527 | (p.Ser177Leu) | c.530C>T | - | 1 | P |
| LDLR | NM_000527 | (p.Glu288Lys) | c.862G>A | - | 1 | P |
| LDLR | NM_000527 | (p.Asp354Asn) | c.1060G>A | - | 1 | LP |
| LDLR | NM_000527 | (p.Arg406Trp) | c.1216C>T | - | 1 | P |
| LDLR | NM_000527 | (p.Arg416Trp) | c.1246C>T | 1/113,496 | 3 | P |
| LDLR | NM_000527 | (p.Val429Met) | c.1285G>A | 1/113,592 | 11 | P |
| LDLR | NM_000527 | (p.Trp490Ter) | c.1470G>A | - | 1 | P |
| LDLR | NM_000527 | (p.Phe530SerfsTer20) | c.1589_1614del | 2 | P | |
| LDLR | NM_000527 | (p.Gly592Glu) | c.1775G>A | 15/129,176 | 7 | P |
| LDLR | NM_000527 | (p.Leu599Ser) | c.1796T>C | - | 1 | LP |
| LDLR | NM_000527 | (p.Glu602Ter) | c.1804G>T | - | 1 | P |
| LDLR | NM_000527 | (p.Phe655Leu) | c.1965C>G | - | 1 | LP |
| LDLR | NM_000527 | (p.Leu658Pro) | c.1973T>C | - | 2 | LP |
| LDLR | NM_000527 | (p.Asp700Gly) | c.2099A>G | - | 1 | LP |
| LDLR | NM_000527 | (p.Leu799PhefsTer127) | c.2395_2404del | 1 | P | |
| LDLR | NM_000527 | (p.Arg814Gln) | c.2441G>A | - | 2 | LP |
| LDLR | NM_000527 | Splicing | c.313+2insT | - | 2 | P |
| LDLR | NM_000527 | Splicing | c.1987+1G>A | - | 1 | P |
| LDLR | NM_000527 | Splicing | c.1988-2A>T | - | 1 | P |
| LDLR | NM_000527 | Splicing | c.2389+4A>G | 1/113,644 | 11 | P |
| LDLR | NM_000527 | Copy number variant | c.2390-2583del | - | 1 | P |
| LDLRAP1 | NM_015627 | p.Ala70ProfsTer19 | c.207delC | - | 1 | P |
| APOB | NM_000384 | (p.Thr1558Ala) | c.4672A>G | - | 1 | VUS |
| APOB | NM_000384 | (p.Asp1908Asn) | c.5722G>A | 2/129,088 | 1 | VUS |
| LDLR | NM_000527 | (p.Asn297His) | c.889A>C | - | 1 | VUS |
| LDLR | NM_000527 | (p.Ala606Ser) | c.1816G>T | 30/129,154 | 1 | VUS |
| LDLR | NM_000527 | (p.Hys656Asn) | c.1966C>A | 3/113,732 | 1 | VUS |
| LDLR | NM_000527 | (p.Arg253Gln) | c.758G>A | - | 1 | VUS |
| Identified Patients (58) | N/Mean | Frequency/SD |
|---|---|---|
| Men | 31 | 53.5% |
| Women | 27 | 46.5% |
| Current age (years), mean ± SD | 51 | ±19 SD |
| Age at dyslipidaemia diagnosis | 29 | ±17 SD |
| Age at definite genetic diagnosis | 48 | ±19 SD |
| Other cardiovascular risk factors | ||
| Previous smoker/ Current smoker | 13 | 22% |
| High BP | 11 | 19% |
| DM | 8 | 14% |
| At least 1 cardiovascular risk factor | 26 | 45% |
