MYD88-Mutated Chronic Lymphocytic Leukaemia/Small Lymphocytic Lymphoma as a Distinctive Molecular Subgroup Is Associated with Atypical Immunophenotypes in Chinese Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Sample Collection
2.2. Flow Cytometry (FC)
2.3. Next-Generation Sequencing (NGS) and Variant Curation
2.4. Immunoglobulin Heavy Chain (IGH) Variable (V) Region Mutational Status Analysis
2.5. Chromosomal Karyotype and Fluorescence In Situ Hybridization (FISH) Analysis
2.6. Statistical Analysis
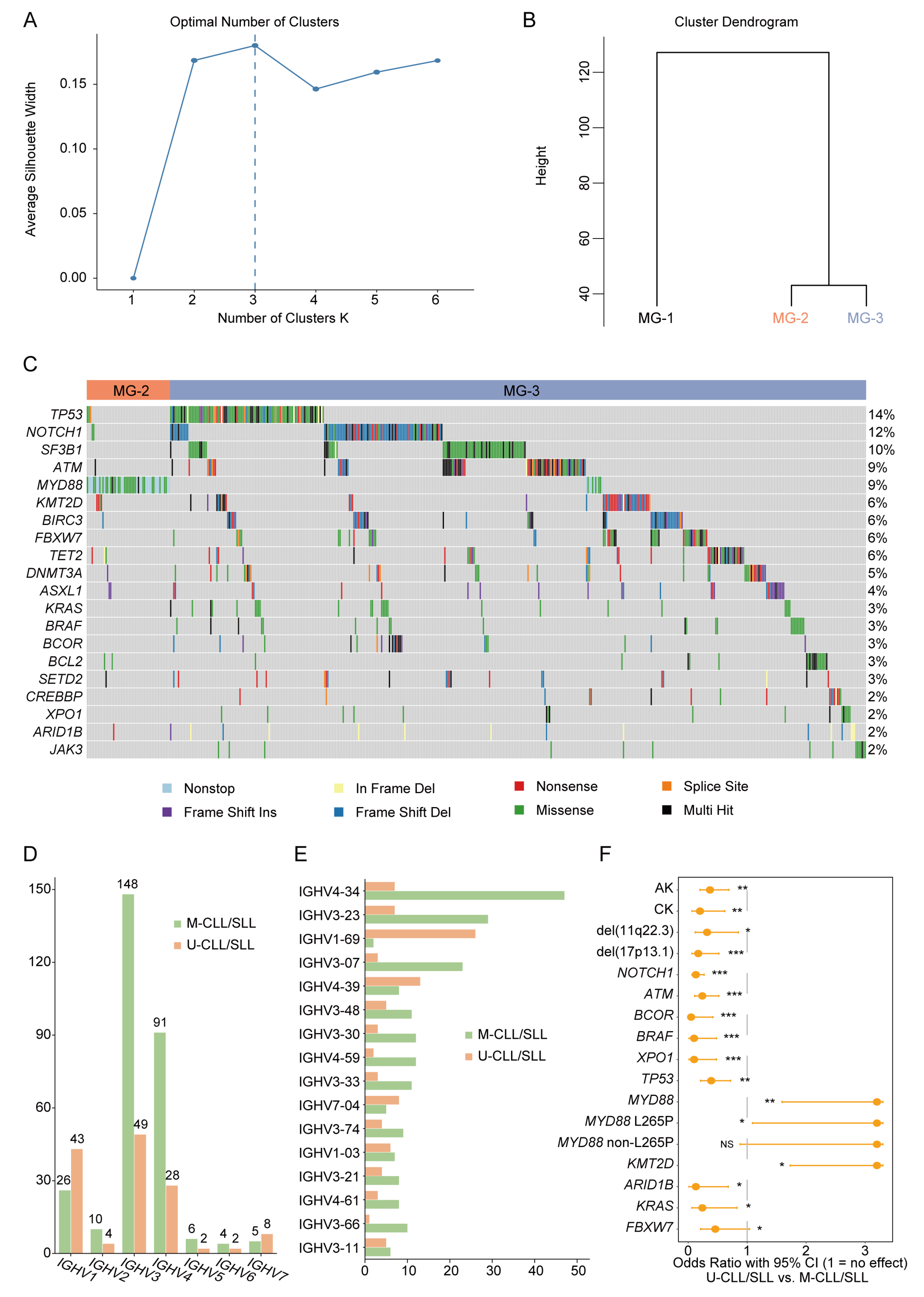
3. Results
3.1. A High Proportion of Chinese CLL/SLL Patients Had Atypical Immunophenotypes
3.2. Mutation Landscape of Chinese CLL/SLL Patients
3.3. MYD88 Variants Related to Hypermutated IGHV Status
3.4. MYD88 Variants Related to Abnormal Karyotypes
3.5. MYD88 Variants Related to Atypical CLL/SLL Immunophenotypes
3.6. MYD88 Variants May Be a Potential Differential Diagnostic Marker between Atypical CLL/SLL and Mantle Cell Lymphoma (MCL)
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bosch, F.; Dalla-Favera, R. Chronic lymphocytic leukaemia: From genetics to treatment. Nat. Rev. Clin. Oncol. 2019, 16, 684–701. [Google Scholar] [CrossRef]
- Eichhorst, B.; Robak, T.; Montserrat, E.; Ghia, P.; Niemann, C.U.; Kater, A.P.; Gregor, M.; Cymbalista, F.; Buske, C.; Hillmen, P.; et al. Chronic lymphocytic leukaemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2021, 32, 23–33. [Google Scholar] [CrossRef]
- Chen, J.; McMillan, N.A. Molecular basis of pathogenesis, prognosis and therapy in chronic lymphocytic leukaemia. Cancer Biol. Ther. 2008, 7, 174–179. [Google Scholar] [CrossRef]
- Hallek, M.; Shanafelt, T.D.; Eichhorst, B. Chronic lymphocytic leukaemia. Lancet 2018, 391, 1524–1537. [Google Scholar] [CrossRef]
- Huang, Y.J.; Kuo, M.C.; Chang, H.; Wang, P.N.; Wu, J.H.; Huang, Y.M.; Ma, M.C.; Tang, T.C.; Kuo, C.Y.; Shih, L.Y. Distinct immunoglobulin heavy chain variable region gene repertoire and lower frequency of del(11q) in Taiwanese patients with chronic lymphocytic leukaemia. Br. J. Haematol. 2019, 187, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Nakahashi, H.; Tsukamoto, N.; Hashimoto, Y.; Koiso, H.; Yokohama, A.; Saitoh, T.; Uchiumi, H.; Handa, H.; Murakami, H.; Nojima, Y.; et al. Characterization of immunoglobulin heavy and light chain gene expression in chronic lymphocytic leukemia and related disorders. Cancer Sci. 2009, 100, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Gale, R.P.; Shi, H.; Liu, Y.; Lai, Y.; Lu, J.; Huang, X. Is there an epidemic of chronic lymphocytic leukaemia (CLL) in China. Leuk. Res. 2018, 73, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Varghese, A.M.; Sood, N.; Chiattone, C.; Akinola, N.O.; Huang, X.; Gale, R.P. Ethnic and geographic diversity of chronic lymphocytic leukaemia. Leukemia 2021, 35, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.M.; Li, J.Y.; Gale, R.P.; Huang, X.J. The mystery of chronic lymphocytic leukemia (CLL): Why is it absent in Asians and what does this tell us about etiology, pathogenesis and biology. Blood Rev. 2015, 29, 205–213. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.; Zheng, W.; Wu, Y.; Qiao, C.; Fan, L.; Xu, W.; Li, J. Distinctive IgVH gene segments usage and mutation status in Chinese patients with chronic lymphocytic leukemia. Leuk. Res. 2008, 32, 1491–1498. [Google Scholar] [CrossRef]
- Tian, Z.; Liu, M.; Fang, X.; Zhou, X.; Li, P.; Li, Y.; Zhang, L.; Liu, F.; Zhang, Y.; Wang, X. Distinct Age-Related Clinical Features and Risk Assessment in Chinese with Chronic Lymphocytic Leukemia. Front. Oncol. 2022, 12, 885150. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.; Yan, Y.; Jin, M.; Xiong, W.; Yu, Z.; Yu, Y.; Cui, R.; Wang, J.; Wang, Y.; Lin, Y.; et al. High incidence of MYD88 and KMT2D mutations in Chinese with chronic lymphocytic leukemia. Leukemia 2021, 35, 2412–2415. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, W.; Deng, Q.; Li, L.; Hsi, E.D.; Young, K.H.; Zhang, M.; Li, Y. MYD88 L265P Mutation in Lymphoid Malignancies. Cancer Res. 2018, 78, 2457–2462. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, W.; Deng, Q.; Liu, H.; Wang, X.; Hu, H.; Cao, Y.; Xu-Monette, Z.Y.; Li, L.; Zhang, M.; et al. MYD88 L265P elicits mutation-specific ubiquitination to drive NF-κB activation and lymphomagenesis. Blood 2021, 137, 1615–1627. [Google Scholar] [CrossRef]
- Bogusz, A.M.; Bagg, A. Genetic aberrations in small B-cell lymphomas and leukemias: Molecular pathology, clinical relevance and therapeutic targets. Leuk. Lymphoma 2016, 57, 1991–2013. [Google Scholar] [CrossRef]
- Ryder, S.P. #CRISPRbabies: Notes on a Scandal. CRISPR J. 2018, 1, 355–357. [Google Scholar] [CrossRef]
- Qin, S.C.; Xia, Y.; Miao, Y.; Zhu, H.Y.; Wu, J.Z.; Fan, L.; Li, J.Y.; Xu, W.; Qiao, C. MYD88 mutations predict unfavorable prognosis in Chronic Lymphocytic Leukemia patients with mutated IGHV gene. Blood Cancer J. 2017, 7, 651. [Google Scholar] [CrossRef]
- Shi, K.; Sun, Q.; Qiao, C.; Zhu, H.; Wang, L.; Wu, J.; Wang, L.; Fu, J.; Young, K.H.; Fan, L.; et al. 98% IGHV gene identity is the optimal cutoff to dichotomize the prognosis of Chinese patients with chronic lymphocytic leukemia. Cancer Med. 2020, 9, 999–1007. [Google Scholar] [CrossRef]
