High-Throughput Sequencing of Oral Microbiota in Candida Carriage Sjögren’s Syndrome Patients: A Pilot Cross-Sectional Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Clinical Examinations
2.3. Whole Saliva Collection
2.4. DNA Isolation, PCR Amplification, and High-Throughput Sequencing
2.5. Data Processing and Statistical Analyses
3. Results
3.1. Patients Characteristics
3.2. General Outline
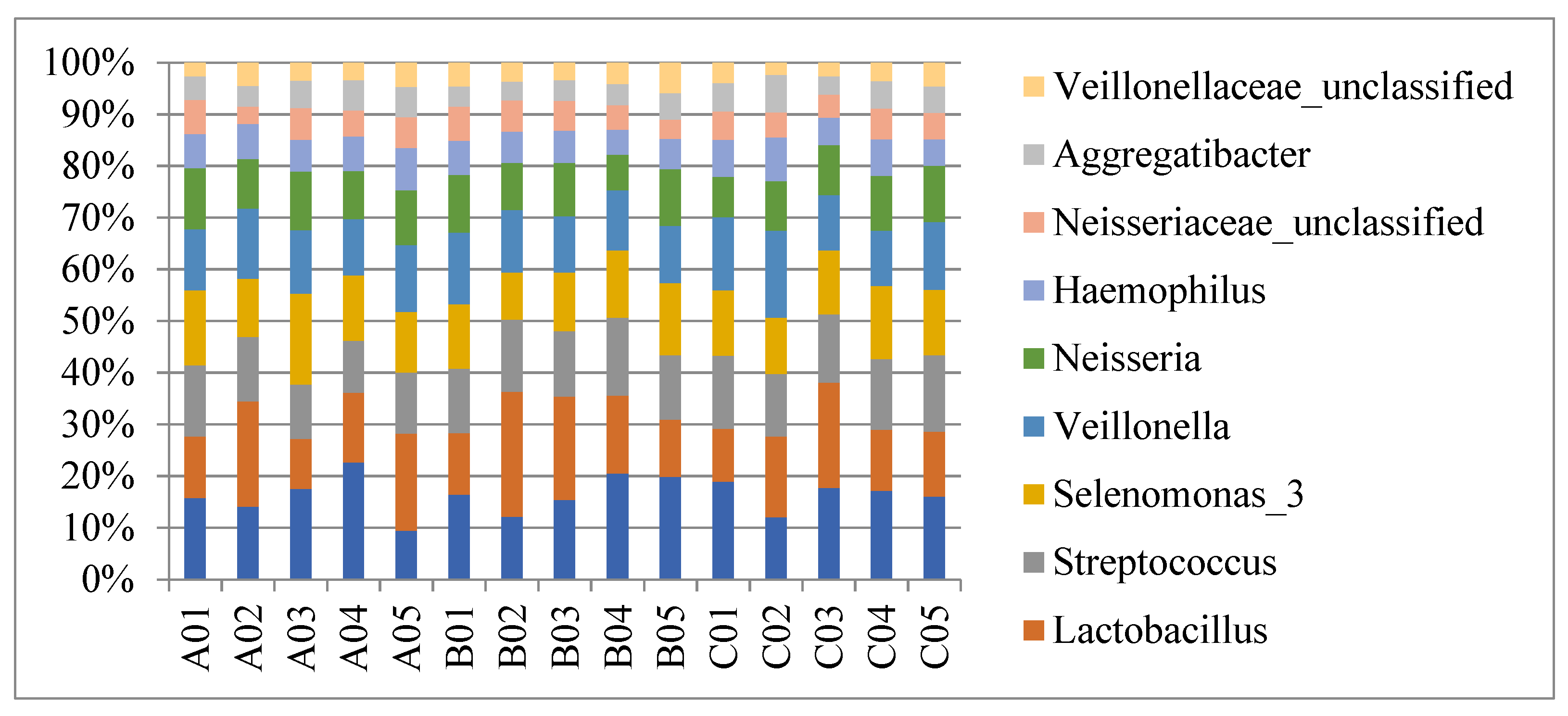
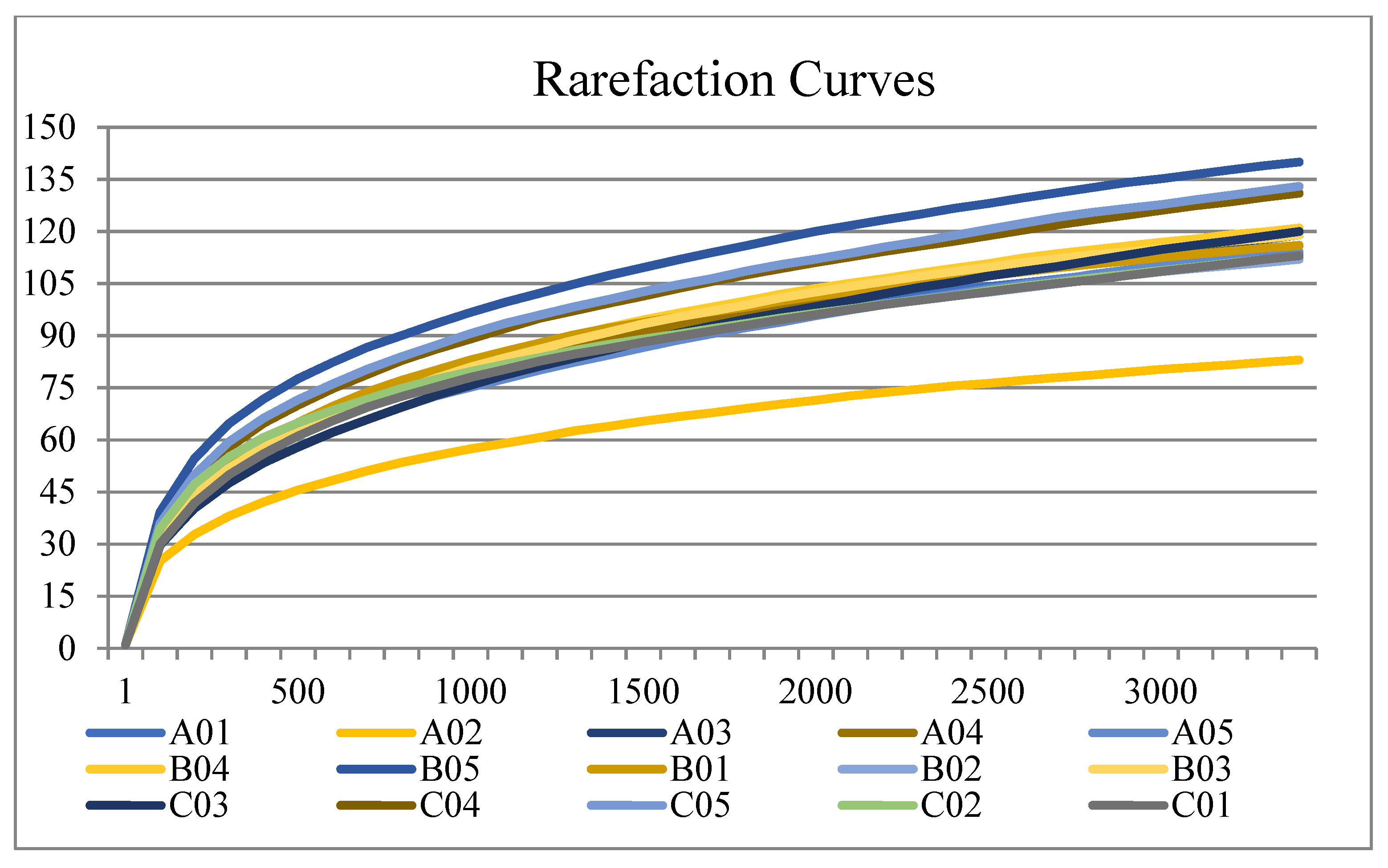
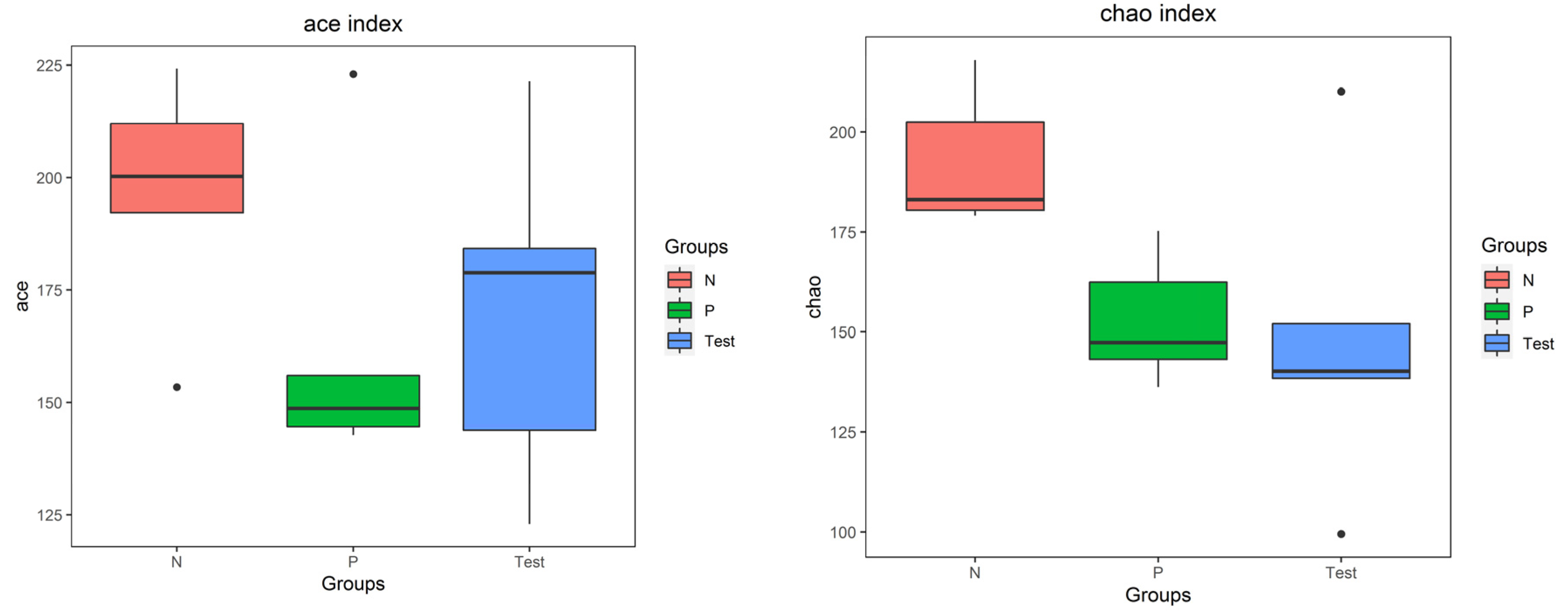
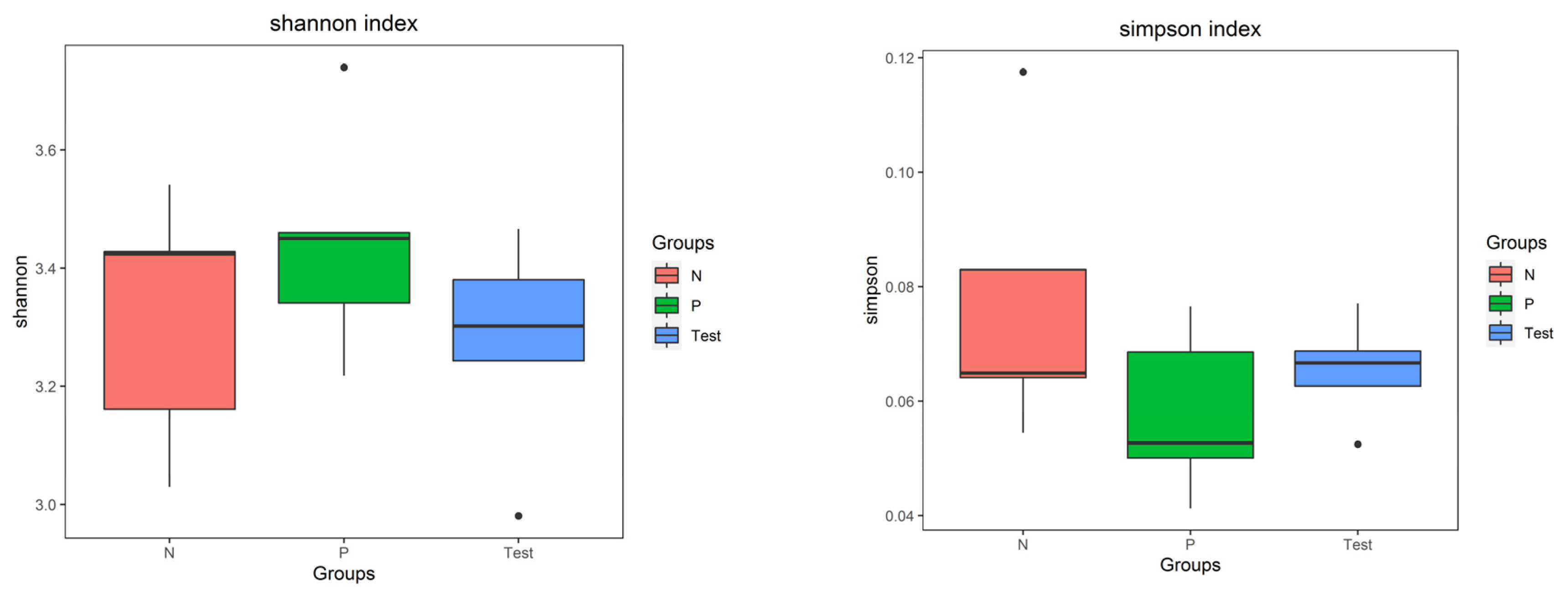
3.3. Biodiversity of the Salivary Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mariette, X.; Criswell, L.A. Primary Sjögren’s Syndrome. N. Engl. J. Med. 2018, 378, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Zunt, S.L. Xerostomia/Salivary Gland Hypofunction: Diagnosis and Management. Compend. Contin. Educ. Dent. 2018, 39, 365. [Google Scholar] [PubMed]
- Kassebaum, N.J.; Smith, A.G.C.; Bernabé, E.; Fleming, T.D.; Reynolds, A.E.; Vos, T.; Murray, C.J.L.; Marcenes, W.; GBD 2015 Oral Health Collaborators. Global, Regional, and National Prevalence, Incidence, and Disability-Adjusted Life Years for Oral Conditions for 195 Countries, 1990–2015: A Systematic Analysis for the Global Burden of Diseases, Injuries, and Risk Factors. J. Dent. Res. 2017, 96, 380–387. [Google Scholar] [CrossRef] [PubMed]
- González, S.; Sung, H.; Sepúlveda, D.; González, M.; Molina, C. Oral manifestations and their treatment in Sjögren′s syndrome. Oral Dis. 2014, 20, 153–161. [Google Scholar] [CrossRef]
- Maarse, F.; Jager, D.J.; Forouzanfar, T.; Wolff, J.; Brand, H. Tooth loss in Sjogren’s syndrome patients compared to age and gender matched controls. Med. Oral Patol. Oral. Cir. Bucal. 2018, 23, e545–e551. [Google Scholar] [CrossRef]
- Zhou, Z.; Ling, G.; Ding, N.; Xun, Z.; Zhu, C.; Hua, H.; Chen, X. Molecular analysis of oral microflora in patients with primary Sjögren’s syndrome by using high-throughput sequencing. PeerJ 2018, 6, e5649. [Google Scholar] [CrossRef]
- Featherstone, J.D.B. The Continuum of Dental Caries—Evidence for a Dynamic Disease Process. J. Dent. Res. 2004, 83 (Suppl. 1), C39–C42. [Google Scholar] [CrossRef]
- Saleh, J.; Figueiredo, M.A.Z.; Cherubini, K.; Salum, F.G. Salivary hypofunction: An update on aetiology, diagnosis and therapeutics. Arch. Oral Biol. 2015, 60, 242–255. [Google Scholar] [CrossRef]
