Metformin Treatment Reduces the Incidence of Rheumatoid Arthritis: A Two-Sample Mendelian Randomized Study
Abstract
:1. Introduction
2. Materials and Methods
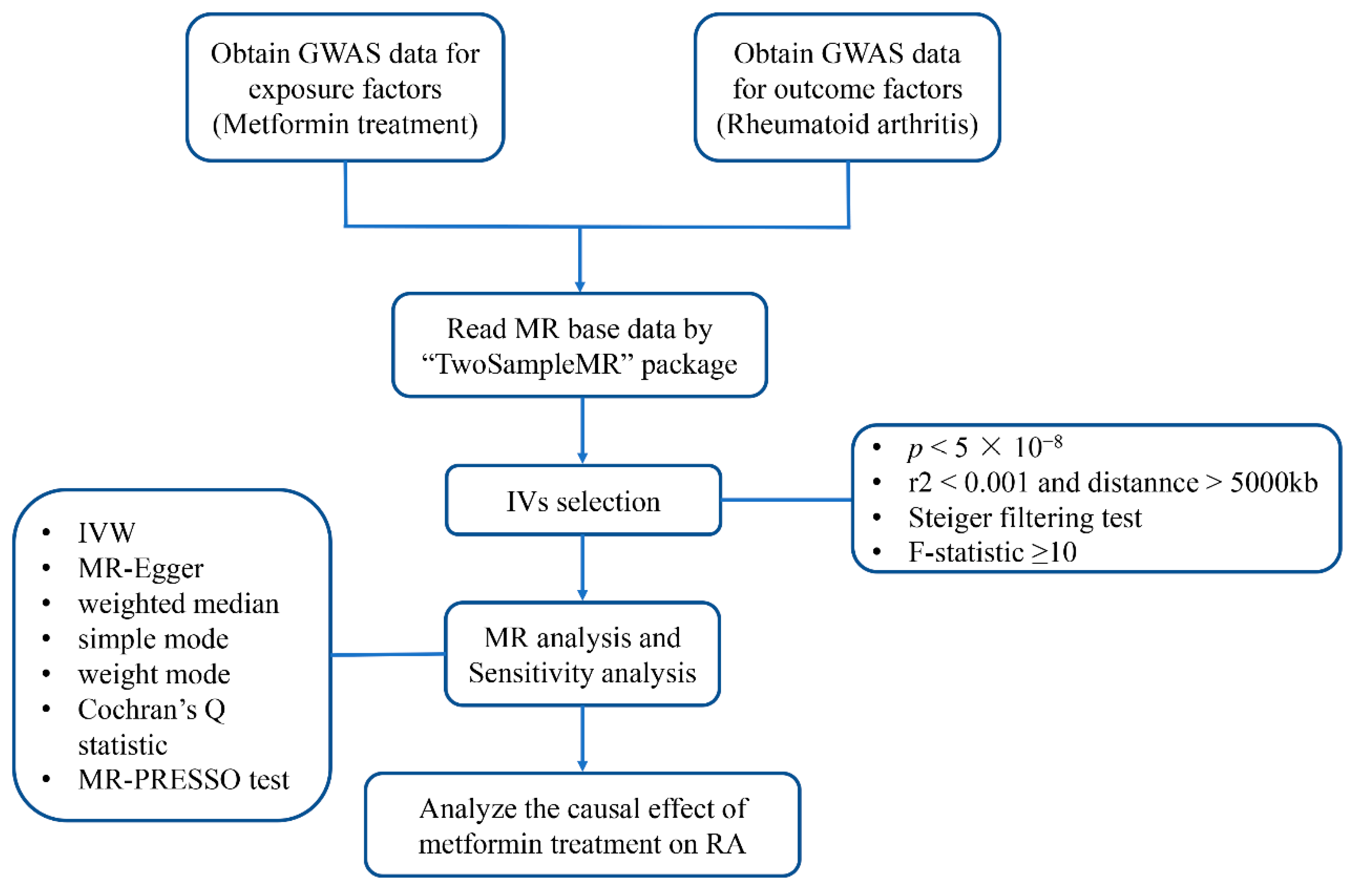
2.1. Study Design
2.2. Data sources and SNPs Selection
2.3. Mendelian Randomization Analysis
2.4. Sensitivity Analysis
2.5. Statistical Analysis
3. Results
3.1. Genetic Variant Selection
3.2. Causal Effects of Metformin Treatment on RA
3.3. Sensitivity Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alamanos, Y.; Voulgari, P.V.; Drosos, A.A. Incidence and prevalence of rheumatoid arthritis, based on the 1987 American College of Rheumatology criteria: A systematic review. Semin. Arthritis Rheum 2006, 36, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Cross, M.; Smith, E.; Hoy, D.; Carmona, L.; Wolfe, F.; Vos, T.; Williams, B.; Gabriel, S.; Lassere, M.; Johns, N.; et al. The global burden of rheumatoid arthritis: Estimates from the global burden of disease 2010 study. Ann. Rheum Dis. 2014, 73, 1316–1322. [Google Scholar] [CrossRef] [PubMed]
- Kitas, G.D.; Gabriel, S.E. Cardiovascular disease in rheumatoid arthritis: State of the art and future perspectives. Ann. Rheum Dis. 2011, 70, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Sokka, T.; Kautiainen, H.; Pincus, T.; Verstappen, S.M.; Aggarwal, A.; Alten, R.; Andersone, D.; Badsha, H.; Baecklund, E.; Belmonte, M.; et al. Work disability remains a major problem in rheumatoid arthritis in the 2000s: Data from 32 countries in the QUEST-RA study. Arthritis Res. Ther. 2010, 12, R42. [Google Scholar] [CrossRef] [Green Version]
- Rossini, M.; Bagnato, G.; Frediani, B.; Iagnocco, A.; La Montagna, G.; Minisola, G.; Caminiti, M.; Varenna, M.; Adami, S. Relationship of focal erosions, bone mineral density, and parathyroid hormone in rheumatoid arthritis. J. Rheumatol. 2011, 38, 997–1002. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Redden, D.T.; McGwin, G., Jr.; Callahan, L.F.; Smith, E.A.; Alarcon, G.S.; Moreland, L.W.; van der Heijde, D.M.; Brown, E.E.; Arnett, D.K.; et al. Generalized bone loss as a predictor of three-year radiographic damage in African American patients with recent-onset rheumatoid arthritis. Arthritis Rheum 2010, 62, 2219–2226. [Google Scholar] [CrossRef]
