Design of Proposed Software System for Prediction of Iliosacral Screw Placement for Iliosacral Joint Injuries Based on X-ray and CT Images
Abstract
1. Introduction
- Multiregional 3D segmentation model of pelvis area from CT images
- Reconstructed DDR projections with virtual iliosacral screw
- Multimodal (X-ray/CT) image registration for optimal CT slice selection according to the reference X-ray image.
2. Recent Work
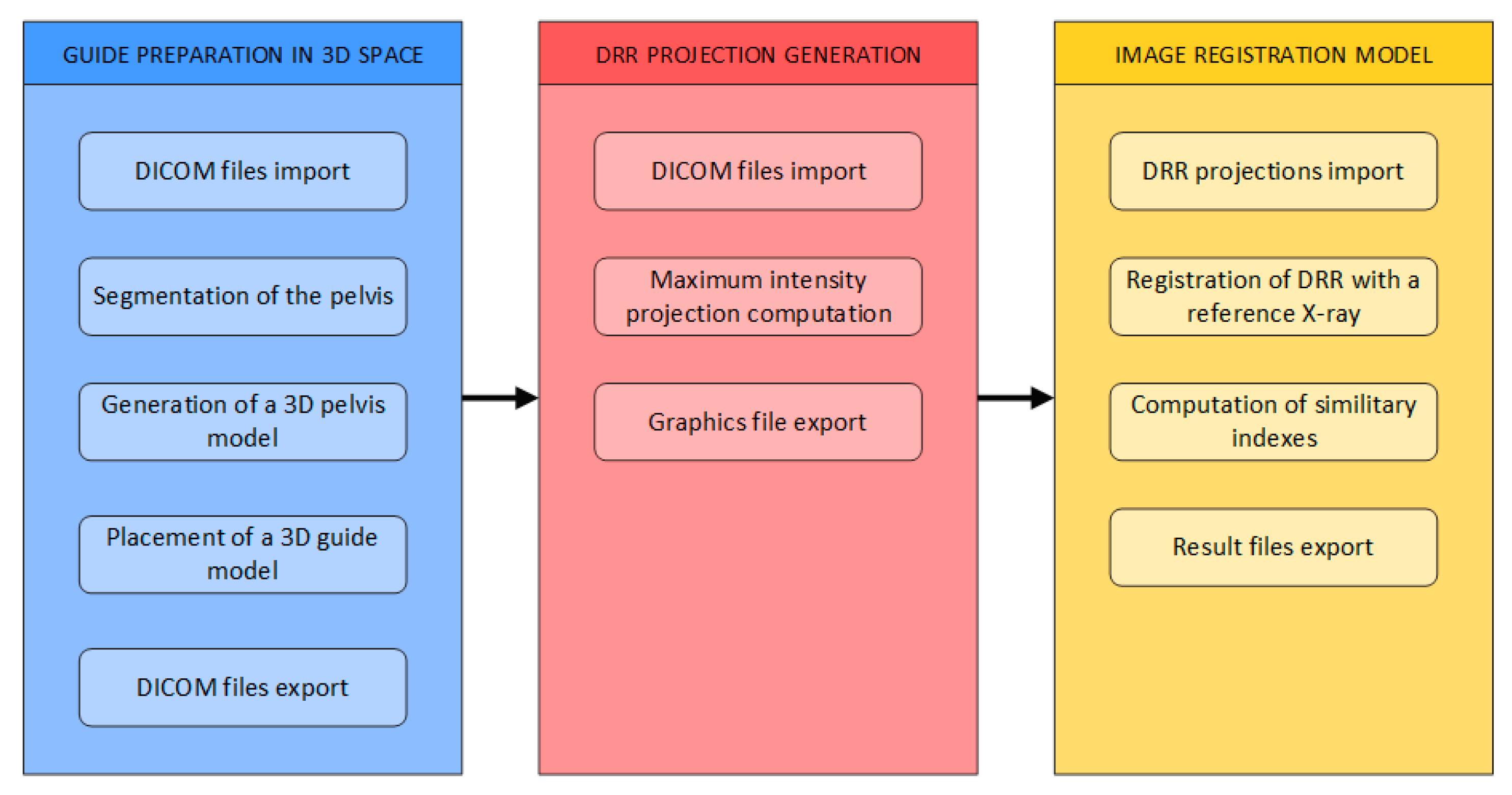
3. Materials and Methods
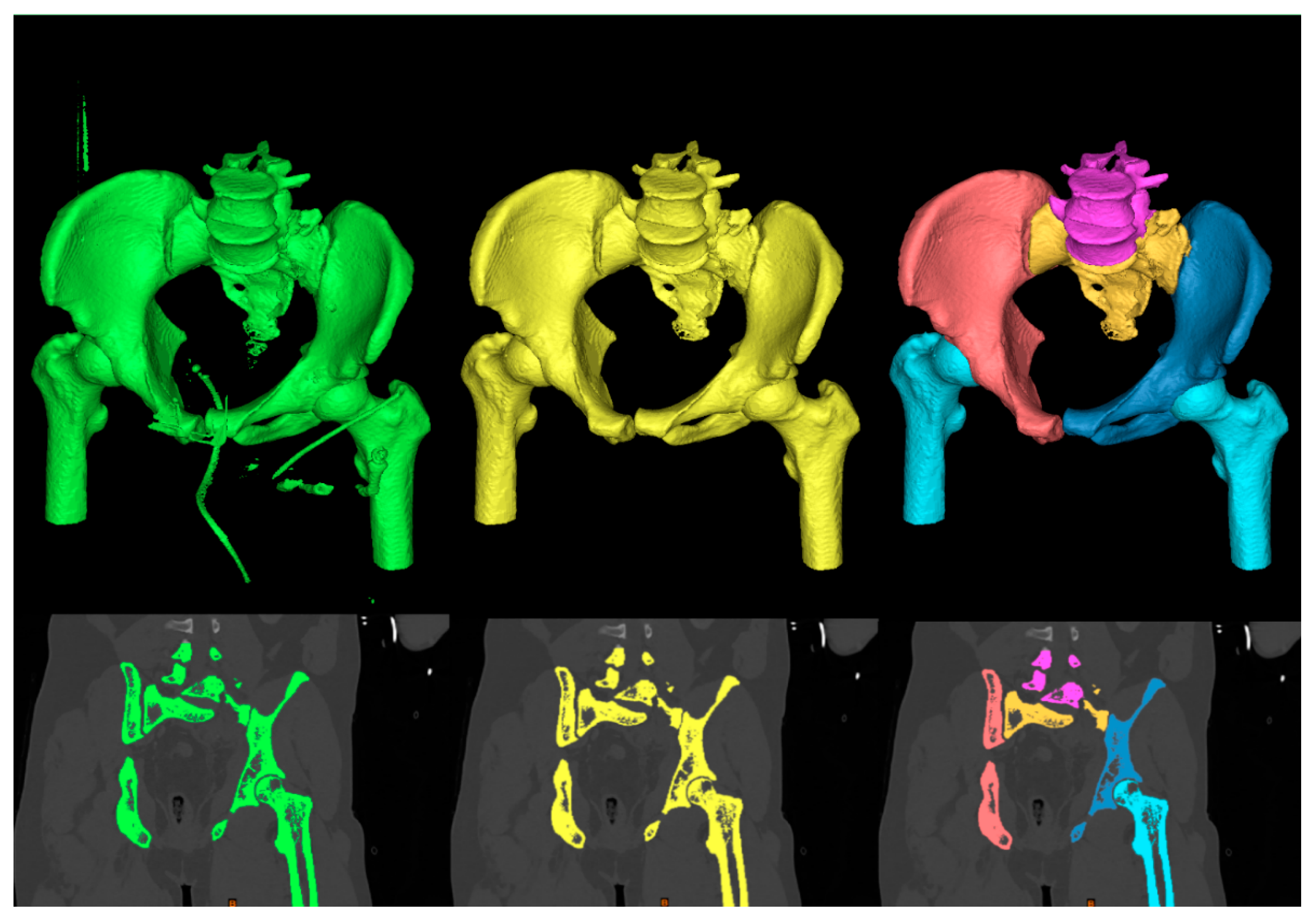
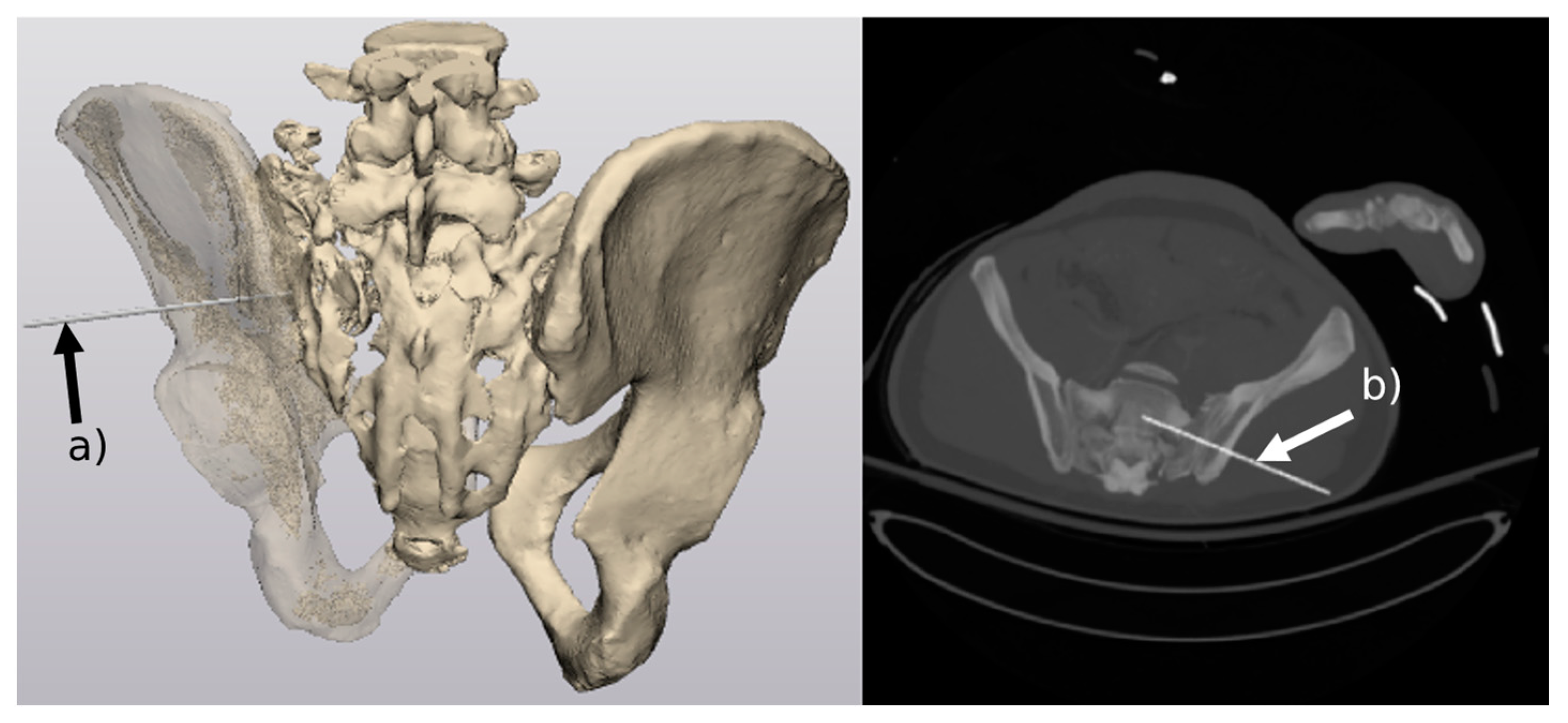
- Generation of 3D models of the pelvis
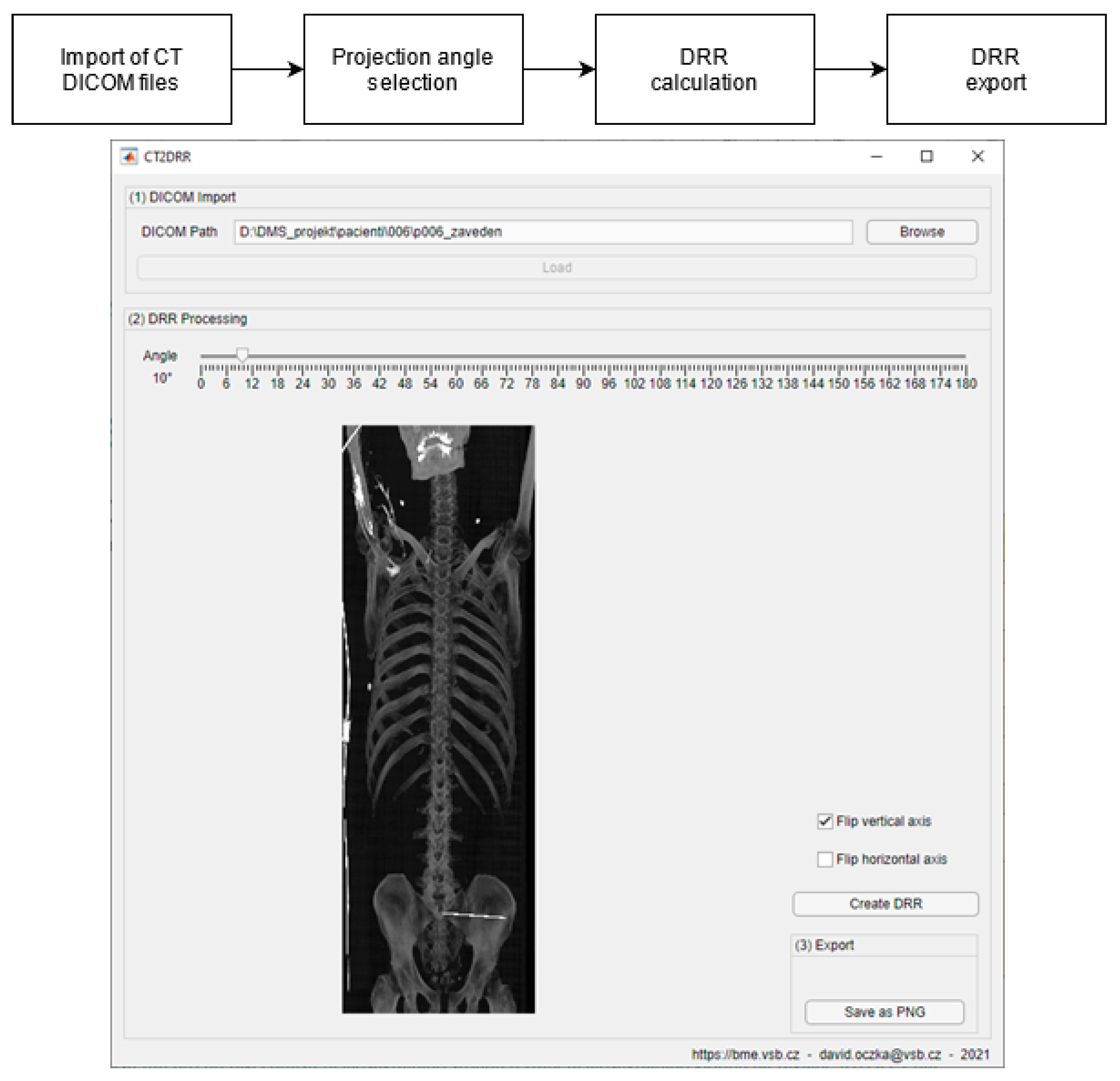
- Generation of digitally reconstructed radiograph (DRR) projections
- Multimodal image registration of DRR projections to a reference X-ray image
3.1. Generation of 3D Models of the Pelvis
3.2. DDR Projection Generation (CT2DDR)
3.3. Image Histogram Pre-Processing
3.4. Image Registration Model
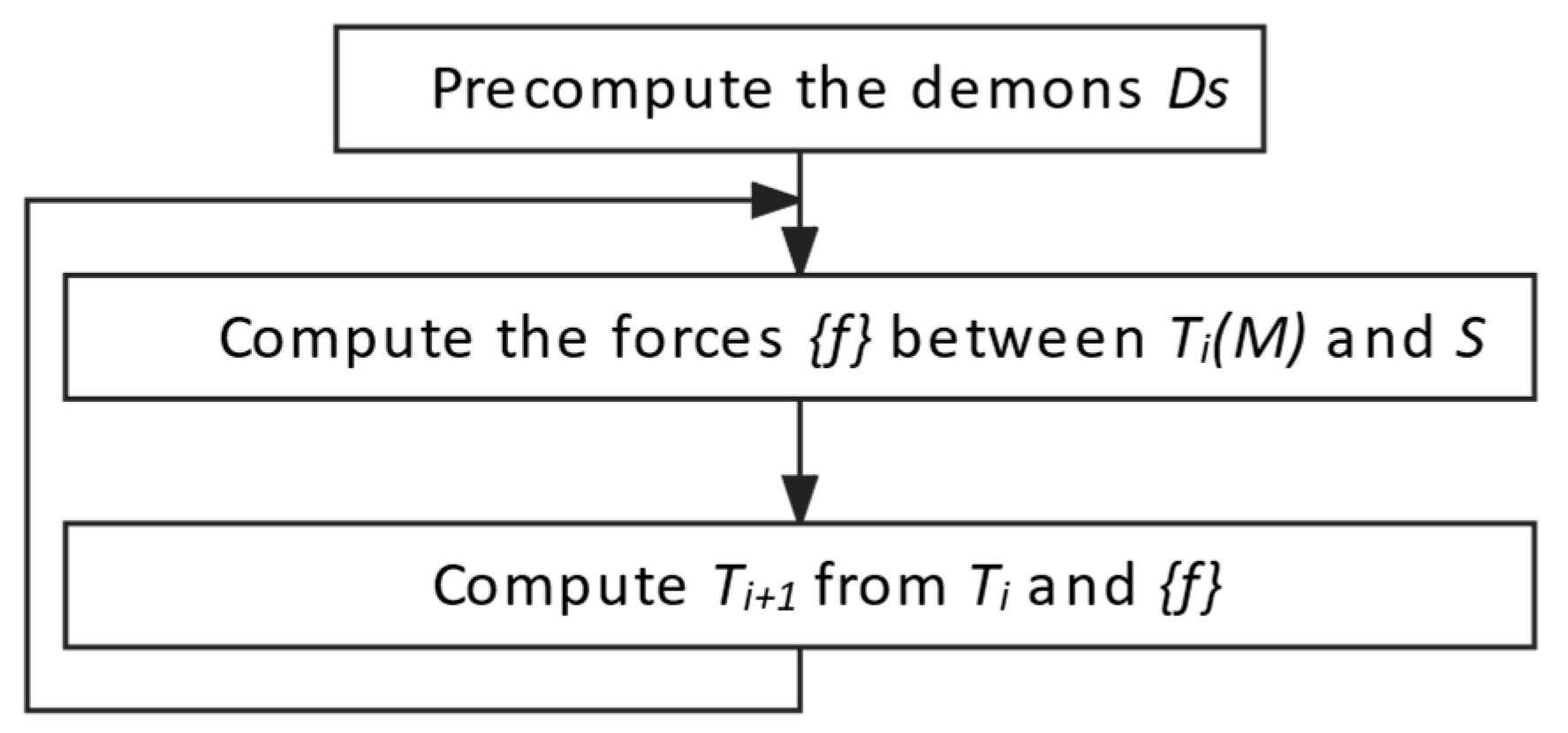
3.4.1. Multimodal image registration algorithm
3.4.2. Statistical Metrics for Registration Evaluation
4. Results
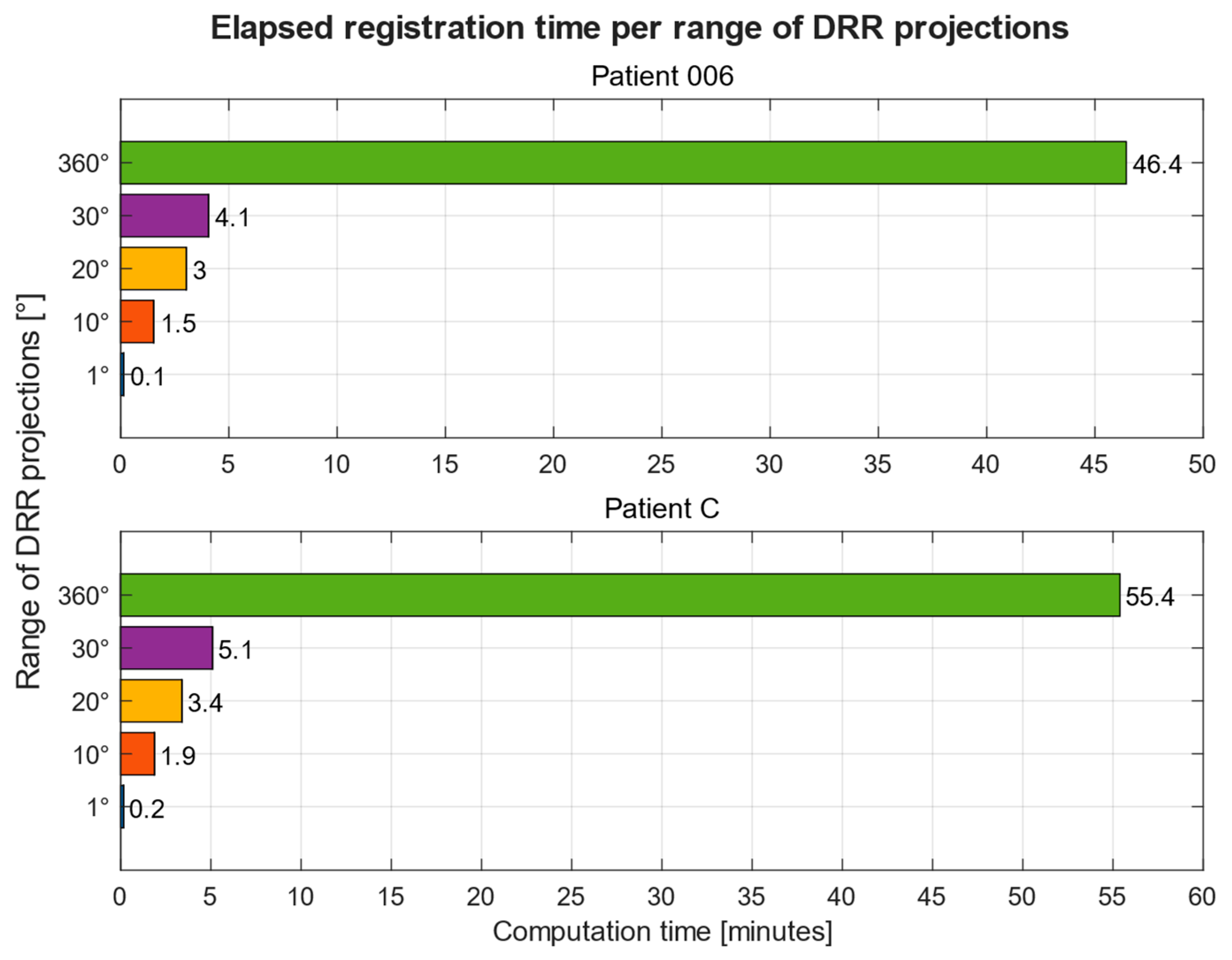
Computation Time
5. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Birkfellner, W.; Burgstaller, W.; Wirth, J.; Baumann, B.; Jacob, A.L.; Bieri, K.; Traud, S.; Strub, M.; Regazzoni, P.; Messmer, P. LORENZ: A system for planning long-bone fracture reduction. In Medical Imaging 2003: Visualization, Image-Guided Procedures, and Display; SPIE—The International Society for Optical Engineering: Bellingham, WA, USA, 2003; Volume 5029, pp. 500–503. [Google Scholar] [CrossRef]