- Mosaddad, S.A.; Tahmasebi, E.; Yazdanian, A.; Rezvani, M.B.; Seifalian, A.; Yazdanian, M.; Tebyanian, H. Oral microbial biofilms: An update. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 2005–2019. [Google Scholar] [CrossRef]
- Dawes, C. Salivary flow patterns and the health of hard and soft oral tissues. J. Am. Dent. Assoc. 2008, 139, 18S–24S. [Google Scholar] [CrossRef]
- Twetman, S. Prevention of dental caries as a non-communicable disease. Eur. J. Oral Sci. 2018, 126 (Suppl. 1), 19–25. [Google Scholar] [CrossRef] [PubMed]
- Contaldo, M.; Lucchese, A.; Lajolo, C.; Rupe, C.; Di Stasio, D.; Romano, A.; Petruzzi, M.; Serpico, R. The Oral Microbiota Changes in Orthodontic Patients and Effects on Oral Health: An Overview. J. Clin. Med. 2021, 10, 780. [Google Scholar] [CrossRef] [PubMed]
- Müller, V.J.; Belibasakis, G.N.; Bosshard, P.P.; Wiedemeier, D.B.; Bichsel, D.; Rücker, M.; Stadlinger, B. Change of saliva composition with radiotherapy. Arch. Oral Biol. 2019, 106, 104480. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, H.; Chen, T.; Aliko, A.; Mydel, P.M.; Jonsson, R.; Olsen, I. Microbiological and bioinformatics analysis of primary Sjögren’s syndrome patients with normal salivation§. J. Oral Microbiol. 2016, 8, 31119. [Google Scholar] [CrossRef] [PubMed]
- Belstrøm, D.; Holmstrup, P.; Fiehn, N.-E.; Rosing, K.; Bardow, A.; Paster, B.; Pedersen, A.L. Bacterial composition in whole saliva from patients with severe hyposalivation—A case-control study. Oral Dis. 2016, 22, 330–337. [Google Scholar] [CrossRef]
- Sembler-Møller, M.L.; Belstrøm, D.; Locht, H.; Enevold, C.; Pedersen, A.M.L. Next-generation sequencing of whole saliva from patients with primary Sjögren’s syndrome and non-Sjögren’s sicca reveals comparable salivary microbiota. J. Oral Microbiol. 2019, 11, 1660566. [Google Scholar] [CrossRef]
- Sharma, D.; Sandhya, P.; Vellarikkal, S.K.; Surin, A.K.; Jayarajan, R.; Verma, A.; Kumar, A.; Ravi, R.; Danda, D.; Sivasubbu, S.; et al. Saliva microbiome in primary Sjögren’s syndrome reveals distinct set of disease-associated microbes. Oral Dis. 2020, 26, 295–301. [Google Scholar] [CrossRef]
- Cunha-Cruz, J.; Scott, J.; Rothen, M.; Mancl, L.; Lawhorn, T.; Brossel, K.; Berg, J. Salivary characteristics and dental caries: Evidence from general dental practices. J. Am. Dent. Assoc. 2013, 144, e31–e40. [Google Scholar] [CrossRef]