- Rheumatoid arthritis. Nat. Rev. Dis. Prim. 2018, 4, 18002. [CrossRef]
- Gelman, M.I.; Ward, J.R. Septic Arthritis: A Complication of Rheumatoid Arthritis. Diagn. Radiol. 1977, 122, 17–23. [Google Scholar] [CrossRef]
- Kaneta, T.; Hakamatsuka, T.; Yamada, T.; Takase, K.; Sato, A.; Higano, S.; Ito, H.; Fukuda, H.; Takahashi, S.; Yamada, S. Atlantoaxial Osteoarthritis in Rheumatoid Arthritis: FDG PET/CT Findings. Clin. Nucl. Med. 2006, 31, 209. [Google Scholar] [CrossRef]
- Lora, V.; Cerroni, L.; Cota, C. Skin manifestations of rheumatoid arthritis. G Ital. Dermatol. Venereol. 2018, 153, 243–255. [Google Scholar] [CrossRef]
- Sayah, A.; English, J.C., III. Rheumatoid arthritis: A review of the cutaneous manifestations. J. Am. Acad. Dermatol. 2005, 53, 191–209; quiz 192–210. [Google Scholar] [CrossRef] [PubMed]
- Nurmohamed, M.T.; Heslinga, M.; Kitas, G.D. Cardiovascular comorbidity in rheumatic diseases. Nat. Rev. Rheumatol. 2015, 11, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Olson, A.L.; Swigris, J.J.; Sprunger, D.B.; Fischer, A.; Fernandez-Perez, E.R.; Solomon, J.; Murphy, J.; Cohen, M.; Raghu, G.; Brown, K.K. Rheumatoid arthritis-interstitial lung disease-associated mortality. Am. J. Respir. Crit. Care Med. 2011, 183, 372–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Behl, T.; Sehgal, A.; Singh, S.; Sharma, N.; Naved, T.; Bhatia, S.; Al-Harrasi, A.; Chakrabarti, P.; Aleya, L.J.I.I. Mechanistic insights into the role of b cells in rheumatoid arthritis. Int. Immunopharmacol. 2021, 99, 108078. [Google Scholar] [CrossRef] [PubMed]
- Kurowska, W.; Kuca-Warnawin, E.H.; Radzikowska, A.; Maśliński, W. The role of anti-citrullinated protein antibodies (ACPA) in the pathogenesis of rheumatoid arthritis. Cent. Eur. J. Immunol. 2017, 42, 390–398. [Google Scholar] [CrossRef]
- Marathe, P.H.; Gao, H.X.; Close, K.L. American Diabetes Association Standards of Medical Care in Diabetes 2017. J. Diabetes 2017, 9, 320–324. [Google Scholar] [CrossRef] [Green Version]
- Koh, S.J.; Kim, J.M.; Kim, I.K.; Ko, S.H.; Kim, J.S. Anti-inflammatory mechanism of metformin and its effects in intestinal inflammation and colitis-associated colon cancer. J. Gastroenterol. Hepatol. 2014, 29, 502–510. [Google Scholar] [CrossRef]