- Jiménez-Delgado, J.J.; Paulano-Godino, F.; PulidoRam-Ramírez, R.; Jiménez-Pérez, J.R. Computer assisted preoperative planning of bone fracture reduction: Simulation techniques and new trends. Med. Image Anal. 2016, 30, 30–45. [Google Scholar] [CrossRef] [PubMed]
- Rambani, R.; Varghese, M. Computer assisted navigation in orthopaedics and trauma surgery. Orthop. Trauma 2014, 28, 50–57. [Google Scholar] [CrossRef]
- Nolte, L.-P.; Zamorano, L.; Visarius, H.; Berlemann, U.; Langlotz, F.; Arm, E.; Schwarzenbach, O. Clinical evaluation of a system for precision enhancement in spine surgery. Clin. Biomech. 1995, 10, 293–303. [Google Scholar] [CrossRef]
- Merloz, P.; Tonetti, J.; Eid, A.; Faure, C.; Lavallee, S.; Troccaz, J.; Sautot, P.; Hamadeh, A.; Cinquin, P. Computer Assisted Spine Surgery. Clin. Orthop. Relat. Res. 1997, 337, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, D.; Garimella, R.; Eltorai, A.E.M.; Daniels, A.H. Computer-assisted Orthopaedic Surgery. Orthop. Surg. 2017, 9, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.-D.; Kim, I.-S.; Bhardwaj, A.M.; Badami, R.N. The Evolution of Computer-Assisted Total Hip Arthroplasty and Relevant Applications. Hip Pelvis 2017, 29, 1–14. [Google Scholar] [CrossRef]
