Machine Learning Identifies New Predictors on Restenosis Risk after Coronary Artery Stenting in 10,004 Patients with Surveillance Angiography
Abstract
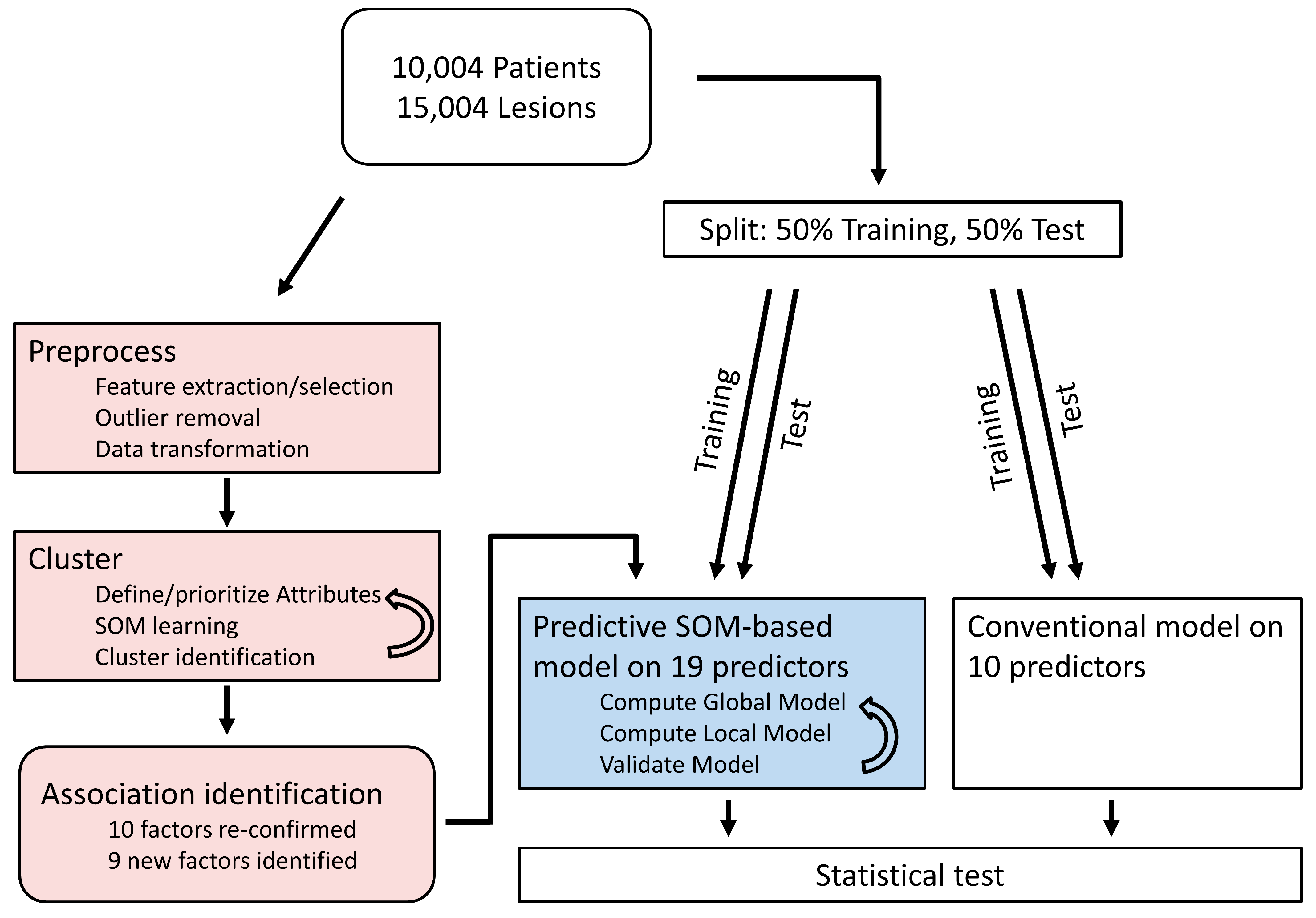
:1. Introduction
2. Methods
2.1. Machine Learning Analysis
2.2. Conventional Data Analysis
2.3. Comparison of Models
3. Results
3.1. General Characteristics
3.2. Identification of Clusters Based on SOM Analysis
3.3. Description and Comparison of the ‘Restenosis’ Cluster
3.4. Identification of Potential Predictors of Restenosis
4. Discussion
4.1. Differences between Both Approaches
4.2. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine Learning in Medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef] [PubMed]
- Fortmeier, V.; Lachmann, M.; Korber, M.I.; Unterhuber, M.; von Scheidt, M.; Rippen, E.; Harmsen, G.; Gercek, M.; Friedrichs, K.P.; Roder, F.; et al. Solving the Pulmonary Hypertension Paradox in Patients With Severe Tricuspid Regurgitation by Employing Artificial Intelligence. JACC Cardiovasc. Interv. 2022, 15, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Fortmeier, V.; Lachmann, M.; Unterhuber, M.; Stolz, L.; Kassar, M.; Ochs, L.; Gercek, M.; Schober, A.R.; Stocker, T.J.; Omran, H.; et al. Epiphenomenon or Prognostically Relevant Interventional Target? A Novel Proportionality Framework for Severe Tricuspid Regurgitation. J. Am. Heart Assoc. 2023, 12, e028737. [Google Scholar] [CrossRef] [PubMed]
- Lachmann, M.; Rippen, E.; Rueckert, D.; Schuster, T.; Xhepa, E.; von Scheidt, M.; Pellegrini, C.; Trenkwalder, T.; Rheude, T.; Stundl, A.; et al. Harnessing feature extraction capacities from a pre-trained convolutional neural network (VGG-16) for the unsupervised distinction of aortic outflow velocity profiles in patients with severe aortic stenosis. Eur. Heart J. Digit. Health 2022, 3, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Lachmann, M.; Rippen, E.; Schuster, T.; Xhepa, E.; von Scheidt, M.; Trenkwalder, T.; Pellegrini, C.; Rheude, T.; Hesse, A.; Stundl, A.; et al. Artificial intelligence-enabled phenotyping of patients with severe aortic stenosis: On the recovery of extra-aortic valve cardiac damage after transcatheter aortic valve replacement. Open Heart 2022, 9, e002068. [Google Scholar] [CrossRef] [PubMed]
- Trenkwalder, T.; Lachmann, M.; Stolz, L.; Fortmeier, V.; Covarrubias, H.A.A.; Rippen, E.; Schurmann, F.; Presch, A.; von Scheidt, M.; Ruff, C.; et al. Machine learning identifies pathophysiologically and prognostically informative phenotypes among patients with mitral regurgitation undergoing transcatheter edge-to-edge repair. Eur. Heart J. Cardiovasc. Imaging 2023, jead013. [Google Scholar] [CrossRef]