- Nath, N.; Khan, M.; Paintlia, M.K.; Singh, I.; Hoda, M.N.; Giri, S. Metformin attenuated the autoimmune disease of the central nervous system in animal models of multiple sclerosis. J. Immunol. 2009, 182, 8005–8014. [Google Scholar] [CrossRef] [Green Version]
- Kang, K.Y.; Kim, Y.K.; Yi, H.; Kim, J.; Jung, H.R.; Kim, I.J.; Cho, J.H.; Park, S.H.; Kim, H.Y.; Ju, J.H. Metformin downregulates Th17 cells differentiation and attenuates murine autoimmune arthritis. Int. Immunopharmacol. 2013, 16, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Elmenshawy, H.; Farouk, H.; Sabri, N.; Ahmed, M. The Impact of Pharmaceutical Care Services on Patients with Active Rheumatoid Arthritis: A Randomized Controlled Study. Arch. Pharm. Sci. Ain. Shams Univ. 2022, 6, 141–155. [Google Scholar] [CrossRef]
- Ala, M.; Ala, M. Metformin for Cardiovascular Protection, Inflammatory Bowel Disease, Osteoporosis, Periodontitis, Polycystic Ovarian Syndrome, Neurodegeneration, Cancer, Inflammation and Senescence: What Is Next? ACS Pharmacol. Transl. Sci. 2021, 4, 1747–1770. [Google Scholar] [CrossRef]
- Chen, Y.; Qiu, F.; Yu, B.; Chen, Y.; Zuo, F.; Zhu, X.; Nandakumar, K.S.; Xiao, C. Metformin, an AMPK Activator, Inhibits Activation of FLSs but Promotes HAPLN1 Secretion. Mol. Ther.–Methods Clin. Dev. 2020, 17, 1202–1214. [Google Scholar] [CrossRef]
- Kim, E.K.; Min, H.K.; Lee, S.Y.; Kim, D.S.; Ryu, J.G.; Na, H.S.; Jung, K.A.; Choi, J.W.; Park, S.H.; Cho, M.L. Metformin rescues rapamycin-induced mitochondrial dysfunction and attenuates rheumatoid arthritis with metabolic syndrome. Arthritis Res. Ther. 2020, 22, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gharib, M.; Elbaz, W.; Darweesh, E.; Sabri, N.A.; Shawki, M.A. Efficacy and Safety of Metformin Use in Rheumatoid Arthritis: A Randomized Controlled Study. Front. Pharm. 2021, 12, 726490. [Google Scholar] [CrossRef]
- Chen, K.; Lin, Z.-W.; He, S.-M.; Wang, C.-Q.; Yang, J.-C.; Lu, Y.; Xie, X.-B.; Li, Q. Metformin inhibits the proliferation of rheumatoid arthritis fibroblast-like synoviocytes through IGF-IR/PI3K/AKT/m-TOR pathway. Biomed. Pharmacother. 2019, 115, 108875. [Google Scholar] [CrossRef] [PubMed]