- Rusthen, S.; Kristoffersen, A.K.; Young, A.; Galtung, H.K.; Petrovski, B.; Palm, Ø.; Enersen, M.; Jensen, J.L. Dysbiotic salivary microbiota in dry mouth and primary Sjögren’s syndrome patients. PLoS ONE 2019, 14, e0218319. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Li, H.; Ni, C.; Du, Z.; Yan, F. Human oral microbiota and its modulation for oral health. Biomed. Pharmacother. 2018, 99, 883–893. [Google Scholar] [CrossRef]
- Tarapan, S.; Matangkasombut, O.; Trachootham, D.; Sattabanasuk, V.; Talungchit, S.; Paemuang, W.; Phonyiam, T.; Chokchaitam, O.; Mungkung, O.; Lam-Ubol, A. Oral Candida colonization in xerostomic postradiotherapy head and neck cancer patients. Oral Dis. 2019, 25, 1798–1808. [Google Scholar] [CrossRef] [PubMed]
- Nikou, S.-A.; Kichik, N.; Brown, R.; Ponde, N.; Ho, J.; Naglik, J.; Richardson, J. Candida albicans Interactions with Mucosal Surfaces during Health and Disease. Pathogens 2019, 8, 53. [Google Scholar] [CrossRef] [PubMed]
- Serrano, J.; López-Pintor, R.; Ramírez, L.; Fernández-Castro, M.; Sanz, M.; Melchor, S.; Peiteado, D.; Hernández, G. Risk factors related to oral candidiasis in patients with primary Sjögren’s syndrome. Med. Oral. Patol. Oral. Cir. Bucal. 2020, 25, e700–e705. [Google Scholar] [CrossRef] [PubMed]
- Xin, W.; Leung, K.C.M.; Lo, E.C.M.; Mok, M.Y.; Leung, M.H. Sicca Symptoms, Oral Health Conditions, Salivary Flow and Oral Candida in Sjögren’s Syndrome Patients. Int. J. Environ. Res. Public Health 2020, 17, 3625. [Google Scholar] [CrossRef]
- Ergun, S.; Cekici, A.; Topcuoglu, N.; Migliari, D.; Kulekci, G.; Tanyeri, H.; Isik, G. Oral status and Candida colonization in patients with Sjogren s Syndrome. Med. Oral. Patol. Oral. Cir. Bucal. 2010, 15, e310–e315. [Google Scholar] [CrossRef]
- Janus, M.M.; Crielaard, W.; Volgenant, C.M.C.; van der Veen, M.H.; Brandt, B.W.; Krom, B.P. Candida albicans alters the bacterial microbiome of early in vitro oral biofilms. J. Oral Microbiol. 2017, 9, 1270613. [Google Scholar] [CrossRef]
- Montelongo-Jauregui, D.; Lopez-Ribot, J.L. Candida Interactions with the Oral Bacterial Microbiota. J. Fungi 2018, 4, 122. [Google Scholar] [CrossRef]
- Dige, I.; Nyvad, B. Candida species in intact in vivo biofilm from carious lesions. Arch. Oral Biol. 2019, 101, 142–146. [Google Scholar] [CrossRef]
- Patel, R.; Shahane, A. The epidemiology of Sjögren’s syndrome. Clin. Epidemiol. 2014, 6, 247–255. [Google Scholar] [CrossRef]
