ESKD Risk Prediction Model in a Multicenter Chronic Kidney Disease Cohort in China: A Derivation, Validation, and Comparison Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Approval and Declaration
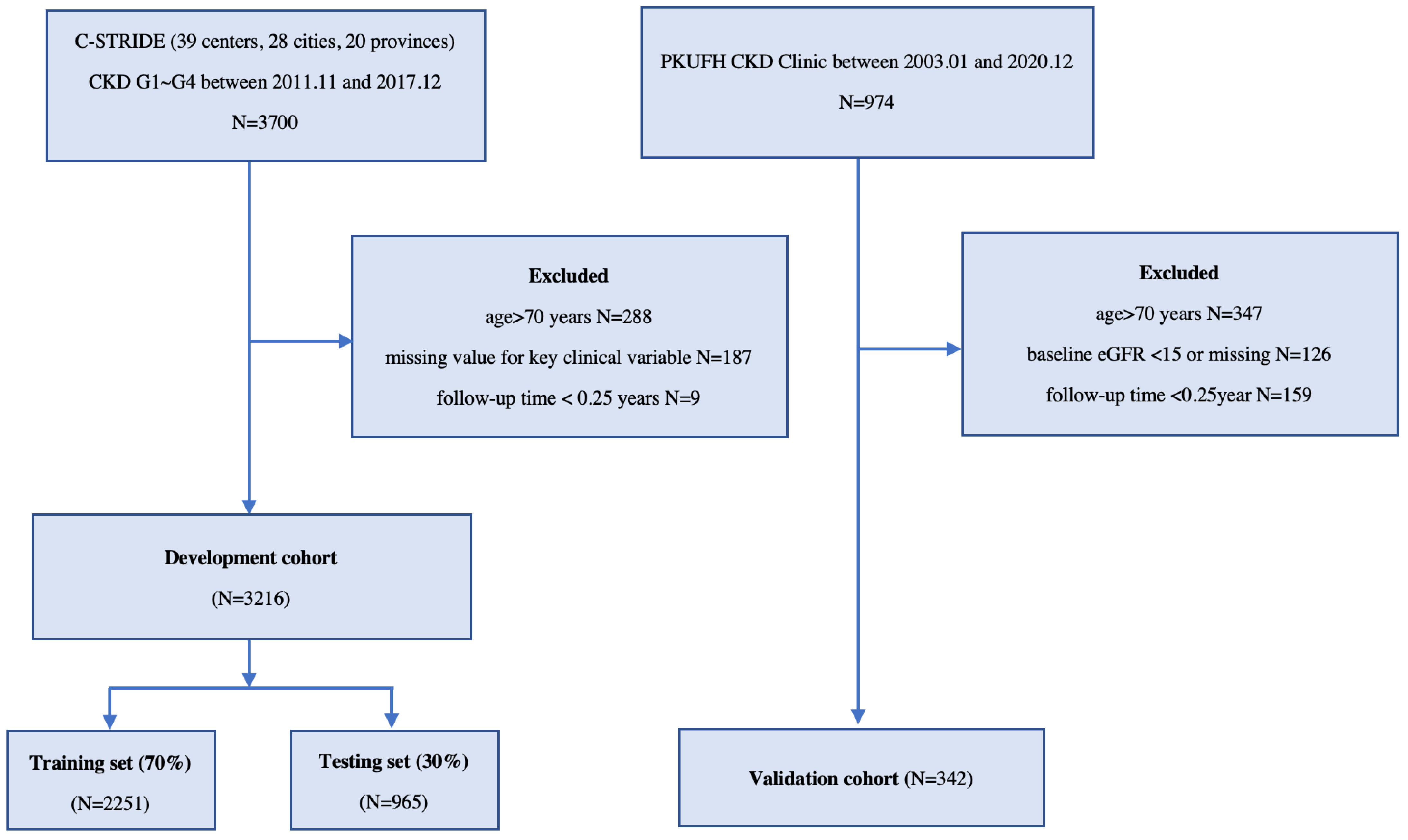
2.2. Data Source and Study Population
2.2.1. Development Cohort
2.2.2. Validation Cohort
2.2.3. Candidate Variables
2.3. Data Preprocessing and Statistical Analysis
2.4. Model Development and Evaluation
Model-Driven Feature Selection
2.5. Model Training
Model Evaluation and Comparison
2.6. Implementation Setup
3. Results
3.1. Cohort Description
3.2. Feature Selection
3.3. Model Performance
3.4. Model Comparison
3.5. Web Application
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bikbov, B.; Purcell, C.A.; Levey, A.S.; Smith, M.; Abdoli, A.; Abebe, M.; Owolabi, M.O. Global, regional, and national burden of chronic kidney disease, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2020, 395, 709–733. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, F.; Wang, L.; Wang, W.; Liu, B.; Liu, J.; Wang, H. Prevalence of chronic kidney disease in China: A cross-sectional survey. Lancet 2012, 379, 815–822. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, M.-H.; Zuo, L.; Wang, Y.; Yu, F.; Zhang, H.; Zhou, Z. China Kidney Disease Network (CK-NET) 2016 Annual Data Report. Kidney Int. Suppl. 2020, 10, e97–e185. [Google Scholar] [CrossRef]
- Yang, C.; Wang, H.; Zhao, X.; Matsushita, K.; Coresh, J.; Zhang, L.; Zhao, M.H. CKD in China: Evolving Spectrum and Public Health Implications. Am. J. Kidney Dis. 2020, 76, 258–264. [Google Scholar] [CrossRef]
- De Nicola, L.; Minutolo, R.; Chiodini, P.; Borrelli, S.; Zoccali, C.; Postorino, M.; Conte, G. The effect of increasing age on the prognosis of non-dialysis patients with chronic kidney disease receiving stable nephrology care. Kidney Int. 2012, 82, 482–488. [Google Scholar] [CrossRef]
- Tangri, N.; Grams, M.E.; Levey, A.S.; Coresh, J.; Appel, L.J.; Astor, B.C.; CKD Prognosis Consortium. Multinational Assessment of Accuracy of Equations for Predicting Risk of Kidney Failure: A Meta-analysis. JAMA 2016, 315, 164–174. [Google Scholar] [CrossRef]
- Tangri, N.; Stevens, L.A.; Griffith, J.; Tighiouart, H.; Djurdjev, O.; Naimark, D.; Levey, A.S. A predictive model for progression of chronic kidney disease to kidney failure. JAMA 2011, 305, 1553–1559. [Google Scholar] [CrossRef]
- Kang, M.W.; Tangri, N.; Kim, Y.C.; An, J.N.; Lee, J.; Li, L.; Lee, J.P. An independent validation of the kidney failure risk equation in an Asian population. Sci. Rep. 2020, 10, 12920. [Google Scholar] [CrossRef]