- Naffaa, M.E.; Rosenberg, V.; Watad, A.; Tiosano, S.; Yavne, Y.; Chodick, G.; Amital, H.; Shalev, V. Adherence to metformin and the onset of rheumatoid arthritis: A population-based cohort study. Scand. J. Rheumatol. 2020, 49, 173–180. [Google Scholar] [CrossRef]
- Lu, C.H.; Chung, C.H.; Lee, C.H.; Su, S.C.; Liu, J.S.; Lin, F.H.; Tsao, C.H.; Hsieh, P.S.; Hung, Y.J.; Hsieh, C.H.; et al. Combination of COX-2 inhibitor and metformin attenuates rate of admission in patients with rheumatoid arthritis and diabetes in Taiwan. Medicine 2019, 98, e17371. [Google Scholar] [CrossRef] [PubMed]
- Zemedikun, D.T.; Gokhale, K.; Chandan, J.S.; Cooper, J.; Lord, J.M.; Filer, A.; Falahee, M.; Nirantharakumar, K.; Raza, K. Type 2 diabetes mellitus, glycaemic control, associated therapies and risk of rheumatoid arthritis: A retrospective cohort study. Rheumatology 2021, 60, 5567–5575. [Google Scholar] [CrossRef]
- Raslan, M.A.; Alshahawey, M.; Shehata, E.M.; Sabri, N.A. Does human leukocyte antigen gene polymorphism affect management of COVID-19 Patients? A review article. Sci. J. Genet. Gene Ther. 2020, 6, 1–3. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, D.; Zhu, Z.; Chen, S.; Lu, M.; Cao, P.; Chen, T.; Li, S.; Xue, S.; Zhang, Y.; et al. Evaluating the impact of metformin targets on the risk of osteoarthritis: A mendelian randomization study. Osteoarthr. Cartil. 2022, 30, 1506–1514. [Google Scholar] [CrossRef]
- Zheng, J.; Xu, M.; Walker, V.; Yuan, J.; Korologou-Linden, R.; Robinson, J.; Huang, P.; Burgess, S.; Au Yeung, S.L.; Luo, S.; et al. Evaluating the efficacy and mechanism of metformin targets on reducing Alzheimer’s disease risk in the general population: A Mendelian randomisation study. Diabetologia 2022, 65, 1664–1675. [Google Scholar] [CrossRef] [PubMed]
- Sekula, P.; Del Greco, M.F.; Pattaro, C.; Kottgen, A. Mendelian Randomization as an Approach to Assess Causality Using Observational Data. J. Am. Soc. Nephrol. 2016, 27, 3253–3265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elsworth, B. IEU Open GWAS Project. Available online: https://gwas.mrcieu.ac.uk/datasets/ukb-b-14609/ (accessed on 20 December 2022).