- Vitali, C.; Bombardieri, S.; Jonsson, R.; Moutsopoulos, H.M.; Alexander, E.L.; Carsons, S.E.; Daniels, T.E.; Fox, P.C.; Fox, R.I.; Kassan, S.S.; et al. Classification criteria for Sjogren’s syndrome: A revised version of the European criteria proposed by the American-European Consensus Group. Ann. Rheum. Dis. 2002, 61, 554–558. [Google Scholar] [CrossRef]
- Shiboski, C.H.; Shiboski, S.C.; Seror, R.; Criswell, L.A.; Labetoulle, M.; Lietman, T.M.; Mariette, X. 2016 ACR-EULAR classification criteria for primary Sjögren’s syndrome: A consensus and data-driven methodology involving three international patient cohorts. Arthritis Rheumatol. 2017, 69, 35–45. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Dyrhovden, R.; Rippin, M.; Øvrebø, K.K.; Nygaard, R.M.; Ulvestad, E.; Kommedal, Ø. Managing Contamination and Diverse Bacterial Loads in 16S rRNA Deep Sequencing of Clinical Samples: Implications of the Law of Small Numbers. mBio 2021, 12, e0059821. [Google Scholar] [CrossRef]
- Zeng, H.; Chan, Y.; Gao, W.; Leung, W.K.; Watt, R.M. Diversity of Treponema denticola and Other Oral Treponeme Lineages in Subjects with Periodontitis and Gingivitis. Microbiol. Spectr. 2021, 9, e0070121. [Google Scholar] [CrossRef] [PubMed]
- van der Meulen, T.A.; Harmsen, H.J.M.; Bootsma, H.; Liefers, S.C.; Vila, A.V.; Zhernakova, A.; Weersma, R.K.; Spijkervet, F.K.L.; Kroese, F.G.M.; Vissink, A. Reduced salivary secretion contributes more to changes in the oral microbiome of patients with primary Sjögren’s syndrome than underlying disease. Ann. Rheum. Dis. 2018, 77, 1542–1544. [Google Scholar] [CrossRef] [PubMed]
- Alam, J.; Lee, A.; Lee, J.; Kwon, D.I.; Park, H.K.; Park, J.-H.; Jeon, S.; Baek, K.; Lee, J.; Park, S.-H.; et al. Dysbiotic oral microbiota and infected salivary glands in Sjögren’s syndrome. PLoS ONE 2020, 15, e0230667. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Cai, Y.; Wang, M.; Yang, W.; Duan, N. Oral microbial flora of patients with Sicca syndrome. Mol. Med. Rep. 2018, 18, 4895–4903. [Google Scholar] [CrossRef]
- Buranarom, N.; Komin, O.; Matangkasombut, O. Hyposalivation, oral health, and Candida colonization in independent dentate elders. PLoS ONE 2020, 15, e0242832. [Google Scholar] [CrossRef]
- Ponde, N.O.; Lortal, L.; Ramage, G.; Naglik, J.R.; Richardson, J.P. Candida albicans biofilms and polymicrobial interactions. Crit. Rev. Microbiol. 2021, 47, 91–111. [Google Scholar] [CrossRef]