- Cro, L.; Ferrario, A.; Lionetti, M.; Bertoni, F.; Zucal, N.N.; Nobili, L.; Fabris, S.; Todoerti, K.; Cortelezzi, A.; Guffanti, A.; et al. The clinical and biological features of a series of immunophenotypic variant of B-CLL. Eur. J. Haematol. 2010, 85, 120–129. [Google Scholar] [CrossRef]
- Li, Y.; Tong, X.; Huang, L.; Li, L.; Wang, C.; He, C.; Liu, S.; Wang, Z.; Xiao, M.; Mao, X.; et al. A new score including CD43 and CD180: Increased diagnostic value for atypical chronic lymphocytic leukemia. Cancer Med. 2021, 10, 4387–4396. [Google Scholar] [CrossRef]
- Matutes, E.; Owusu-Ankomah, K.; Morilla, R.; Garcia Marco, J.; Houlihan, A.; Que, T.H.; Catovsky, D. The immunological profile of B-cell disorders and proposal of a scoring system for the diagnosis of CLL. Leukemia 1994, 8, 1640–1645. [Google Scholar] [PubMed]
- Moreau, E.J.; Matutes, E.; A’Hern, R.P.; Morilla, A.M.; Morilla, R.M.; Owusu-Ankomah, K.A.; Seon, B.K.; Catovsky, D. Improvement of the chronic lymphocytic leukemia scoring system with the monoclonal antibody SN8 (CD79b). Am. J. Clin. Pathol. 1997, 108, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Baliakas, P.; Hadzidimitriou, A.; Agathangelidis, A.; Rossi, D.; Sutton, L.A.; Kminkova, J.; Scarfo, L.; Pospisilova, S.; Gaidano, G.; Stamatopoulos, K.; et al. Prognostic relevance of MYD88 mutations in CLL: The jury is still out. Blood 2015, 126, 1043–1044. [Google Scholar] [CrossRef] [PubMed]
- Baliakas, P.; Hadzidimitriou, A.; Sutton, L.A.; Rossi, D.; Minga, E.; Villamor, N.; Larrayoz, M.; Kminkova, J.; Agathangelidis, A.; Davis, Z.; et al. Recurrent mutations refine prognosis in chronic lymphocytic leukemia. Leukemia 2015, 29, 329–336. [Google Scholar] [CrossRef]
- Shuai, W.; Lin, P.; Strati, P.; Patel, K.P.; Routbort, M.J.; Hu, S.; Wei, P.; Khoury, J.D.; You, M.J.; Loghavi, S.; et al. Clinicopathological characterization of chronic lymphocytic leukemia with MYD88 mutations: L265P and non-L265P mutations are associated with different features. Blood Cancer J. 2020, 10, 86. [Google Scholar] [CrossRef]
- Jang, M.A.; Yoo, E.H.; Kim, K.; Kim, W.S.; Jung, C.W.; Kim, S.H.; Kim, H.J. Chronic lymphocytic leukemia in Korean patients: Frequent atypical immunophenotype and relatively aggressive clinical behavior. Int. J. Hematol. 2013, 97, 403–408. [Google Scholar] [CrossRef]
- Puente, X.S.; Pinyol, M.; Quesada, V.; Conde, L.; Ordóñez, G.R.; Villamor, N.; Escaramis, G.; Jares, P.; Beà, S.; González-Díaz, M.; et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011, 475, 101–105. [Google Scholar] [CrossRef]
- Mauerer, K.; Zahrieh, D.; Gorgun, G.; Li, A.; Zhou, J.; Ansén, S.; Rassenti, L.Z.; Gribben, J.G. Immunoglobulin gene segment usage, location and immunogenicity in mutated and unmutated chronic lymphocytic leukaemia. Br. J. Haematol. 2005, 129, 499–510. [Google Scholar] [CrossRef]