- Okada, Y.; Wu, D.; Trynka, G.; Raj, T.; Terao, C.; Ikari, K.; Kochi, Y.; Ohmura, K.; Suzuki, A.; Yoshida, S.; et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 2014, 506, 376–381. [Google Scholar] [CrossRef] [Green Version]
- Arnett, F.C.; Edworthy, S.M.; Bloch, D.A.; McShane, D.J.; Fries, J.F.; Cooper, N.S.; Healey, L.A.; Kaplan, S.R.; Liang, M.H.; Luthra, H.S.; et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum 1988, 31, 315–324. [Google Scholar] [CrossRef]
- Myers, T.A.; Chanock, S.J.; Machiela, M.J. LDlinkR: An R Package for Rapidly Calculating Linkage Disequilibrium Statistics in Diverse Populations. Front. Genet. 2020, 11, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgess, S.; Small, D.S.; Thompson, S.G. A review of instrumental variable estimators for Mendelian randomization. Stat. Methods Med. Res. 2017, 26, 2333–2355. [Google Scholar] [CrossRef] [Green Version]
- Luo, S.; Au Yeung, S.L.; Zuber, V.; Burgess, S.; Schooling, C.M. Impact of Genetically Predicted Red Blood Cell Traits on Venous Thromboembolism: Multivariable Mendelian Randomization Study Using UK Biobank. J. Am. Heart Assoc. 2020, 9, e016771. [Google Scholar] [CrossRef]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Davey Smith, G.; Sheehan, N.; Thompson, J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat. Med. 2017, 36, 1783–1802. [Google Scholar] [CrossRef] [Green Version]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Zhao, Q.; Lawlor, D.A.; Sheehan, N.A.; Thompson, J.; Davey Smith, G. Improving the accuracy of two-sample summary-data Mendelian randomization: Moving beyond the NOME assumption. Int. J. Epidemiol. 2019, 48, 728–742. [Google Scholar] [CrossRef] [Green Version]
- Burgess, S.; Butterworth, A.; Thompson, S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013, 37, 658–665. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.; Demetriou, M.; Gillen, D.L. Genome-Wide Analysis of Gene-Gene and Gene-Environment Interactions Using Closed-Form Wald Tests. Genet. Epidemiol. 2015, 39, 446–455. [Google Scholar] [CrossRef] [Green Version]
- Qi, G.; Chatterjee, N. Mendelian randomization analysis using mixture models for robust and efficient estimation of causal effects. Nat. Commun. 2019, 10, 1941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowden, J.; Davey Smith, G.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J.F.; Chalumeau, M.; Cohen, R.; Korevaar, D.A.; Khoshnood, B.; Bossuyt, P.M. Cochran’s Q test was useful to assess heterogeneity in likelihood ratios in studies of diagnostic accuracy. J. Clin. Epidemiol. 2015, 68, 299–306. [Google Scholar] [CrossRef]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef] [Green Version]
- Verbanck, M.; Chen, C.Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Hemani, G.Z.J.; Elsworth, B.; Wade, K.H.; Baird, D.; Haberland, V.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; Tan, V.Y.; et al. Mendelian Randomization with GWAS Summary Data. Available online: https://mrcieu.github.io/TwoSampleMR/ (accessed on 20 December 2022).