- Feldner-Busztin, D.; Firbas Nisantzis, P.; Edmunds, S.J.; Boza, G.; Racimo, F.; Gopalakrishnan, S.; Limborg, M.T.; Lahti, L.; de Polavieja, G.G. Dealing with dimensionality: The application of machine learning to multi-omics data. Bioinformatics 2023, 39, btad021. [Google Scholar] [CrossRef]
- Jin, H.; Goossens, P.; Juhasz, P.; Eijgelaar, W.; Manca, M.; Karel, J.M.H.; Smirnov, E.; Sikkink, C.; Mees, B.M.E.; Waring, O.; et al. Integrative multiomics analysis of human atherosclerosis reveals a serum response factor-driven network associated with intraplaque hemorrhage. Clin. Transl. Med. 2021, 11, e458. [Google Scholar] [CrossRef]
- Reel, P.S.; Reel, S.; Pearson, E.; Trucco, E.; Jefferson, E. Using machine learning approaches for multi-omics data analysis: A review. Biotechnol. Adv. 2021, 49, 107739. [Google Scholar] [CrossRef]
- Lachmann, M.; Rippen, E.; Schuster, T.; Xhepa, E.; von Scheidt, M.; Pellegrini, C.; Trenkwalder, T.; Rheude, T.; Stundl, A.; Thalmann, R.; et al. Subphenotyping of Patients With Aortic Stenosis by Unsupervised Agglomerative Clustering of Echocardiographic and Hemodynamic Data. JACC Cardiovasc. Interv. 2021, 14, 2127–2140. [Google Scholar] [CrossRef] [PubMed]
- Sarker, I.H. Machine Learning: Algorithms, Real-World Applications and Research Directions. SN Comput. Sci. 2021, 2, 160. [Google Scholar] [CrossRef] [PubMed]
- Obermeyer, Z.; Emanuel, E.J. Predicting the Future—Big Data, Machine Learning, and Clinical Medicine. N. Engl. J. Med. 2016, 375, 1216–1219. [Google Scholar] [CrossRef] [PubMed]
- Jansen, C.; Ramirez, R.N.; El-Ali, N.C.; Gomez-Cabrero, D.; Tegner, J.; Merkenschlager, M.; Conesa, A.; Mortazavi, A. Building gene regulatory networks from scATAC-seq and scRNA-seq using Linked Self Organizing Maps. PLoS Comput. Biol. 2019, 15, e1006555. [Google Scholar] [CrossRef] [PubMed]
- Janssen, D.J.A.; Rechberger, S.; Wouters, E.F.M.; Schols, J.; Johnson, M.J.; Currow, D.C.; Curtis, J.R.; Spruit, M.A. Clustering of 27,525,663 Death Records from the United States Based on Health Conditions Associated with Death: An Example of big Health Data Exploration. J. Clin. Med. 2019, 8, 922. [Google Scholar] [CrossRef]
- Mortazavi, A.; Pepke, S.; Jansen, C.; Marinov, G.K.; Ernst, J.; Kellis, M.; Hardison, R.C.; Myers, R.M.; Wold, B.J. Integrating and mining the chromatin landscape of cell-type specificity using self-organizing maps. Genome Res. 2013, 23, 2136–2148. [Google Scholar] [CrossRef]
- Kurth, C.; Wegerer, V.; Reulbach, U.; Lewczuk, P.; Kornhuber, J.; Steinhoff, B.J.; Bleich, S. Analysis of hippocampal atrophy in alcoholic patients by a Kohonen feature map. Neuroreport 2004, 15, 367–371. [Google Scholar] [CrossRef]
- Iglesias-Rozas, J.R.; Hopf, N. Histological heterogeneity of human glioblastomasinvestigated with an unsupervised neural network (SOM). Histol. Histopathol. 2005, 20, 351–356. [Google Scholar]