- Orlandi, E.M.; Zibellini, S.; Pascutto, C.; Picone, C.; Giardini, I.; Pochintesta, L.; Lazzarino, M. IGHV unmutated status influences outcome more than IGHV1-69 gene usage per se in patients with chronic lymphocytic leukemia. Clin. Lymphoma Myeloma 2009, 9, 390–393. [Google Scholar] [CrossRef]
- Urbanova, R.; Humplikova, L.; Drimalova, H.; Prochazka, V.; Turcsanyi, P.; Pikalova, Z.; Holzerova, M.; Kruzova, L.; Jarosova, M.; Urban, J.; et al. Biological and clinical characteristics of patients with chronic lymphocytic leukemia with the IGHV3-21 and IGHV1-69; analysis of data from a single center. Neoplasma 2015, 62, 618–626. [Google Scholar] [CrossRef]
- Tucci, F.A.; Kitanovski, S.; Johansson, P.; Klein-Hitpass, L.; Kahraman, A.; Dürig, J.; Hoffmann, D.; Küppers, R. Biased IGH VDJ gene repertoire and clonal expansions in B cells of chronically hepatitis C virus-infected individuals. Blood 2018, 131, 546–557. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions, and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |

| Patient Characteristics | CLL/SLL |
|---|---|
| Total patients, n | 761 |
| Age, median (range) | 64.8 (20–92) |
| Sex, n | |
| Female | 261 (of 761, 34.3%) |
| Male | 500 (of 761, 65.7%) |
| RMH, n | |
| RMH ≥ 4 points (classic group) | 519 (of 618, 84.0%) |
| RMH < 4 points (atypical group) | 99 (of 618, 16.0%) |
| CD, n | |
| CD5 (pos) | 724 (of 726, 99.7%) |
| CD10 (neg) | 724 (of 725, 99.9%) |
| CD23 (pos) | 709 (of 720, 98.5%) |
| FMC7 (neg) | 658 (of 720, 91.4%) |
| CD22 (neg/dim) | 145 (of 360, 40.3%) |
| CD79b (neg/dim) | 427 (of 600, 71.2%) |
| sIg (neg/dim) | 241 (of 724, 33.3%) |
| CD200 (pos) | 558 (of 587, 95.1%) |
| 2008 (n = 65) [10] | 2009 (n = 46) [16] | 2017 (n = 145) [17] | 2019 (n = 194) [5] | 2020 (n = 595) [18] | Present (n = 410) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | n | % | n | % | |
| IGHV4-39 | 2 | 3.1% | 3 | 6.5% | 5 | 3.4% | 11 | 5.7% | 39 | 6.6% | 21 | 5.1% |
| IGHV3-74 | 3 | 4.6% | 0 | 0% | <3 | <2.0% | 14 | 7.2% | <19 | <3.0% | 13 | 3.2% |
| IGHV1-69 | 1 | 1.5% | 1 | 2.2% | <3 | <2.0% | 11 | 5.7% | 37 | 6.2% | 28 | 6.8% |
| IGHV3-21 | 2 | 3.1% | 2 | 4.3% | 4 | 2.8% | <11 | <5.0% | 19 | 3.2% | 12 | 2.9% |
| IGHV3-30 | 4 | 6.2% | 1 | 2.2% | <3 | <2.0% | 12 | 6.2% | 34 | 5.7% | 15 | 3.7% |
| IGHV3-07 | 3 | 4.6% | 1 | 2.2% | 20 | 13.8% | 18 | 9.3% | 45 | 7.6% | 26 | 6.3% |
| IGHV4-34 | 8 | 12.3% | 10 | 21.7% | 28 | 19.3% | 11 | 5.7% | 59 | 9.9% | 54 | 13.2% |
| IGHV4-59 | 7 | 10.8% | 1 | 2.2% | 5 | 3.4% | 13 | 6.7% | 26 | 4.4% | 14 | 3.4% |