- Xiao, J.; Grier, A.; Faustoferri, R.; Alzoubi, S.; Gill, A.; Feng, C.; Liu, Y.; Quivey, R.; Kopycka-Kedzierawski, D.; Koo, H.; et al. Association between Oral Candida and Bacteriome in Children with Severe ECC. J. Dent. Res. 2018, 97, 1468–1476. [Google Scholar] [CrossRef] [PubMed]
- Fujinami, W.; Nishikawa, K.; Ozawa, S.; Hasegawa, Y.; Takebe, J. Correlation between the relative abundance of oral bacteria and Candida albicans in denture and dental plaques. J. Oral Biosci. 2021, 63, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Young, T.; Alshanta, O.-A.; Kean, R.; Bradshaw, D.; Pratten, J.; Williams, C.; Woodall, C.; Ramage, G.; Brown, J.L. Candida albicans as an Essential “Keystone” Component within Polymicrobial Oral Biofilm Models? Microorganisms 2020, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Nishimaki, F.; Yamada, S.-I.; Kawamoto, M.; Sakurai, A.; Hayashi, K.; Kurita, H. Relationship Between the Quantity of Oral Candida and Systemic Condition/Diseases of the Host: Oral Candida Increases with Advancing Age and Anemia. Mycopathologia 2019, 184, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Eidt, G.; Waltermann, E.D.M.; Hilgert, J.B.; Arthur, R.A. Candida and dental caries in children, adolescents and adults: A systematic review and meta-analysis. Arch. Oral Biol. 2020, 119, 104876. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Ren, B.; He, J.; Peng, X.; Guo, Q.; Zheng, L.; Li, J.; Dai, H.; Chen, V.; Zhang, L.; et al. Candida albicans promotes tooth decay by inducing oral microbial dysbiosis. ISME J. 2020, 15, 894–908. [Google Scholar] [CrossRef]
- Dame-Teixeira, N.; de Lima, A.K.A.; Do, T.; Stefani, C.M. Meta-Analysis Using NGS Data: The Veillonella Species in Dental Caries. Front. Oral Health 2021, 2, 770917. [Google Scholar] [CrossRef]
- Periasamy, S.; Kolenbrander, P.E.; Zeng, L.; Das, S.; Burne, R.A. Central Role of the Early Colonizer Veillonella sp. in Establishing Multispecies Biofilm Communities with Initial, Middle, and Late Colonizers of Enamel. J. Bacteriol. 2010, 192, 2965–2972. [Google Scholar] [CrossRef]
- Luppens SBI, K.D.; Bandounas, L.; Jonker, M.J.; Wittink, F.R.A.; Bruning, O.; Breit, T.M.; ten Cate, J.M.; Crielaard, W. Effect of Veillonella parvula on the antimicrobial resistance and gene expression of Streptococcus mutans grown in a dual-species biofilm. Oral Microbiol. Immunol. 2008, 23, 183–189. [Google Scholar] [CrossRef]