- Marie Verbanck, C.-Y.C.; Benjamin Neale, R.D. Github. Available online: https://github.com/rondolab/MR-PRESSO (accessed on 20 December 2022).
- Abdallah, M.S.; Alarfaj, S.J.; Saif, D.S.; El-Naggar, M.E.; Elsokary, M.A.; Elsawah, H.K.; Abdelsattar Zaki, S.; Wahsh, E.A.; Abo Mansour, H.E.; Mosalam, E.M. The AMPK modulator metformin as adjunct to methotrexate in patients with rheumatoid arthritis: A proof-of-concept, randomized, double-blind, placebo-controlled trial. Int. Immunopharmacol. 2021, 95, 107575. [Google Scholar] [CrossRef]
- Salvatore, T.; Pafundi, P.C.; Galiero, R.; Gjeloshi, K.; Masini, F.; Acierno, C.; Di Martino, A.; Albanese, G.; Alfano, M.; Rinaldi, L.; et al. Metformin: A Potential Therapeutic Tool for Rheumatologists. Pharmaceuticals 2020, 13, 234. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Choe, J.Y.; Park, S.H. Metformin and its therapeutic applications in autoimmune inflammatory rheumatic disease. Korean J. Intern. Med. 2022, 37, 13–26. [Google Scholar] [CrossRef]
- Fan, K.J.; Wu, J.; Wang, Q.S.; Xu, B.X.; Zhao, F.T.; Wang, T.Y. Metformin inhibits inflammation and bone destruction in collagen-induced arthritis in rats. Ann. Transl. Med. 2020, 8, 1565. [Google Scholar] [CrossRef]
- Kim, E.K.; Lee, S.H.; Lee, S.Y.; Kim, J.K.; Jhun, J.Y.; Na, H.S.; Kim, S.Y.; Choi, J.Y.; Yang, C.W.; Park, S.H.; et al. Metformin ameliorates experimental-obesity-associated autoimmune arthritis by inducing FGF21 expression and brown adipocyte differentiation. Exp. Mol. Med. 2018, 50, e432. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.C.; Schneeweiss, S.; Glynn, R.J.; Doherty, M.; Goldfine, A.B.; Solomon, D.H. Dipeptidyl peptidase-4 inhibitors in type 2 diabetes may reduce the risk of autoimmune diseases: A population-based cohort study. Ann. Rheum. Dis. 2015, 74, 1968–1975. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Han, J.Y.; Shin, J.Y.; Kim, S.I.; Lee, J.M.; Hong, S.; Kim, S.H.; Nam, M.S.; Kim, Y.S. Metformin-associated lactic acidosis: Predisposing factors and outcome. Endocrinol Metab 2015, 30, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Knowler, W.C.; Barrett-Connor, E.; Fowler, S.E.; Hamman, R.F.; Lachin, J.M.; Walker, E.A.; Nathan, D.M.; Watson, P.G.; Mendoza, J.T.; Smith, K.A.; et al. Reduction in the Incidence of Type 2 Diabetes with Lifestyle Intervention or Metformin. N. Engl. J. Med. 2002, 346, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Radu, A.F.; Bungau, S.G. Management of Rheumatoid Arthritis: An Overview. Cells 2021, 10, 2857. [Google Scholar] [CrossRef]
- Park, M.H.; Hong, J.T. Roles of NF-kappaB in Cancer and Inflammatory Diseases and Their Therapeutic Approaches. Cells 2016, 5, 15. [Google Scholar] [CrossRef] [PubMed]
- Hattori, Y.; Suzuki, K.; Hattori, S.; Kasai, K. Metformin inhibits cytokine-induced nuclear factor kappaB activation via AMP-activated protein kinase activation in vascular endothelial cells. Hypertension 2006, 47, 1183–1188. [Google Scholar] [CrossRef] [Green Version]
- Saisho, Y. Metformin and Inflammation: Its Potential Beyond Glucose-lowering Effect. Endocr. Metab. Immune Disord. Drug Targets 2015, 15, 196–205. [Google Scholar] [CrossRef]