- Picard, F.; Deakin, A.H.; Riches, P.E.; Deep, K.; Baines, J. Computer assisted orthopaedic surgery: Past, present and future. Med. Eng. Phys. 2019, 72, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Nolte, L.-P. Computer-Aided Orthopaedic Surgery: State-of-the-Art and Future Perspectives. Adv. Exp. Med. Biol. 2018, 1093, 1–20. [Google Scholar] [CrossRef]
- Pan, W.-B.; Liang, J.-B.; Wang, B.; Chen, G.-F.; Hong, H.-X.; Li, Q.-Y.; Chen, H.-X. The invention of an iliosacral screw fixation guide and its preliminary clinical application. Orthop. Surg. 2012, 4, 55–59. [Google Scholar] [CrossRef]
- Gandhi, G.; Vijayvargiya, M.; Shetty, V.; Agashe, V.; Maheshwari, S.; Monteiro, J. CT-guided percutaneous sacroiliac stabilization in unstable pelvic fractures: A safe and accurate technique. Rev. Bras. Ortop. 2018, 53, 323–331. [Google Scholar] [CrossRef]
- Richter, P.; Gebhard, F.; Dehner, C.; Scola, A. Accuracy of computer-assisted iliosacral screw placement using a hybrid operating room. Injury 2016, 47, 402–407. [Google Scholar] [CrossRef] [PubMed]
- Tonetti, J.; Boudissa, M.; Kerschbaumer, G.; Seurat, O. Role of 3D intraoperative imaging in orthopedic and trauma surgery. Orthop. Traumatol. Surg. Res. 2020, 106, S19–S25. [Google Scholar] [CrossRef]
- Xu, P.; Wang, H.; Liu, Z.-Y.; Mu, W.-D.; Xu, S.-H.; Wang, L.-B.; Chen, C.; Cavanaugh, J.M. An evaluation of three-dimensional image–guided technologies in percutaneous pelvic and acetabular lag screw placement. J. Surg. Res. 2013, 185, 338–346. [Google Scholar] [CrossRef]