| IGHV3-23 | 5 | 7.7% | 4 | 8.7% | 20 | 13.8% | 20 | 10.3% | 61 | 10.3% | 36 | 8.8% |
| IGHV4-61 | 5 | 7.7% | 1 | 2.2% | <3 | <2.0% | <11 | <5.0% | <19 | <3.0% | 11 | 2.7% |
| IGHV3-11 | 5 | 7.7% | 1 | 2.2% | <3 | <2.0% | <11 | <5.0% | <19 | <3.0% | 11 | 2.7% |
| IGHV3-48 | 1 | 1.5% | 4 | 8.7% | <3 | <2.0% | <11 | <5.0% | 23 | 3.9% | 16 | 3.9% |
| MG-1 | MG-2 | MG-3 | p Value | |
|---|---|---|---|---|
| Age, median | 63.7% | 64.1% | 65.3% | 0.20 |
| Sex, male/female ratio | 1.7 | 3.5 | 1.9 | 0.11 |
| AK (pos, %) | 6.9% | 8.1% | 23.1% | <0.001 |
| CK (pos, %) | 0.8% | 5.4% | 5.8% | 0.06 |
| Trisomy 12 (pos, %) | 11.9% | 11.1% | 26.3% | 0.053 |
| Del(11q22.3) (pos, %) | 5.5% | 0.0% | 14.6% | 0.07 |
| Del(17p13.1) (pos, %) | 0.0% | 0.0% | 12.6% | <0.001 |
| Del(13q14.2) (pos, %) | 30.6% | 25.0% | 17.8% | 0.26 |
| CD5 (pos, %) | 100.0% | 98.1% | 99.8% | 0.06 |
| CD10 (neg, %) | 100.0% | 100.0% | 99.8% | 0.78 |
| CD23 (pos, %) | 99.5% | 94.3% | 98.5% | 0.03 |
| FMC7 (neg, %) | 93.0% | 66.0% | 93.5% | <0.001 |
| CD22 (neg/dim, %) | 42.5% | 35.5% | 40.1% | 0.79 |
| CD79b (neg/dim, %) | 77.4% | 53.3% | 70.7% | 0.007 |
| sIg (neg/dim, %) | 36.9% | 29.6% | 32.3% | 0.44 |
| CD200 (pos, %) | 97.5% | 73.2% | 96.4% | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mu, Y.; Fan, X.; Chen, T.; Meng, Y.; Lin, J.; Yuan, J.; Yu, S.; Chen, Y.; Liu, L. MYD88-Mutated Chronic Lymphocytic Leukaemia/Small Lymphocytic Lymphoma as a Distinctive Molecular Subgroup Is Associated with Atypical Immunophenotypes in Chinese Patients. J. Clin. Med. 2023, 12, 2667. https://doi.org/10.3390/jcm12072667
Mu Y, Fan X, Chen T, Meng Y, Lin J, Yuan J, Yu S, Chen Y, Liu L. MYD88-Mutated Chronic Lymphocytic Leukaemia/Small Lymphocytic Lymphoma as a Distinctive Molecular Subgroup Is Associated with Atypical Immunophenotypes in Chinese Patients. Journal of Clinical Medicine. 2023; 12(7):2667. https://doi.org/10.3390/jcm12072667
Chicago/Turabian StyleMu, Yafei, Xijie Fan, Tao Chen, Yuhuan Meng, Junwei Lin, Jiecheng Yuan, Shihui Yu, Yuxin Chen, and Lingling Liu. 2023. "MYD88-Mutated Chronic Lymphocytic Leukaemia/Small Lymphocytic Lymphoma as a Distinctive Molecular Subgroup Is Associated with Atypical Immunophenotypes in Chinese Patients" Journal of Clinical Medicine 12, no. 7: 2667. https://doi.org/10.3390/jcm12072667
APA StyleMu, Y., Fan, X., Chen, T., Meng, Y., Lin, J., Yuan, J., Yu, S., Chen, Y., & Liu, L. (2023). MYD88-Mutated Chronic Lymphocytic Leukaemia/Small Lymphocytic Lymphoma as a Distinctive Molecular Subgroup Is Associated with Atypical Immunophenotypes in Chinese Patients. Journal of Clinical Medicine, 12(7), 2667. https://doi.org/10.3390/jcm12072667