- Wu, H.; Esteve, E.; Tremaroli, V.; Khan, M.T.; Caesar, R.; Manneras-Holm, L.; Stahlman, M.; Olsson, L.M.; Serino, M.; Planas-Felix, M.; et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 2017, 23, 850–858. [Google Scholar] [CrossRef]
- de la Cuesta-Zuluaga, J.; Mueller, N.T.; Corrales-Agudelo, V.; Velasquez-Mejia, E.P.; Carmona, J.A.; Abad, J.M.; Escobar, J.S. Metformin Is Associated with Higher Relative Abundance of Mucin-Degrading Akkermansia muciniphila and Several Short-Chain Fatty Acid-Producing Microbiota in the Gut. Diabetes Care 2017, 40, 54–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Exposure/Outcome | Year | Author | Participants | Number of SNPs | Web Source if Publicly Available |
|---|---|---|---|---|---|
| Metformin | 2018 | Ben Elsworth [33] | 462,933 individuals (11,552 metformin use cases and 451,381 controls) of European ancestry | 9,851,867 | https://gwas.mrcieu.ac.uk/datasets/ukb-b-14609/ (accessed on 20 December 2022) |
| RA | 2014 | Okada Y [34] | 58,284 individuals (14,361 RA cases and 43,923 controls) of European ancestry | 8,747,963 | https://gwas.mrcieu.ac.uk/datasets/ieu-a-832/ (accessed on 20 December 2022) |
| SNP | chr | EA | OA | EAF | F | SNP-Metformin Association | SNP-RA Association | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | p-Value | Beta | SE | p-Value | ||||||
| rs10195252 | 2 | C | T | 0.4051 | 33.8179 | −0.0019 | 0.0003 | 6.100 × 10−9 | 0.0101 | 0.0136 | 0.460 |
| rs10420309 | 19 | G | A | 0.4375 | 33.4369 | −0.0019 | 0.0003 | 7.400 × 10−9 | 0.0202 | 0.0187 | 0.280 |
| rs10965246 | 9 | C | T | 0.1767 | 102.4984 | −0.0043 | 0.0004 | 4.300 × 10−24 | 0.0408 | 0.0186 | 0.028 |
| rs11257655 | 10 | T | C | 0.2082 | 47.1683 | 0.0027 | 0.0004 | 6.500 × 10−12 | −0.0202 | 0.0216 | 0.350 |
| rs11708067 | 3 | G | A | 0.2424 | 32.9477 | −0.0022 | 0.0004 | 9.500 × 10−9 | 0.0202 | 0.0199 | 0.310 |
| rs1215468 | 13 | G | A | 0.2914 | 66.9935 | −0.0029 | 0.0004 | 2.700 × 10−16 | −0.0100 | 0.0340 | 0.770 |
| rs13266634 | 8 | T | C | 0.3096 | 52.7446 | −0.0025 | 0.0004 | 3.800 × 10−13 | 0.0392 | 0.0183 | 0.032 |
| rs1421085 | 16 | C | T | 0.4035 | 114.2529 | 0.0035 | 0.0003 | 1.100 × 10−26 | −0.0100 | 0.0233 | 0.670 |
| rs1496653 | 3 | G | A | 0.2034 | 52.8812 | −0.0029 | 0.0004 | 3.500 × 10−13 | 0.0101 | 0.0418 | 0.810 |
| rs17036160 | 3 | T | C | 0.1175 | 38.7915 | −0.0031 | 0.0005 | 4.700 × 10−10 | −0.0619 | 0.0280 | 0.027 |
| rs17513135 | 1 | T | C | 0.2275 | 34.7648 | 0.0023 | 0.0004 | 3.700 × 10−9 | −0.0101 | 0.0442 | 0.820 |
| rs1800961 | 20 | T | C | 0.0310 | 33.0454 | 0.0054 | 0.0009 | 9.000 × 10−9 | 0.0583 | 0.0465 | 0.210 |
| rs2796441 | 9 | A | G | 0.4185 | 31.0899 | −0.0018 | 0.0003 | 2.500 × 10−8 | −0.0101 | 0.0203 | 0.620 |
| rs34744311 | 10 | T | C | 0.3773 | 72.5694 | −0.0028 | 0.0003 | 1.600 × 10−17 | −0.0202 | 0.0161 | 0.210 |
| rs34872471 | 10 | C | T | 0.2918 | 577.9145 | 0.0086 | 0.0004 | 1.099 × 10−127 | −0.0488 | 0.0188 | 0.009 |
| rs459193 | 5 | G | A | 0.7466 | 40.5767 | 0.0024 | 0.0004 | 1.900 × 10−10 | −0.0392 | 0.0228 | 0.086 |