| Kidney failure | 1 | 2% |
| Peripheral vascular disease | 1 | 2% |
| Personal history of PCVD | 3 | 5% |
| Family history of PCVD | 24 | 41% |
| Family history of hypercholesteremia | 53 | 91% |
| Corneal arcus and/or tendon xanthomas | 9 | 15.5% |
| Lipid profile | ||
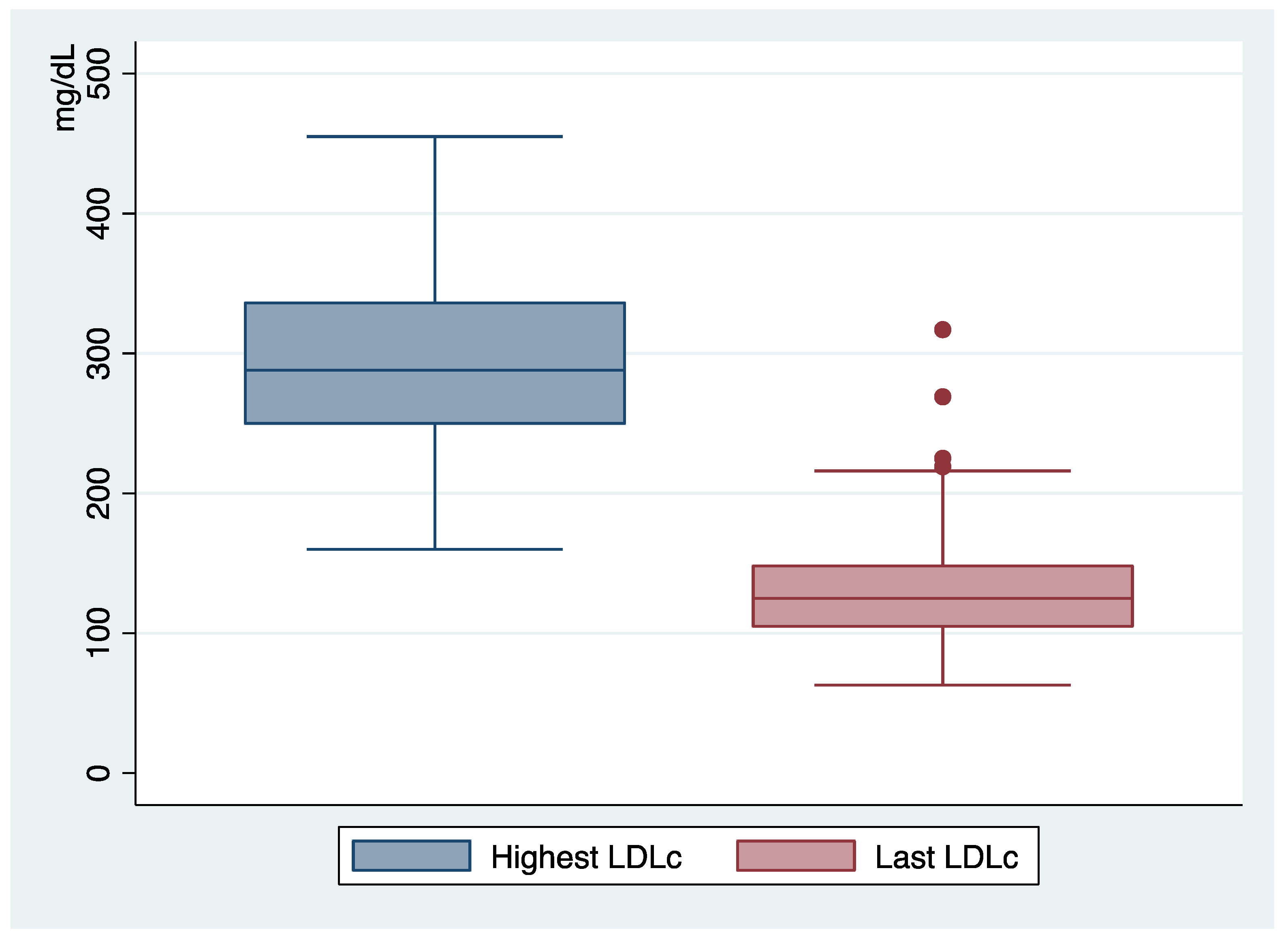
| Highest LDLc level (mg/dL) | 294 | ±65 SD |
| Last LDLc level (mg/dL) | 133 | ±50 SD |
| LpA (nmol/L) | 34 | ±74 SD |
| Medical treatment | ||
| None | 3 | 5% |
| Statins | 9 | 15.5% |
| Statins + ezetimibe | 39 | 67% |
| Statins + ezetimibe + IPCSK9 | 7 | 12% |
| DLCN criteria | ||
| <3 | 6 | 10% |
| 3–5 possible | 8 | 14% |
| 6–8 probable | 21 | 36% |
| >8 definite | 23 | 40% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aparicio, A.; Villazón, F.; Suárez-Gutiérrez, L.; Gómez, J.; Martínez-Faedo, C.; Méndez-Torre, E.; Avanzas, P.; Álvarez-Velasco, R.; Cuesta-Llavona, E.; García-Lago, C.; et al. Clinical Evaluation of Patients with Genetically Confirmed Familial Hypercholesterolemia. J. Clin. Med. 2023, 12, 1030. https://doi.org/10.3390/jcm12031030
Aparicio A, Villazón F, Suárez-Gutiérrez L, Gómez J, Martínez-Faedo C, Méndez-Torre E, Avanzas P, Álvarez-Velasco R, Cuesta-Llavona E, García-Lago C, et al. Clinical Evaluation of Patients with Genetically Confirmed Familial Hypercholesterolemia. Journal of Clinical Medicine. 2023; 12(3):1030. https://doi.org/10.3390/jcm12031030
Chicago/Turabian StyleAparicio, Andrea, Francisco Villazón, Lorena Suárez-Gutiérrez, Juan Gómez, Ceferino Martínez-Faedo, Edelmiro Méndez-Torre, Pablo Avanzas, Rut Álvarez-Velasco, Elías Cuesta-Llavona, Claudia García-Lago, and et al. 2023. "Clinical Evaluation of Patients with Genetically Confirmed Familial Hypercholesterolemia" Journal of Clinical Medicine 12, no. 3: 1030. https://doi.org/10.3390/jcm12031030
APA StyleAparicio, A., Villazón, F., Suárez-Gutiérrez, L., Gómez, J., Martínez-Faedo, C., Méndez-Torre, E., Avanzas, P., Álvarez-Velasco, R., Cuesta-Llavona, E., García-Lago, C., Neuhalfen, D., Coto, E., & Lorca, R. (2023). Clinical Evaluation of Patients with Genetically Confirmed Familial Hypercholesterolemia. Journal of Clinical Medicine, 12(3), 1030. https://doi.org/10.3390/jcm12031030