- Nikitakis, N.G.; Papaioannou, W.; I Sakkas, L.; Kousvelari, E. The autoimmunity-oral microbiome connection. Oral Dis. 2016, 23, 828–839. [Google Scholar] [CrossRef]
- Singh, M.; Teles, F.; Uzel, N.G.; Papas, A. Characterizing Microbiota from Sjogren’s Syndrome Patients. JDR Clin. Trans. Res. 2020, 6, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and Qualitative β Diversity Measures Lead to Different Insights into Factors That Structure Microbial Communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Palmer, R.J.; Cotton, S.L.; Kokaras, A.S.; Gardner, P.; Grisius, M.; Pelayo, E.; Warner, B.; Paster, B.J.; Alevizos, I. Analysis of oral bacterial communities: Comparison of HOMINGS with a tree-based approach implemented in QIIME. J. Oral Microbiol. 2019, 11, 1586413. [Google Scholar] [CrossRef] [PubMed]
- de Paiva, C.S.; Jones, D.B.; Stern, M.E.; Bian, F.; Moore, Q.L.; Corbiere, S.; Streckfus, C.F.; Hutchinson, D.S.; Ajami, N.J.; Petrosino, J.F.; et al. Altered Mucosal Microbiome Diversity and Disease Severity in Sjögren Syndrome. Sci. Rep. 2016, 6, 23561. [Google Scholar] [CrossRef]
- Shen, L.; He, J.; Kramer, J.M.; Bunya, V.Y. Sjögren’s Syndrome: Animal Models, Etiology, Pathogenesis, Clinical Subtypes, and Diagnosis. J. Immunol. Res. 2019, 2019, 8101503. [Google Scholar] [CrossRef]
- Zorba, M.; Melidou, A.; Patsatsi, A.; Ioannou, E.; Kolokotronis, A. The possible role of oral microbiome in autoimmunity. Int. J. Women’s Dermatol. 2020, 6, 357–364. [Google Scholar] [CrossRef]
- Szymula, A.; Rosenthal, J.; Szczerba, B.M.; Bagavant, H.; Fu, S.M.; Deshmukh, U.S. T cell epitope mimicry between Sjögren’s syndrome Antigen A (SSA)/Ro60 and oral, gut, skin and vaginal bacteria. Clin. Immunol. 2014, 152, 1–9. [Google Scholar] [CrossRef]
- della Vella, F.; Lauritano, D.; Lajolo, C.; Lucchese, A.; Di Stasio, D.; Contaldo, M.; Serpico, R.; Petruzzi, M. The pseudolesions of the oral mucosa: Differential diagnosis and related systemic conditions. Appl. Sci. 2019, 9, 2412. [Google Scholar] [CrossRef]
- Ames, N.J.; Sulima, P.; Ngo, T.; Barb, J.; Munson, P.J.; Paster, B.J.; Hart, T.C. A Characterization of the Oral Microbiome in Allogeneic Stem Cell Transplant Patients. PLoS ONE 2012, 7, e47628. [Google Scholar] [CrossRef]
- Li, M.; Zou, Y.; Jiang, Q.; Jiang, L.; Yu, Q.; Ding, X.; Yu, Y. A preliminary study of the oral microbiota in Chinese patients with Sjögren’s syndrome. Arch. Oral Biol. 2016, 70, 143–148. [Google Scholar] [CrossRef]