- Kwek, J.L.; Pang, H.Q.J.; Li, H.; Lim, W.W.L.; Choo, J.C.J.; Choong, H.L.; Chan, C.M. Validation of the kidney failure risk equation in predicting the risk of progression to kidney failure in a multi-ethnic Singapore chronic kidney disease cohort. Singap. Med. J. 2020, 63, 313. [Google Scholar] [CrossRef]
- Bai, Q.; Su, C.; Tang, W.; Li, Y. Machine learning to predict end stage kidney disease in chronic kidney disease. Sci. Rep. 2022, 12, 1–8. [Google Scholar] [CrossRef]
- Xiao, J.; Ding, R.; Xu, X.; Guan, H.; Feng, X.; Sun, T.; Ye, Z. Comparison and development of machine learning tools in the prediction of chronic kidney disease progression. J. Transl. Med. 2019, 17, 119. [Google Scholar] [CrossRef]
- Zhao, J.; Gu, S.P.; McDermaid, A. Predicting outcomes of chronic kidney disease from EMR data based on Random Forest Regression. Math. Biosci. 2019, 310, 24–30. [Google Scholar] [CrossRef]
- Sheer, R.; Nair, R.; Pasquale, M.K.; Evers, T.; Cockrell, M.; Gay, A.; Schmedt, N. Predictive Risk Models to Identify Patients at High-Risk for Severe Clinical Outcomes With Chronic Kidney Disease and Type 2 Diabetes. J. Prim. Care Community Health 2022, 13, 21501319211063726. [Google Scholar] [CrossRef]
- Niel, O.; Bastard, P. Artificial Intelligence in Nephrology: Core Concepts, Clinical Applications, and Perspectives. Am. J. Kidney Dis. 2019, 74, 803–810. [Google Scholar] [CrossRef]
- Beam, A.L.; Kohane, I.S. Big Data and Machine Learning in Health Care. JAMA 2018, 319, 1317–1318. [Google Scholar] [CrossRef]
- Gao, B.; Zhang, L.; Wang, H.; Zhao, M. Chinese cohort study of chronic kidney disease: Design and methods. Chin. Med. J. 2014, 127, 2180–2185. [Google Scholar]
- Yuan, J.; Zou, X.-R.; Han, S.-P.; Cheng, H.; Wang, L.; Wang, J.W.; Wang, X.Q. Prevalence and risk factors for cardiovascular disease among chronic kidney disease patients: Results from the Chinese cohort study of chronic kidney disease (C-STRIDE). BMC Nephrol. 2017, 18, 23. [Google Scholar] [CrossRef]
- Levey, A.S.; Stevens, L.A.; Schmid, C.H.; Zhang, Y.; Castro, A.F., III; Feldman, H.I. A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 2009, 150, 604–612. [Google Scholar] [CrossRef]
- De Nicola, L.; Chiodini, P.; Zoccali, C.; Borrelli, S.; Cianciaruso, B.; Di Iorio, B.; SIN-TABLE CKD Study Group. Prognosis of CKD patients receiving outpatient nephrology care in Italy. Clin. J. Am. Soc. Nephrol. CJASN 2011, 6, 2421–2428. [Google Scholar] [CrossRef]
- Ferguson, T.; Ravani, P.; Sood, M.M.; Clarke, A.; Komenda, P.; Rigatto, C.; Tangri, N. Development and External Validation of a Machine Learning Model for Progression of CKD. Kidney Int. Rep. 2022, 7, 1772–1781. [Google Scholar] [CrossRef]