| rs4686471 | 3 | C | T | 0.6101 | 32.1655 | 0.0019 | 0.0003 | 1.400 × 10−8 | 0.0101 | 0.0228 | 0.660 |
| rs4752792 | 11 | A | G | 0.5445 | 41.5855 | 0.0021 | 0.0003 | 1.100 × 10−10 | 0.0296 | 0.0236 | 0.210 |
| rs4932264 | 15 | C | T | 0.7296 | 36.9493 | −0.0022 | 0.0004 | 1.200 × 10−9 | −0.0198 | 0.0240 | 0.410 |
| rs67232546 | 11 | T | C | 0.2125 | 32.6216 | 0.0023 | 0.0004 | 1.100 × 10−8 | −0.0202 | 0.0212 | 0.340 |
| rs6769511 | 3 | C | T | 0.3158 | 83.6027 | 0.0032 | 0.0003 | 6.001 × 10−20 | −0.0198 | 0.0216 | 0.360 |
| rs7177055 | 15 | A | G | 0.7175 | 39.2887 | 0.0022 | 0.0004 | 3.700 × 10−10 | −0.0101 | 0.0252 | 0.690 |
| rs72802357 | 16 | T | C | 0.0781 | 44.5496 | −0.0040 | 0.0006 | 2.500 × 10−11 | 0.0677 | 0.0283 | 0.017 |
| rs73188924 | 22 | A | C | 0.2248 | 31.1092 | 0.0022 | 0.0004 | 2.400 × 10−8 | −0.0202 | 0.0322 | 0.530 |
| rs7482891 | 11 | G | A | 0.6221 | 42.3038 | −0.0022 | 0.0003 | 7.800 × 10−11 | 0.0101 | 0.0252 | 0.690 |
| rs7756992 | 6 | G | A | 0.2664 | 76.4924 | 0.0032 | 0.0004 | 2.200 × 10−18 | 0.0101 | 0.0122 | 0.410 |
| rs780093 | 2 | C | T | 0.6152 | 38.6220 | 0.0021 | 0.0003 | 5.100 × 10−10 | 0.0101 | 0.0133 | 0.450 |
| rs849142 | 7 | C | T | 0.5051 | 54.9143 | −0.0024 | 0.0003 | 1.300 × 10−13 | 0.0202 | 0.0179 | 0.260 |
| rs8756 | 12 | A | C | 0.5176 | 33.9234 | 0.0019 | 0.0003 | 5.700 × 10−9 | −0.0305 | 0.0164 | 0.063 |
| rs947791 | 11 | A | G | 0.2176 | 34.0158 | 0.0023 | 0.0004 | 5.500 × 10−9 | −0.0202 | 0.0230 | 0.380 |
| rs987237 | 6 | G | A | 0.1796 | 33.1730 | 0.0024 | 0.0004 | 8.400 × 10−9 | 0.0305 | 0.0300 | 0.310 |
| rs9957264 | 18 | A | C | 0.1665 | 36.4231 | −0.0026 | 0.0004 | 1.600 × 10−9 | 0.0198 | 0.0191 | 0.300 |
| Method | p-Value | OR | LCI | UCI |
|---|---|---|---|---|
| MR Egger | 0.086 | 0.0034 | 6.4266 × 10−6 | 1.7990 |
| WM | 0.003 | 0.0035 | 8.8251 × 10−5 | 0.1423 |
| IVW | 0.006 | 0.0232 | 1.6046 × 10−3 | 0.3368 |
| Simple mode | 0.094 | 0.0009 | 2.9682 × 10−7 | 2.6077 |
| Weighted mode | 0.004 | 0.0017 | 3.0310 × 10−5 | 0.0975 |
| MR-Egger | MR-PRESSO | Cochran’s Q Test | |||
|---|---|---|---|---|---|
| Estimates | SE | p-Value | Global Test p-Value | Q | p-Value |
| 0.0065 | 0.0098 | 0.511 | 0.120 | 40.812 | 0.112 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, J.; Cai, Y.; Zhang, J.; Jing, Z.; Lv, L.; Zhang, G.; Zhang, R.; Liu, R.; Nan, K.; Dang, X. Metformin Treatment Reduces the Incidence of Rheumatoid Arthritis: A Two-Sample Mendelian Randomized Study. J. Clin. Med. 2023, 12, 2461. https://doi.org/10.3390/jcm12072461
Liang J, Cai Y, Zhang J, Jing Z, Lv L, Zhang G, Zhang R, Liu R, Nan K, Dang X. Metformin Treatment Reduces the Incidence of Rheumatoid Arthritis: A Two-Sample Mendelian Randomized Study. Journal of Clinical Medicine. 2023; 12(7):2461. https://doi.org/10.3390/jcm12072461
Chicago/Turabian StyleLiang, Jialin, Yuanqing Cai, Jianan Zhang, Zhaopu Jing, Leifeng Lv, Guangyang Zhang, Rupeng Zhang, Ruiyu Liu, Kai Nan, and Xiaoqian Dang. 2023. "Metformin Treatment Reduces the Incidence of Rheumatoid Arthritis: A Two-Sample Mendelian Randomized Study" Journal of Clinical Medicine 12, no. 7: 2461. https://doi.org/10.3390/jcm12072461