- Sampedro-Gomez, J.; Dorado-Diaz, P.I.; Vicente-Palacios, V.; Sanchez-Puente, A.; Jimenez-Navarro, M.; San Roman, J.A.; Galindo-Villardon, P.; Sanchez, P.L.; Fernandez-Aviles, F. Machine Learning to Predict Stent Restenosis Based on Daily Demographic, Clinical, and Angiographic Characteristics. Can J. Cardiol. 2020, 36, 1624–1632. [Google Scholar] [CrossRef]
- Erdmann, J.; Kessler, T.; Munoz Venegas, L.; Schunkert, H. A decade of genome-wide association studies for coronary artery disease: The challenges ahead. Cardiovasc. Res. 2018, 114, 1241–1257. [Google Scholar] [CrossRef]
- Aragam, K.G.; Jiang, T.; Goel, A.; Kanoni, S.; Wolford, B.N.; Atri, D.S.; Weeks, E.M.; Wang, M.; Hindy, G.; Zhou, W. Discovery and systematic characterization of risk variants and genes for coronary artery disease in over a million participants. Nat. Genet. 2022, 54, 1803–1815. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S.; Eigenmann, J.; Zhao, Y.; Fleig, J.; Hawe, J.S.; Pan, C.; Bongiovanni, D.; Wengert, S.; Ma, A.; Lusis, A.J. Identification of the Transcription Factor ATF3 as a Direct and Indirect Regulator of the LDLR. Metabolites 2022, 12, 840. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chen, Z.; von Scheidt, M.; Li, S.; Steiner, A.; Güldener, U.; Koplev, S.; Ma, A.; Hao, K.; Pan, C. Transcriptome-wide association study of coronary artery disease identifies novel susceptibility genes. Basic Res. Cardiol. 2022, 117, 6. [Google Scholar] [CrossRef] [PubMed]
- Neiburga, K.D.; Vilne, B.; Bauer, S.; Bongiovanni, D.; Ziegler, T.; Lachmann, M.; Wengert, S.; Hawe, J.S.; Güldener, U.; Westerlund, A.M. Vascular tissue specific miRNA profiles reveal novel correlations with risk factors in coronary artery disease. Biomolecules 2021, 11, 1683. [Google Scholar] [CrossRef]
- Schunkert, H.; von Scheidt, M.; Kessler, T.; Stiller, B.; Zeng, L.; Vilne, B. Genetics of coronary artery disease in the light of genome-wide association studies. Clin. Res. Cardiol. 2018, 107, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Schupke, S.; Neumann, F.J.; Menichelli, M.; Mayer, K.; Bernlochner, I.; Wohrle, J.; Richardt, G.; Liebetrau, C.; Witzenbichler, B.; Antoniucci, D.; et al. Ticagrelor or Prasugrel in Patients with Acute Coronary Syndromes. N. Engl. J. Med. 2019, 381, 1524–1534. [Google Scholar] [CrossRef]
- Sharifi, A.; Wierer, M.; Dang, T.A.; Milic, J.; Moggio, A.; Sachs, N.; von Scheidt, M.; Hinterdobler, J.; Müller, P.; Werner, J. The novel coronary artery disease risk factor ADAMTS-7 modulates atherosclerotic plaque formation by degradation of TIMP-1. bioRxiv 2023. [Google Scholar] [CrossRef]