- Ramspek, C.L.; Evans, M.; Wanner, C.; Drechsler, C.; Chesnaye, N.C.; Szymczak, M.; EQUAL Study Investigators. Kidney Failure Prediction Models: A Comprehensive External Validation Study in Patients with Advanced CKD. J. Am. Soc. Nephrol. 2021, 32, 1174–1186. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: San Francisco, CA, USA, 2016; pp. 785–794. [Google Scholar]
- Widodo, A.; Yang, B.-S. Machine health prognostics using survival probability and support vector machine. Expert Syst. Appl. 2011, 38, 8430–8437. [Google Scholar] [CrossRef]
- Moncada-Torres, A.; van Maaren, M.C.; Hendriks, M.P.; Siesling, S.; Geleijnse, G. Explainable machine learning can outperform Cox regression predictions and provide insights in breast cancer survival. Sci. Rep. 2021, 11, 6968. [Google Scholar] [CrossRef]
- Uno, H.; Cai, T.; Pencina, M.J.; D’Agostino, R.B.; Wei, L.J. On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat. Med. 2011, 30, 1105–1117. [Google Scholar] [CrossRef]
- Kang, L.; Chen, W.; Petrick, N.A.; Gallas, B.D. Comparing two correlated C indices with right-censored survival outcome: A one-shot nonparametric approach. Stat. Med. 2015, 34, 685–703. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: San Francisco, CA, USA, 2016; pp. 1135–1144. [Google Scholar]
- Zhang, X.; Luo, F.; Chen, R.; Shen, J.; Liu, X.; Shi, Y.; Hou, F.F. Use of Histologic Parameters to Predict Glomerular Disease Progression: Findings From the China Kidney Biopsy Cohort Study. Am. J. Kidney Dis. Off. J. Natl. Kidney Found. 2022. [Google Scholar] [CrossRef]
- Grams, M.E.; Sang, Y.; Ballew, S.H.; Carrero, J.J.; Djurdjev, O.; Heerspink, H.J.; Coresh, J. Predicting timing of clinical outcomes in patients with chronic kidney disease and severely decreased glomerular filtration rate. Kidney Int. 2018, 93, 1442–1451. [Google Scholar] [CrossRef]
- Niel, O.; Bastard, P.; Boussard, C.; Hogan, J.; Kwon, T.; Deschênes, G. Artificial intelligence outperforms experienced nephrologists to assess dry weight in pediatric patients on chronic hemodialysis. Pediatr. Nephrol. 2018, 33, 1799–1803. [Google Scholar] [CrossRef]
- Weng, S.F.; Reps, J.; Kai, J.; Garibaldi, J.M.; Qureshi, N. Can machine-learning improve cardiovascular risk prediction using routine clinical data? PLoS ONE 2017, 12, e0174944. [Google Scholar] [CrossRef]
- Lee, H.; Yun, D.; Yoo, J.; Yoo, K.; Kim, Y.C.; Kim, D.K.; Han, S.S. Deep Learning Model for Real-Time Prediction of Intradialytic Hypotension. Clin. J. Am. Soc. Nephrol. 2021, 16, 396–406. [Google Scholar] [CrossRef]