- Takao, M.; Nishii, T.; Sakai, T.; Yoshikawa, H.; Sugano, N. Iliosacral screw insertion using CT-3D-fluoroscopy matching navigation. Injury 2014, 45, 988–994. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Dai, J.; Zhang, J.; Ma, Y.; Zhu, G.; Shen, J.; Niu, G. Individualized 3D printing navigation template for pedicle screw fixation in upper cervical spine. PLoS ONE 2017, 12, e0171509. [Google Scholar] [CrossRef]
- Liu, F.; Yu, J.; Yang, H.; Cai, L.; Chen, L.; Lei, Q.; Lei, P. Iliosacral screw fixation of pelvic ring disruption with tridimensional patient-specific template guidance. Orthop. Traumatol. Surg. Res. 2022, 108, 103210. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, N.; Dubey, V.N.; Wainwright, T.W.; Middleton, R.G. A review of virtual reality based training simulators for orthopaedic surgery. Med. Eng. Phys. 2016, 38, 59–71. [Google Scholar] [CrossRef]
- Tonetti, J.; Vadcard, L.; Girard, P.; Dubois, M.; Merloz, P.; Troccaz, J. Assessment of a percutaneous iliosacral screw insertion simulator. Orthop. Traumatol. Surg. Res. 2009, 95, 471–477. [Google Scholar] [CrossRef]
- Yang, Q.; Feng, S.; Song, J.; Cheng, C.; Liang, C.; Wang, Y. Computer-aided automatic planning and biomechanical analysis of a novel arc screw for pelvic fracture internal fixation. Comput. Methods Programs Biomed. 2022, 220, 106810. [Google Scholar] [CrossRef]
- Zakariaee, R.; Schlosser, C.L.; Baker, D.R.; Meek, R.N.; Coope, R.J. A feasibility study of pelvic morphology for curved implants. Injury 2016, 47, 2195–2202. [Google Scholar] [CrossRef]