- Von Scheidt, M.; Zhao, Y.; de Aguiar Vallim, T.Q.; Che, N.; Wierer, M.; Seldin, M.M.; Franzén, O.; Kurt, Z.; Pang, S.; Bongiovanni, D. Transcription factor MAFF (MAF basic leucine zipper transcription factor F) regulates an atherosclerosis relevant network connecting inflammation and cholesterol metabolism. Circulation 2021, 143, 1809–1823. [Google Scholar] [CrossRef]
- Cassese, S.; Byrne, R.A.; Tada, T.; Pinieck, S.; Joner, M.; Ibrahim, T.; King, L.A.; Fusaro, M.; Laugwitz, K.L.; Kastrati, A. Incidence and predictors of restenosis after coronary stenting in 10 004 patients with surveillance angiography. Heart 2014, 100, 153–159. [Google Scholar] [CrossRef]
- Kohonen, T. Self-Oraganizing Maps, 3rd ed.; Springer: Berlin/Heidelberg, Germany, 2001. [Google Scholar]
- Kohonen, T. Essentials of the self-organizing map. Neural Netw 2013, 37, 52–65. [Google Scholar] [CrossRef]
- Ward, J.H., Jr. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Zeger, S.L.; Liang, K.Y.; Albert, P.S. Models for longitudinal data: A generalized estimating equation approach. Biometrics 1988, 44, 1049–1060. [Google Scholar] [CrossRef] [PubMed]
- Sing, T.; Sander, O.; Beerenwinkel, N.; Lengauer, T. ROCR: Visualizing classifier performance in R. Bioinformatics 2005, 21, 3940–3941. [Google Scholar] [CrossRef] [PubMed]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Muller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Goff, D.C., Jr.; Lloyd-Jones, D.M.; Bennett, G.; Coady, S.; D’Agostino, R.B.; Gibbons, R.; Greenland, P.; Lackland, D.T.; Levy, D.; O’Donnell, C.J.; et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: A report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation 2014, 129, S49–S73. [Google Scholar] [CrossRef]
- Hippisley-Cox, J.; Coupland, C.; Vinogradova, Y.; Robson, J.; Minhas, R.; Sheikh, A.; Brindle, P. Predicting cardiovascular risk in England and Wales: Prospective derivation and validation of QRISK2. BMJ 2008, 336, 1475–1482. [Google Scholar] [CrossRef]
- Ridker, P.M.; Buring, J.E.; Rifai, N.; Cook, N.R. Development and validation of improved algorithms for the assessment of global cardiovascular risk in women: The Reynolds Risk Score. JAMA 2007, 297, 611–619. [Google Scholar] [CrossRef]
- Uszko-Lencer, N.; Janssen, D.J.A.; Gaffron, S.; Vanfleteren, L.; Janssen, E.; Werter, C.; Franssen, F.M.E.; Wouters, E.F.M.; Rechberger, S.; Brunner La Rocca, H.P.; et al. Clustering based on comorbidities in patients with chronic heart failure: An illustration of clinical diversity. ESC Heart Fail 2022, 9, 614–626. [Google Scholar] [CrossRef]
- Vanfleteren, L.; Weidner, J.; Franssen, F.M.E.; Gaffron, S.; Reynaert, N.L.; Wouters, E.F.M.; Spruit, M.A. Biomarker-based clustering of patients with chronic obstructive pulmonary disease. ERJ Open Res. 2023, 9. [Google Scholar] [CrossRef]