- Di Bella, J.M.; Bao, Y.; Gloor, G.B.; Burton, J.P.; Reid, G. High throughput sequencing methods and analysis for microbiome research. J. Microbiol. Methods 2013, 95, 401–414. [Google Scholar] [CrossRef]
- Sanschagrin, S.; Yergeau, E. Next-generation sequencing of 16S ribosomal RNA gene amplicons. J. Vis. Exp. 2014. [Google Scholar] [CrossRef]
- Martínez-Ceballos, M.A.; Aguilera, N.; Garzón-González, K.C.; Cajamarca-Baron, J.; Alzate-Granados, J.P.; Rojas-Villarraga, A. Unstimulated whole salivary flow in Sjögren’s Syndrome: Systematic literature review and meta-analysis. Hortic. Bras. 2021, 61, 12. [Google Scholar] [CrossRef]

| Test Group | Group P | Group N | p-Value | |
|---|---|---|---|---|
| Number of cases | 5 | 5 | 5 | NA |
| Mean age (years) # | 59.8 ± 7.5 | 57.4 ± 15.2 | 45.0 ± 9.4 | 0.122 |
| Women (n, %) | 5 (100%) | 5 (100%) | 5 (100%) | NA |
| Smoker (n) | 0 | 0 | 0 | NA |
| Autoimmune diseases other than pSS (n) | 0 | 0 | 0 | NA |
| Received DMARDs in last 3 months (n) | 3 | 0 | 0 | NA |
| Duration of clinically apparent xerostomia (median months, range) | 24 (6–36) | 0 (0–0) | 0 (0–0) | NA |
| Ocular dryness (n) | 3 | 0 | 0 | NA |
| Positivity of anti-SSA (n) | 3 | 0 | 0 | NA |
| LSG biopsy focus score ≥ 1(n) | 2 | 0 | 0 | NA |
| Candida carriage (cfu/mL) | >200 | >200 | 0 | NA |
| Wearing removable denture (n) | 1 | 1 | 0 | NA |
| DMFT # | 21.6 ± 6.7 | 17.4 ± 6.5 | 13.6 ± 3.8 | 0.137 |
| DMFS # | 67.6 ± 12.9 | 58.0 ± 27.9 | 34.0 ± 19.7 | 0.069 |
| Total incisal caries UWSF (mL/min) | 3 0 | 0 >0.1 | 0 >0.1 | 0.148 0.047 * |
| (a) | ||||
|---|---|---|---|---|
| OTU | Taxonomy (Genera) | Mean ± SD Test Group | Mean ± SD Group P | Metastat p-Value |
| Increase in SS | ||||
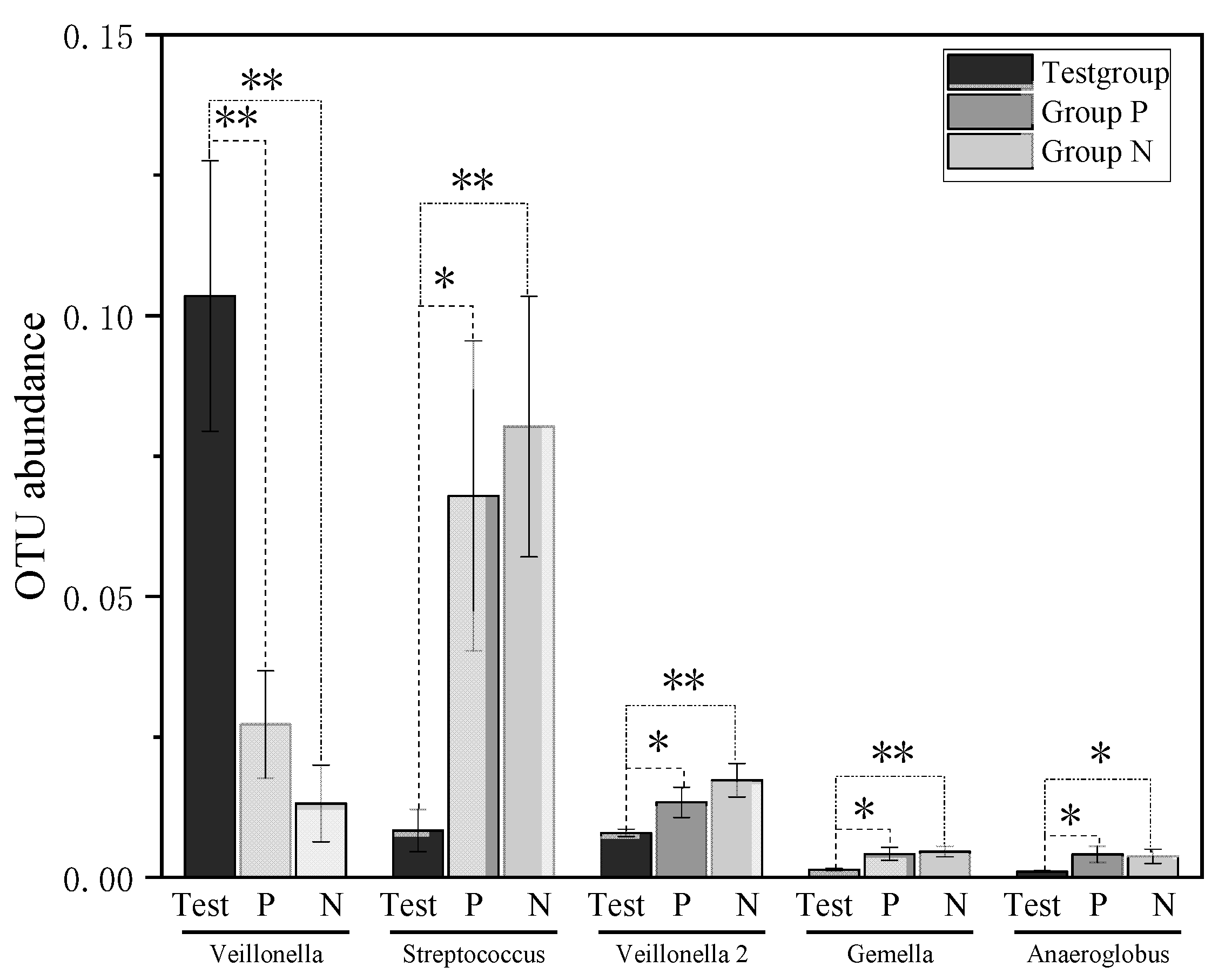
| OTU001 | Veillonella | 0.103472 ± 0.024092 | 0.027183 ± 0.00953 | 0.007 ** |
| OTU038 | Gemella | 0.01297 ± 0.002196 | 0.006591 ± 0.000599 | 0.008 ** |
| OTU042 | Neisseria | 0.019715 ± 0.008202 | 0.002831 ± 0.000635 | 0.039 * |
| OTU066 | Lactobacillus | 0.005269 ± 0.001321 | 0.00035 ± 0.0003 | 0.002 ** |
| OTU102 | Selenomonas | 0.001091 ± 0.000422 | 0.000218 ± 0.000096 | 0.042 * |
| Decrease in SS | ||||
| OTU008 | Neisseria | 0.017735 ± 0.007067 | 0.053774 ± 0.015188 | 0.030 * |
| OTU011 | Streptococcus | 0.008269 ± 0.003757 | 0.067866 ± 0.027617 | 0.032 * |
| OTU013 | Firmicutes | 0.003479 ± 0.001823 | 0.021472 ± 0.00627 | 0.009 ** |