- Alambeigi, F.; Wang, Y.; Sefati, S.; Gao, C.; Murphy, R.J.; Iordachita, I.; Taylor, R.H.; Khanuja, H.; Armand, M. A Curved-Drilling Approach in Core Decompression of the Femoral Head Osteonecrosis Using a Continuum Manipulator. IEEE Robot. Autom. Lett. 2017, 2, 1480–1487. [Google Scholar] [CrossRef]
- Qi, J.; Hu, Y.; Yang, Z.; Dong, Y.; Zhang, X.; Hou, G.; Lv, Y.; Guo, Y.; Zhou, F.; Liu, B.; et al. Incidence, Risk Factors, and Outcomes of Symptomatic Bone Cement Displacement following Percutaneous Kyphoplasty for Osteoporotic Vertebral Compression Fracture: A Single Center Study. J. Clin. Med. 2022, 11, 7530. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Wang, T.; Xu, X.; Ran, G.; Zhang, H.; Dong, Q.; Zhang, Q.; Guo, J.; Hou, Z. Risk Factors and Outcomes of Extended Length of Stay in Older Adults with Intertrochanteric Fracture Surgery: A Retrospective Cohort Study of 2132 Patients. J. Clin. Med. 2022, 11, 7366. [Google Scholar] [CrossRef] [PubMed]
- Bashiri, F.S.; Baghaie, A.; Rostami, R.; Yu, Z.; D’Souza, R.M. Multi-Modal Medical Image Registration with Full or Partial Data: A Manifold Learning Approach. J. Imaging 2018, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Du, J.; Zhang, Y.; Gong, Y.; Zhang, B.; Qu, Z.; Hao, D.; He, B.; Yan, L. Predictive Factors for Bone Cement Displacement following Percutaneous Vertebral Augmentation in Kümmell’s Disease. J. Clin. Med. 2022, 11, 7479. [Google Scholar] [CrossRef] [PubMed]