- Tada, H.; Fujino, N.; Nomura, A.; Nakanishi, C.; Hayashi, K.; Takamura, M.; Kawashiri, M.A. Personalized medicine for cardiovascular diseases. J. Hum. Genet 2020, 66, 67–74. [Google Scholar] [CrossRef]
- Leon-Mimila, P.; Wang, J.; Huertas-Vazquez, A. Relevance of Multi-Omics Studies in Cardiovascular Diseases. Front. Cardiovasc. Med. 2019, 6, 91. [Google Scholar] [CrossRef] [PubMed]
- Pencina, M.J.; Goldstein, B.A.; D’Agostino, R.B. Prediction Models—Development, Evaluation, and Clinical Application. N. Engl. J. Med. 2020, 382, 1583–1586. [Google Scholar] [CrossRef] [PubMed]
- Farzadfar, F. Cardiovascular disease risk prediction models: Challenges and perspectives. Lancet Glob. Health 2019, 7, e1288–e1289. [Google Scholar] [CrossRef] [PubMed]


| Baseline Clinical Characteristics | Angiographic Restenosis | ||
|---|---|---|---|
| Yes (n = 2643) | No (n = 7361) | p-Value | |
| Age, years | 65.8 (58.5; 73.1) | 66.1 (57.8; 73.8) | 0.58 |
| Female gender, n (%) | 606 (22.9) | 1831 (24.8) | 0.045 |
| BMI (kg/m2) | 26.8 (24.5; 29.4) | 26.8 (24.5; 29.6) | 0.96 |
| Diabetes type 2, n (%) | 758 (28.7) | 1643 (22.3) | <0.001 |
| Insulin treated, n (%) | 229 (8.6) | 446 (6.0) | <0.001 |
| Current smoker, n (%) | 567 (21.4) | 1610 (21.8) | 0.65 |
| Arterial hypertension, n (%) | 1817 (68.7) | 4959 (67.3) | 0.19 |
| Hypercholesterolemia, n (%) | 1612 (60.9) | 4488 (60.9) | 0.98 |
| History of myocardial infarction, n (%) | 649 (24.5) | 1751 (23.7) | 0.42 |
| History of bypass surgery, n (%) | 377 (14.2) | 824 (11.2) | <0.001 |
| History of coronary angioplasty | 518 (21.6) | 1593 (19.6) | 0.028 |
| Clinical presentation, n (%) | |||
| Stable angina | 1488 (56.3) | 4071 (55.3) | 0.37 |
| NSTEMI | 635 (24.0) | 1978 (26.8) | 0.004 |
| STEMI | 520 (19.6) | 1312 (17.8) | 0.034 |
| Multivessel disease, n (%) | <0.001 | ||
| 2 vessel disease | 694 (26.3) | 2327 (31.6) | |
| 3 vessel disease | 1410 (53.3) | 3005 (40.8) | |
| LVEF, n (%) | 57 (47; 63) | 56 (46; 64) | 0.89 |
| Procedural Characteristics | Angiographic Restenosis | ||
|---|---|---|---|
| Yes (n = 3098) | No (n = 11,906) | p-Value | |
| Target vessel, n (%) | |||
| Left main | 71 (2.3) | 473 (3.9) | <0.001 |
| Left anterior descending coronary artery | 1310 (42.2) | 5091 (42.7) | 0.72 |
| Left circumflex coronary artery | 771 (24.8) | 2646 (22.2) | 0.001 |
| Right coronary artery | 850 (27.4) | 3393 (28.5) | 0.1 |
| Bypass graft | 96 (3.1) | 303 (2.5) | 0.12 |
| Lesion-to-patient ratio | 1.75 ± 0.95 | 1.41 ± 0.97 | <0.001 |
| Complex (type B2/C) lesion, n (%) | 2595 (83.7) | 8989 (75.5) | <0.001 |
| Chronic occlusion, n (%) | 214 (6.9) | 471 (3.9) | <0.001 |