| OTU021 | Veillonella | 0.007866 ± 0.000652 | 0.013299 ± 0.002685 | 0.049 * |
| OTU045 | Gemella | 0.001301 ± 0.000211 | 0.004119 ± 0.001153 | 0.019 * |
| OTU047 | Anaeroglobus | 0.000994 ± 0.00015 | 0.004037 ± 0.001439 | 0.035 * |
| OTU060 | Cardiobacterium | 0.000349 ± 0.000087 | 0.005424 ± 0.002308 | 0.029 * |
| (b) | ||||
| OTU | Taxonomy (Genera) | Mean ± SD Test Group | Mean ± SD Group N | Metastat p-Value |
| Increase in SS | ||||
| OTU001 | Veillonella | 0.103472 ± 0.024092 | 0.013111 ± 0.006831 | 0.002 ** |
| Decrease In SS | ||||
| OTU002 | Streptococcus | 0.082284 ± 0.006397 | 0.182442 ± 0.028316 | 0.003 ** |
| OTU011 | Streptococcus | 0.008269 ± 0.003757 | 0.080205 ± 0.023212 | 0.006 ** |
| OTU012 | Veillonella | 0.001973 ± 0.000453 | 0.006733 ± 0.001754 | 0.014 * |
| OTU015 | Neisseriaceae_unclassified | 0.011237 ± 0.002864 | 0.03873 ± 0.010131 | 0.015 * |
| OTU021 | Veillonella | 0.007866 ± 0.000652 | 0.017235 ± 0.002976 | 0.006 ** |
| OTU045 | Gemella | 0.001301 ± 0.000211 | 0.004557 ± 0.00096 | 0.004 ** |
| OTU047 | Anaeroglobus | 0.000994 ± 0.00015 | 0.003697 ± 0.001258 | 0.037 * |
| OTU050 | Megasphaera | 0.002128 ± 0.00047 | 0.007184 ± 0.002469 | 0.048 * |
| OTU058 | Selenomonas | 0.000647 ± 0.000179 | 0.01557 ± 0.005949 | 0.018 * |
| OTU061 | Veillonella | 0.001773 ± 0.000745 | 0.008027 ± 0.00244 | 0.020 * |
| OTU086 | Veillonella | 0.000267 ± 0.00011 | 0.003788 ± 0.001247 | 0.010 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, H.; Liu, H.; Pan, J. High-Throughput Sequencing of Oral Microbiota in Candida Carriage Sjögren’s Syndrome Patients: A Pilot Cross-Sectional Study. J. Clin. Med. 2023, 12, 1559. https://doi.org/10.3390/jcm12041559
Xing H, Liu H, Pan J. High-Throughput Sequencing of Oral Microbiota in Candida Carriage Sjögren’s Syndrome Patients: A Pilot Cross-Sectional Study. Journal of Clinical Medicine. 2023; 12(4):1559. https://doi.org/10.3390/jcm12041559
Chicago/Turabian StyleXing, Haixia, Hongwei Liu, and Jie Pan. 2023. "High-Throughput Sequencing of Oral Microbiota in Candida Carriage Sjögren’s Syndrome Patients: A Pilot Cross-Sectional Study" Journal of Clinical Medicine 12, no. 4: 1559. https://doi.org/10.3390/jcm12041559
APA StyleXing, H., Liu, H., & Pan, J. (2023). High-Throughput Sequencing of Oral Microbiota in Candida Carriage Sjögren’s Syndrome Patients: A Pilot Cross-Sectional Study. Journal of Clinical Medicine, 12(4), 1559. https://doi.org/10.3390/jcm12041559