- Barbieri, C.; Mari, F.; Stopper, A.; Gatti, E.; Escandell-Montero, P.; Martínez-Martínez, J.M.; Martín-Guerrero, J.D. A new machine learning approach for predicting the response to anemia treatment in a large cohort of End Stage Renal Disease patients undergoing dialysis. Comput. Biol. Med. 2015, 61, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Li, X.; Li, Y.; Xia, E.; Qin, Y.; Liang, S.; Liu, Z. Prediction and Risk Stratification of Kidney Outcomes in IgA Nephropathy. Am. J. Kidney Dis. 2019, 74, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Schena, F.P.; Anelli, V.W.; Trotta, J.; Di Noia, T.; Manno, C.; Tripepi, G.; Madio, D. Development and testing of an artificial intelligence tool for predicting end-stage kidney disease in patients with immunoglobulin A nephropathy. Kidney Int. 2021, 99, 1179–1188. [Google Scholar] [CrossRef]
| Characteristics | C-STRIDE (N = 3216) | Missing Value | PKUFH CKD (N = 342) | Missing Value |
|---|---|---|---|---|
| Age, years | 48 (13) | 0 | 55 (11) | 0 |
| Male, n (%) | 1909 (59.4) | 0 | 133 (38.9) | 0 |
| Smoker, n (%) | 1123 (34.9) | 284 | 11 (3.2) | 320 |
| Hypertension, n (%) | 2363 (73.5) | 218 | 83 (24.3) | 245 |
| T2DM, n (%) | 641 (19.9) | 433 | 99 (28.9) | 182 |
| CVD, n (%) | 270 (8.4) | 0 | 7 (2.0) | 269 |
| Cause of CKD, n (%) | 253 | 72 | ||
| DKD | 385 (12.0) | 88 (25.7) | ||
| GN | 1853 (57.6) | 80 (23.4) | ||
| Other | 725 (22.5) | 102 (29.8) | ||
| Systolic BP, mmHg | 129.39 (17.50) | 462 | 132.39 (18.62) | 132 |
| Diastolic BP, mmHg | 80.97 (10.80) | 462 | 77.65 (11.06) | 132 |
| ALB, g/L | 38.54 (7.46) | 425 | 42.55 (4.69) | 35 |
| HGB, mg/L | 128.75 (21.87) | 360 | 134.09 (20.18) | 14 |
| Creatinine, μmol/L | 154.27 (72.17) | 0 | 196.56 (168.08) | 0 |
| eGFR, mL/min/1.73 m2 | 52.97 (29.50) | 0 | 50.83 (35.04) | 0 |
| >60, n (%) 30~59, n (%) | 1057 (32.8) 1334 (41.5) | 98 (29.0) 154 (45.6) | ||
| 15~29, n (%) | 825 (25.7) | 86 (25.4) | ||
| UACR, mg/g | 376.40 (90.80, 911.45) | 329 | 214.37 (43.55, 1058.50) | 138 |
| <30 | 392 (13.6) | 43 (21.1) | ||
| 30~300 | 889 (30.8) | 64 (31.4) | ||
| ≥300 | 1606 (55.6) | 97 (47.5) | ||
| FBG, mmol/L | 5.30 (1.69) | 504 | 6.11 (1.56) | 32 |
| Uric acid, mmol/L | 404.68 (117.17) | 210 | 396.58 (101.87) | 9 |
| Serum phosphorus, mmol/L | 1.21 (0.37) | 453 | 1.22 (0.31) | 19 |
| Serum calcium, mmol/L | 2.23 (0.20) | 409 | 2.31 (0.15) | 17 |
| Serum potassium, mmol/L | 4.44 (0.74) | 446 | 4.43 (0.56) | 8 |
| Triglyceride, mmol/L | 2.16 (1.41) | 654 | 1.90 (1.21) | 39 |
| TC, mmol/L | 5.23 (2.23) | 644 | 4.54 (1.05) | 38 |