- Cahill, N.; Noble, J.A.; Hawkes, D.J.; Noble, A. A Demons Algorithm for Image Registration with Locally Adaptive Regularization. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2009; Springer: Berlin/Heidelberg, Germany, 2009; pp. 574–581. [Google Scholar] [CrossRef]
- Thirion, J.-P. Image matching as a diffusion process: An analogy with Maxwell′s demons. Med. Image Anal. 1998, 2, 243–260. [Google Scholar] [CrossRef]
- Abidi, A.I.; Singh, S. Deformable Registration Techniques for Thoracic CT Images: An Insight into Medical Image Registration; Springer: Singapore, 2020. [Google Scholar] [CrossRef]
- El-Baz, A.S.; Acharya, U.R.; Laine, A.F.; Suri, J.S. (Eds.) Multi Modality State-of-the-Art Medical Image Segmentation and Registration Methodologies: Volume II; Springer: New York, NY, USA, 2011. [Google Scholar] [CrossRef]
- Wang, Z.; Simoncelli, E.P.; Bovik, A.C. Multiscale structural similarity for image quality assessment. In Proceedings of the Thrity-Seventh Asilomar Conference on Signals, Systems & Computers, Pacific Grove, CA, USA, 9–12 November 2003; Volune 2, pp. 1398–1402. [Google Scholar] [CrossRef]
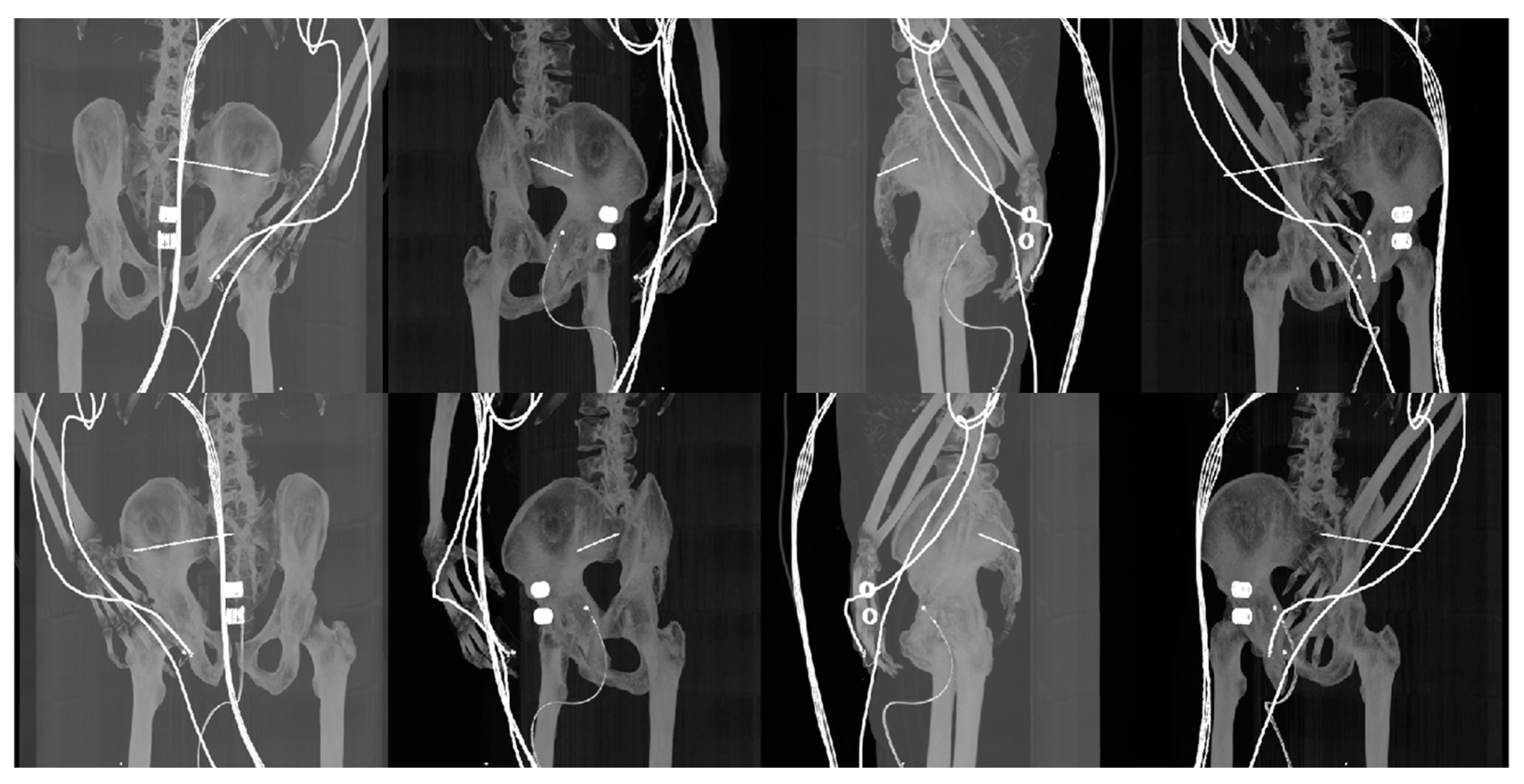
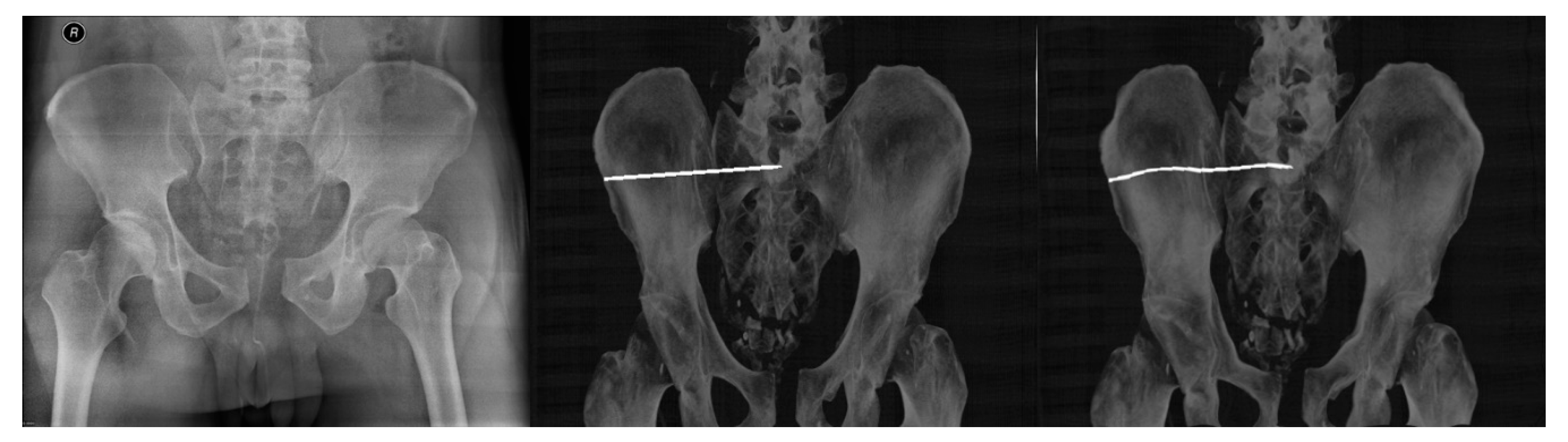
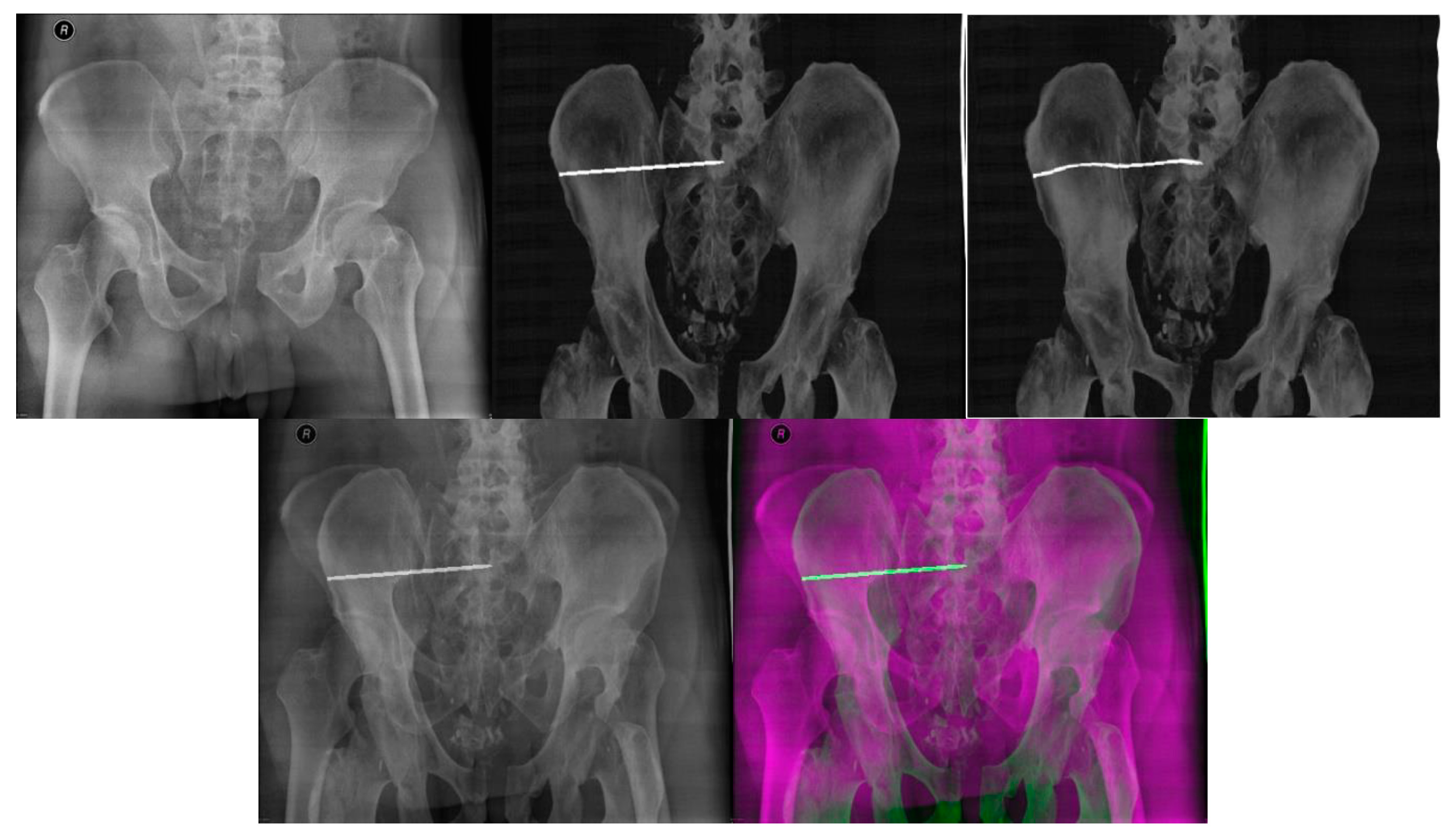
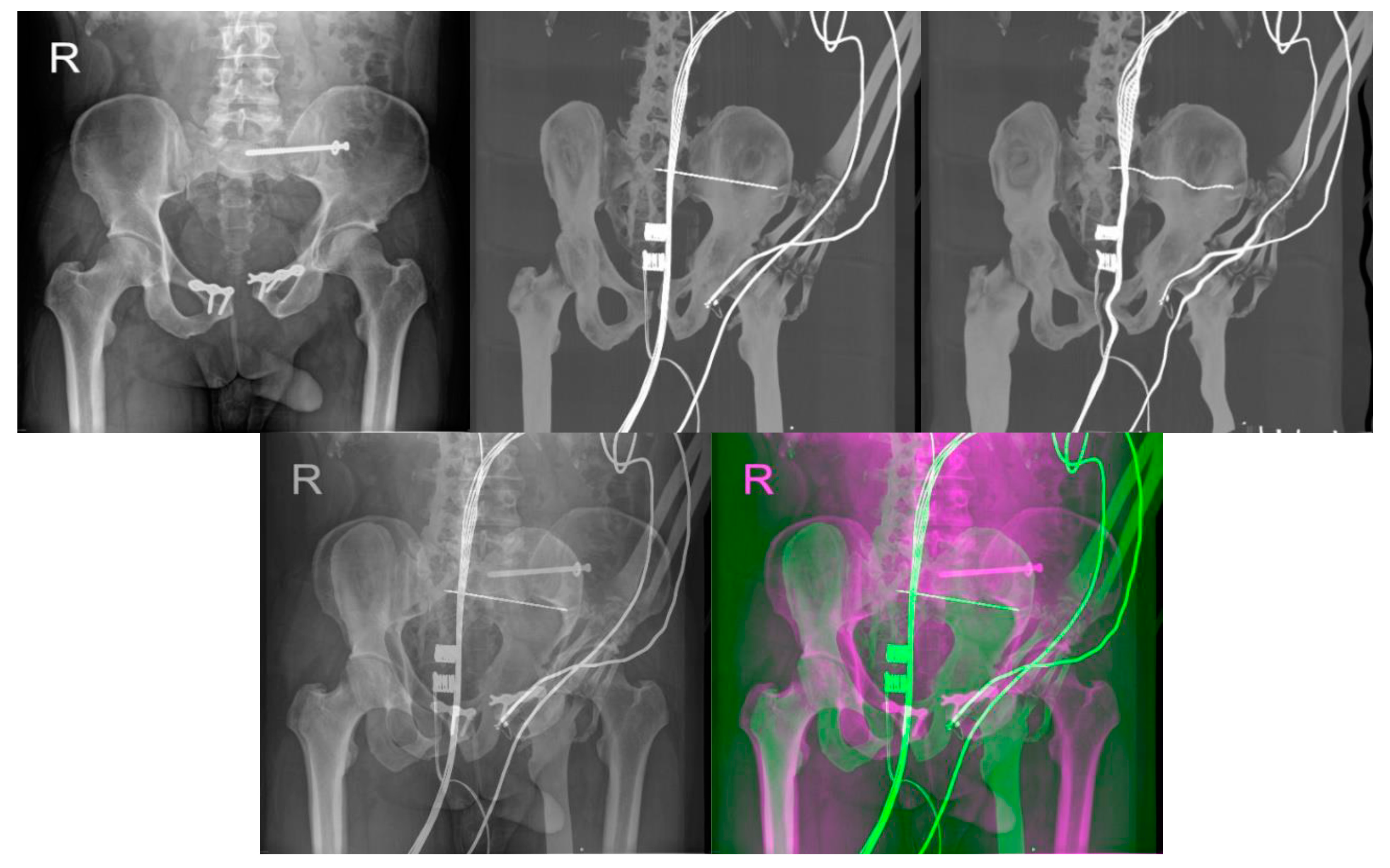
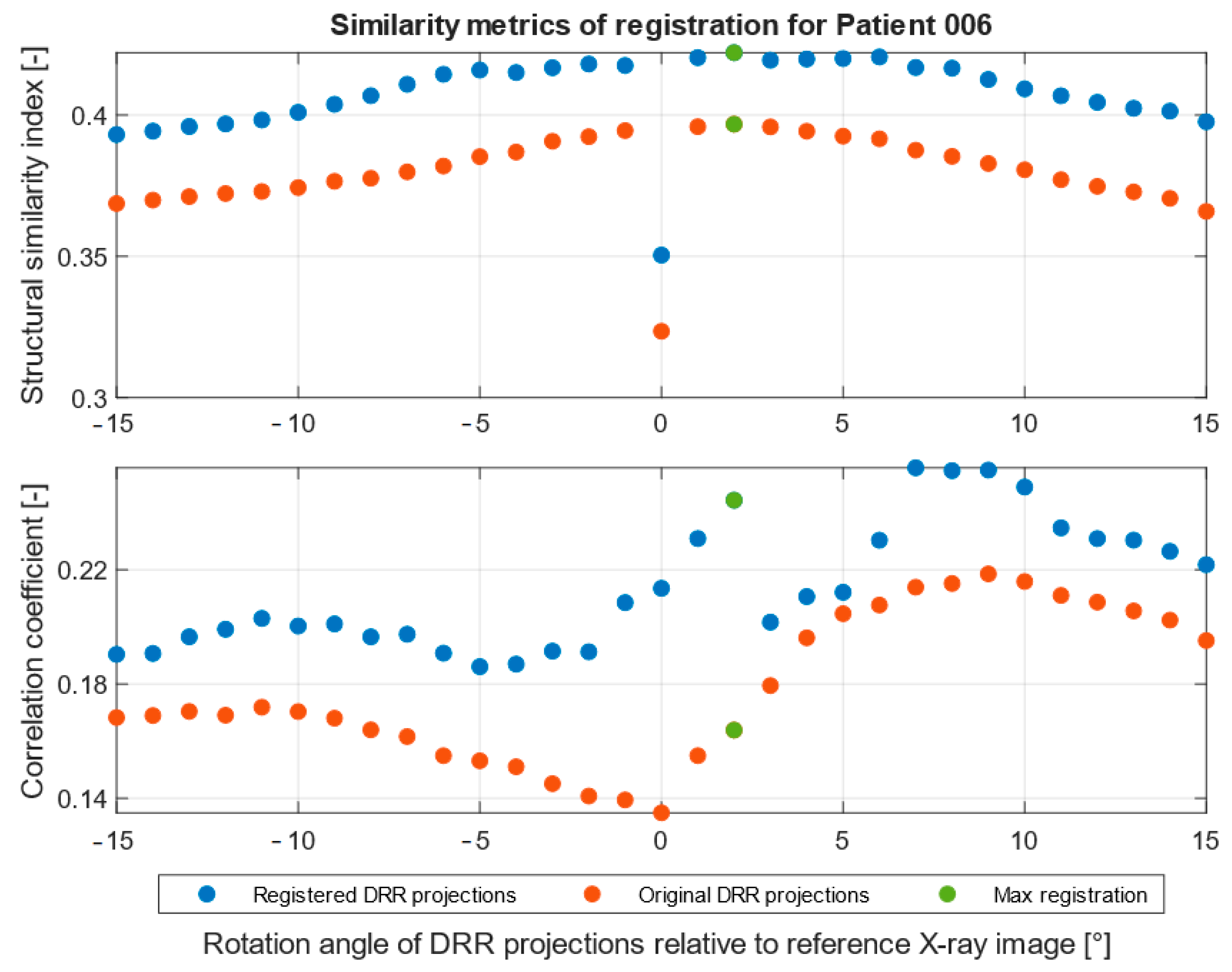
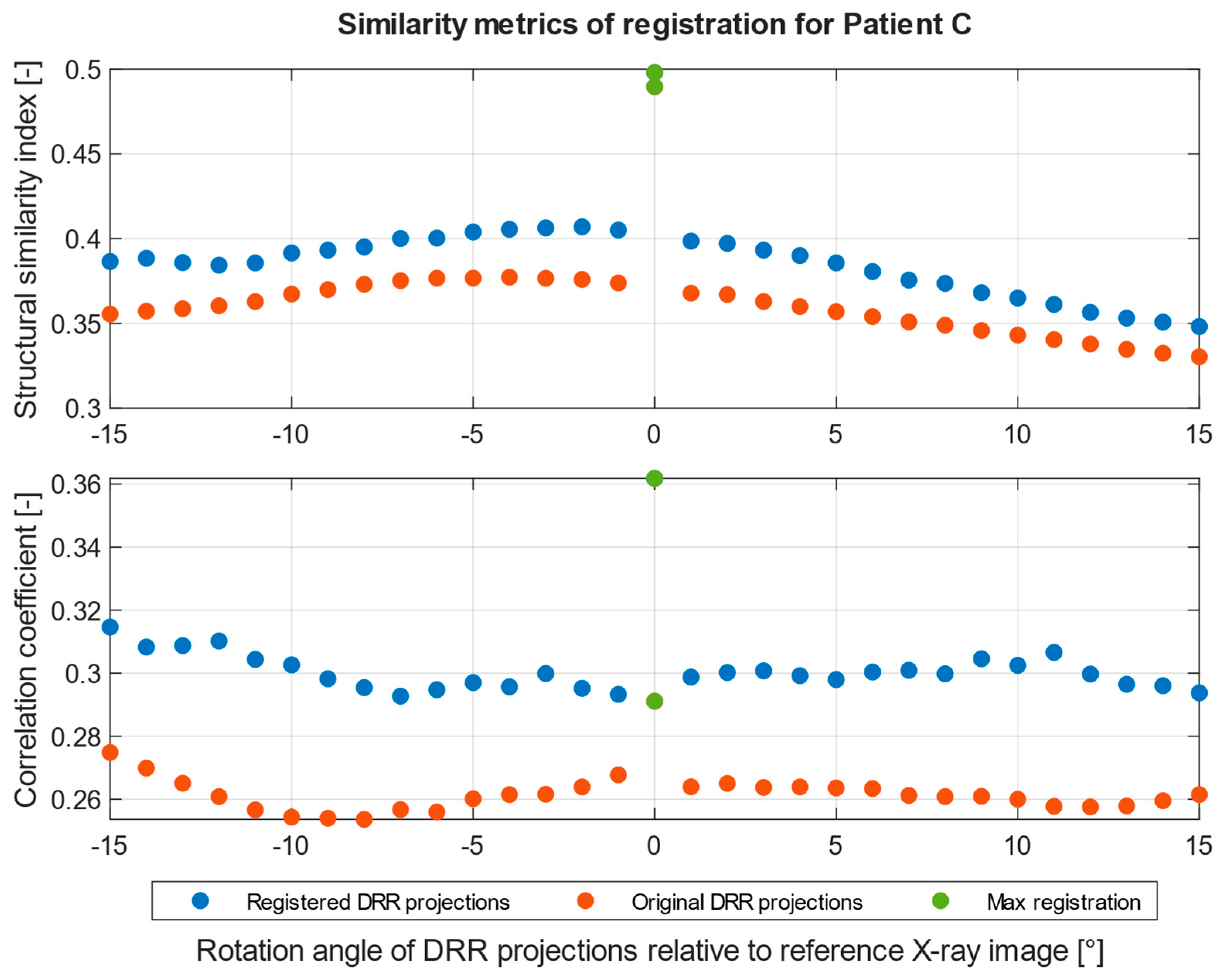