| HDL-C, mmol/L | 1.12 (0.33) | 767 | 1.14 (0.33) | 40 |
| LDL-C, mmol/L | 2.78 (1.04) | 761 | 2.58 (1.44) | 38 |
| KRT, n (%) | 411 (12.8) | - | 25 (7.3) | - |
| Follow-up time, years | 4.45 (1.34) | - | 3.37 (2.92) | - |
| Cox Model | |||
| Training | Testing | Validation | |
| Harrell’s C | 0.841 (0.811~0.871) | 0.834 (0.803~0.865) | 0.761 (0.696~0.824) |
| Uno’s C | 0.807 (0.760~0.850) | 0.833 (0.804~0.865) | 0.796 (0.732~0.850) |
| 2-yr TD-AUC | 0.888 (0.847~0.927) | 0.841 (0.786~0.886) | 0.777 (0.689~0.858) |
| 5-yr TD-AUC | 0.696 (0.235~0.929) | 0.873 (0.780~0.915) | 0.905 (0.824~0.987) |
| Brier score | 0.059 (0.051~0.068) | 0.065 (0.055~0.074) | 0.029 (0.022~0.037) |
| XGBoost model | |||
| Training | Testing | Validation | |
| Harrell’s C | 0.864 (0.836~0.891) | 0.826 (0.793~0.856) | 0.796 (0.738~0.852) |
| Uno’s C | 0.836 (0.797~0.873) | 0.825 (0.792~0.856) | 0.822 (0.766~0.868) |
| 2-yr TD-AUC | 0.902 (0.863~0.938) | 0.839 (0.788~0.886) | 0.833 (0.762~0.900) |
| 5-yr TD-AUC | 0.754 (0.380~0.939) | 0.861 (0.729~0.919) | 0.912 (0.850~0.976) |
| Brier score | 0.055 (0.047~0.064) | 0.066 (0.056~0.077) | 0.028 (0.021~0.035) |
| SSVM model | |||
| Training | Testing | Validation | |
| Harrell’s C | 0.754 (0.715~0.791) | 0.748 (0.709~0.788) | 0.745 (0.674~0.811) |
| Uno’s C | 0.732 (0.685~0.780) | 0.747 (0.702~0.790) | 0.753 (0.693~0.805) |
| 2-yr TD-AUC | 0.787 (0.731~0.842) | 0.751 (0.692~0.812) | 0.820 (0.715~0.909) |
| 5-yr TD-AUC | 0.602 (0.187~0.879) | 0.728 (0.619~0.816) | 0.849 (0.791~0.896) |
| Brier score | 0.068 (0.685~0.780) | 0.070 (0.060~0.081) | 0.031 (0.024~0.039) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hui, M.; Ma, J.; Yang, H.; Gao, B.; Wang, F.; Wang, J.; Lv, J.; Zhang, L.; Yang, L.; Zhao, M. ESKD Risk Prediction Model in a Multicenter Chronic Kidney Disease Cohort in China: A Derivation, Validation, and Comparison Study. J. Clin. Med. 2023, 12, 1504. https://doi.org/10.3390/jcm12041504
Hui M, Ma J, Yang H, Gao B, Wang F, Wang J, Lv J, Zhang L, Yang L, Zhao M. ESKD Risk Prediction Model in a Multicenter Chronic Kidney Disease Cohort in China: A Derivation, Validation, and Comparison Study. Journal of Clinical Medicine. 2023; 12(4):1504. https://doi.org/10.3390/jcm12041504
Chicago/Turabian StyleHui, Miao, Jun Ma, Hongyu Yang, Bixia Gao, Fang Wang, Jinwei Wang, Jicheng Lv, Luxia Zhang, Li Yang, and Minghui Zhao. 2023. "ESKD Risk Prediction Model in a Multicenter Chronic Kidney Disease Cohort in China: A Derivation, Validation, and Comparison Study" Journal of Clinical Medicine 12, no. 4: 1504. https://doi.org/10.3390/jcm12041504