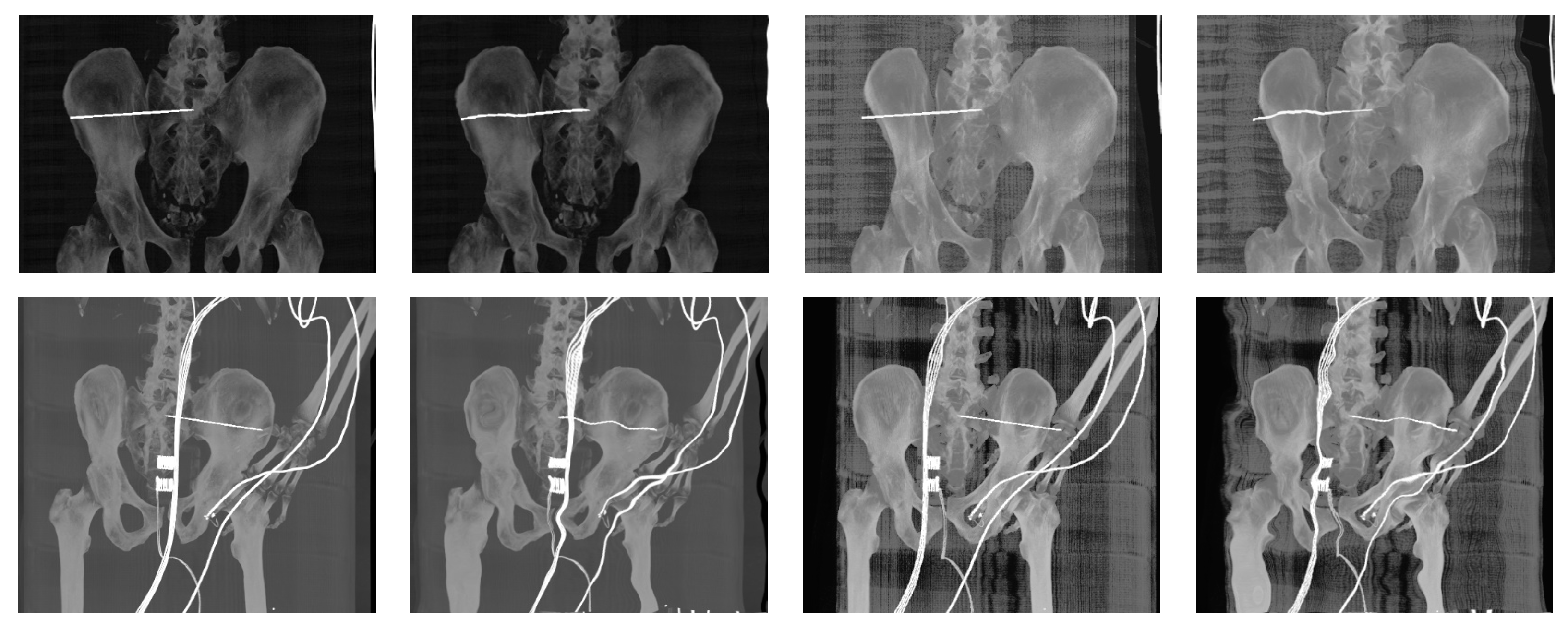
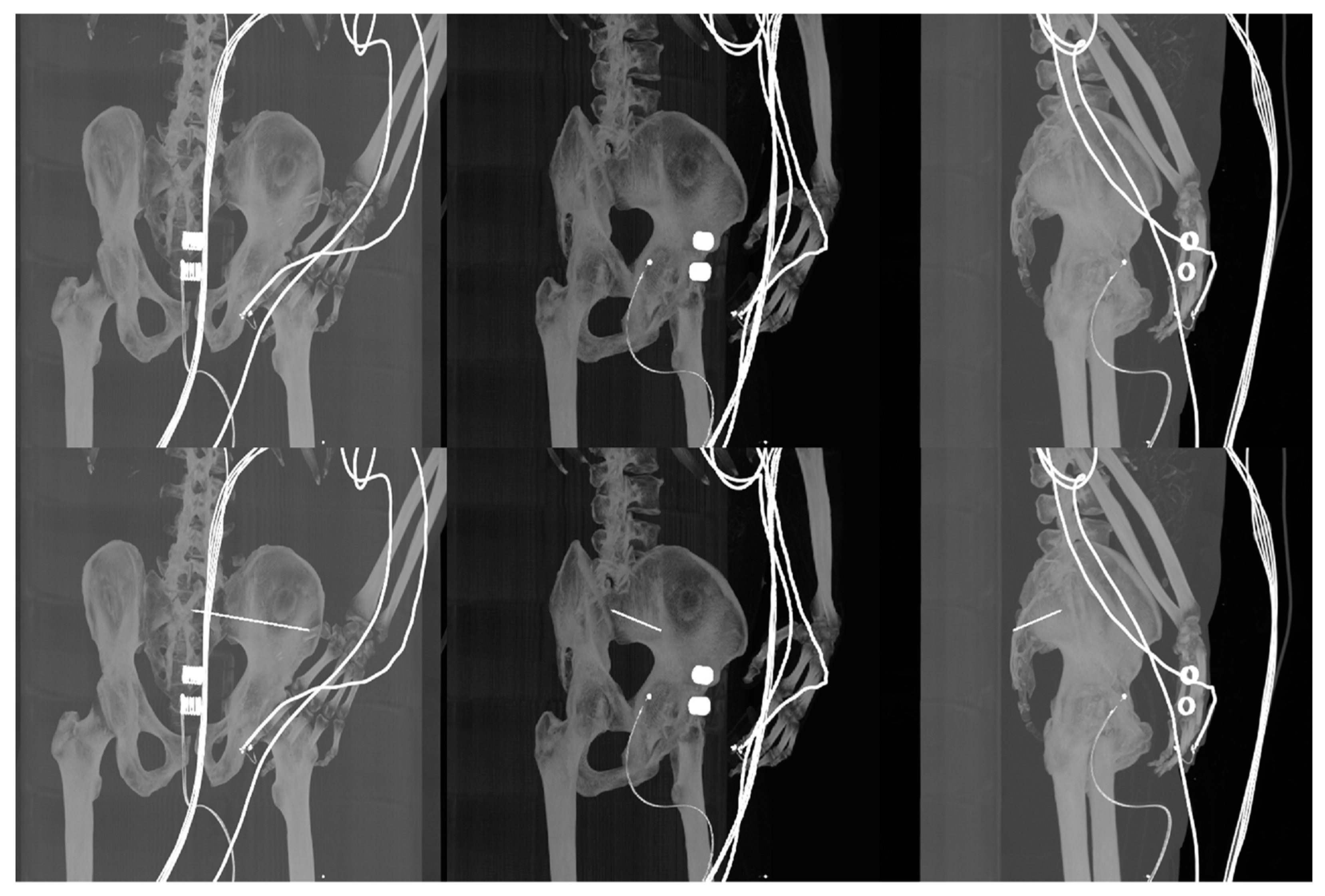
| Imaging Device | Modality | Bit-Depth | Image Resolution [Pixels] | Pixel Spacing [mm] | View Position | Body Part Examined | |
|---|---|---|---|---|---|---|---|
| Patient 1 | Kodak Elite CR | CR | 16 | 2048 × 2500 | 0.17/0.17 | AP | Pelvis |
| Patient 2 | Samsung GC85 | DX | 16 | 2994 × 2990 | 0.13/0.13 | AP | Pelvis |
| Imaging Device | Modality | Bit-Depth | Image Resolution [Pixels] | Pixel Spacing [mm] | |
|---|---|---|---|---|---|
| Patient 1 | Siemens Definition AS | CT | 16 | 512 × 512 | 0.81/0.81 |
| Patient 2 | Siemens Somatom Force | CT | 16 | 512 × 512 | 0.94/0.94 |
| Convolution kernel | Pitch factor [mm] | Number of slices | Slice thickness [mm] | Body part examined | |
| B20f | 1.05 | 1561 | 0.6 | Abdomen | |
| Br40d/2 | 1.4 | 779 | 0.75 | Abdomen |
| Patient 1 | Patient 2 | |
|---|---|---|
| SSIM [-] | 0.42 | 0.50 |
| CORR [-] | 0.26 | 0.36 |
| DRR projection | 2° | 0° |
| Computation Times on PC 1 | Computation Times on PC 2 | |||
|---|---|---|---|---|
| DRR Projections | Patient 1 | Patient 2 | Patient 1 | Patient 2 |
| 1° | 0.13 | 0.15 | 0.11 | 0.13 |
| 10° | 1.53 | 1.87 | 1.32 | 1.36 |
| 20° | 3.02 | 3.40 | 2.23 | 2.64 |
| 30° | 3.93 | 4.64 | 3.26 | 3.89 |
| 360° | 46.45 | 55.39 | 38.24 | 45.15 |
| Without Histogram Matching | With Histogram Matching | |||||||
|---|---|---|---|---|---|---|---|---|
| SSIM [-] | CORR [-] | Mean Time [Seconds] | DRR Projection | SSIM [-] | CORR [-] | Mean Time [Seconds] | DRR Projection | |
| Patient 1 | 0.42 | 0.26 | 7.87 | 2° | 0.51 | 0.42 | 8.17 | 15° |
| Patient 2 | 0.50 | 0.36 | 9.86 | 0° | 0.40 | 0.42 | 9.68 | 345° |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benda, V.; Kubicek, J.; Madeja, R.; Oczka, D.; Cerny, M.; Dostalova, K. Design of Proposed Software System for Prediction of Iliosacral Screw Placement for Iliosacral Joint Injuries Based on X-ray and CT Images. J. Clin. Med. 2023, 12, 2138. https://doi.org/10.3390/jcm12062138
Benda V, Kubicek J, Madeja R, Oczka D, Cerny M, Dostalova K. Design of Proposed Software System for Prediction of Iliosacral Screw Placement for Iliosacral Joint Injuries Based on X-ray and CT Images. Journal of Clinical Medicine. 2023; 12(6):2138. https://doi.org/10.3390/jcm12062138
Chicago/Turabian StyleBenda, Vojtech, Jan Kubicek, Roman Madeja, David Oczka, Martin Cerny, and Kamila Dostalova. 2023. "Design of Proposed Software System for Prediction of Iliosacral Screw Placement for Iliosacral Joint Injuries Based on X-ray and CT Images" Journal of Clinical Medicine 12, no. 6: 2138. https://doi.org/10.3390/jcm12062138
APA StyleBenda, V., Kubicek, J., Madeja, R., Oczka, D., Cerny, M., & Dostalova, K. (2023). Design of Proposed Software System for Prediction of Iliosacral Screw Placement for Iliosacral Joint Injuries Based on X-ray and CT Images. Journal of Clinical Medicine, 12(6), 2138. https://doi.org/10.3390/jcm12062138