| Lesion length, mm | 13.6 (8.9; 20.1) | 12.4 (8.5; 18.1) | <0.001 |
| Vessel size, mm | 2.68 (2.36; 3.02) | 2.86 (2.49; 3.27) | <0.001 |
| Initial diameter stenosis, (%) | 69.0 (57.0; 85.8) | 64.3 (54.0; 77.0) | <0.001 |
| Drug eluting stents implanted, n (%) | 1130 (36.8) | 7353 (61.7) | <0.001 |
| First generation | 559 (18.4) | 3255 (27.3) | |
| Second generation | 571 (18.4) | 4098 (34.4) | |
| TIMI flow pre angiography | |||
| 0 | 462 (14.9) | 1001 (8.4) | <0.001 |
| 1 | 170 (5.5) | 439 (3.7) | <0.001 |
| 2 | 382 (12.3) | 1332 (11.2) | 0.038 |
| 3 | 1941 (62.7) | 8786 (73.8) | <0.001 |
| Maximal balloon diameter, mm | 3.04 (2.66; 3.38) | 3.16 (2.84; 3.58) | <0.001 |
| Maximal balloon pressure, atm | 14 (12; 16) | 14 (12; 16) | <0.001 |
| Balloon-to-vessel ratio | 1.11 (1.05; 1.19) | 1.10 (1.04; 1.17) | <0.001 |
| Stented length, mm | 24 (18; 32) | 20 (16; 28) | <0.001 |
| Final diameter stenosis, (%) | 8.9 (4.5; 13.1) | 8.7 (4.9; 13.1) | 0.75 |
| Parameter Name | Cluster ‘High Restenosis’ | Other Clusters | p-Value | Weight in SOM Ordering | ||
|---|---|---|---|---|---|---|
| Mean | Std. Deviation | Mean | Std. Deviation | |||
| Lesion with restenosis 180d (%) | 62 | 0.486 | 0 | 0 | <0.001 | 0.2 |
| Lesion with high-grade restenosis 180d (%) | 25 | 0.431 | 0 | 0 | <0.001 | 0.6 |
| Grade of stenosis 180d (%) | 58.9 | 18.9 | 17.4 | 9.6 | <0.001 | 1 |
| Late lumen loss (mm) | 1.546 | 0.564 | 0.282 | 0.376 | <0.001 | 1 |
| Minimal lumen diameter 180d (mm) | 1.124 | 0.575 | 2.457 | 0.541 | <0.001 | 1 |
| Conventional Analysis | SOM-Based Analysis | |
|---|---|---|
| DES1 vs. BMS | + | + |
| DES2 vs. DES1 | + | + |
| Diabetes | + | + |
| History Bypass | + | + |
| STEMI/NSTEMI | - | CLIN_PRESENT: numeric by severity: + |
| NSTEACS + | ||
| STEMI: + | ||
| STAP: + | ||
| Left main (LCA) | + | + |
| Complex lesion | + | + |
| Chronic occlusion | - | + |
| Lesion length (10 mm) | - | + |
| Vessel size reduction (−0.5 mm) | + | + |
| Stenosis severity (5% DS increase) | + | + |
| Balloon-to-vessel ratio (for 0.1 +) | + | + |
| Stented Length (+10 mm) | + | + |
| SOM-Based Analysis | |
|---|---|
| Age | + |
| BMI | + |
| Hypercholesterolemia | + |
| History of PCI | + |
| TIMI-flow pre PCI | + |
| Stenosis post PCI | + |
| Pearson Correlation Coefficient | Regression Coefficient Classical Model | Regression Coefficient SOM-Based Model | p-Value | |
|---|---|---|---|---|
| Total Stented Length | 0.0923 | 0.02101 | 0.0853 | <0.0001 |
| Reference pre Vessel size | −0.1324 | −0.8731 | −0.1389 | <0.0001 |
| Stent Type: BMS | 0.2063 | 1.1673 | 0.2284 | <0.0001 |
| Stenosis post PCI | −0.0062 | 0.0539 | <0.0001 | |
| Diabetes | 0.0579 | 0.2400 | 0.0441 | <0.0001 |
| Stent Type: DES1 | −0.0863 | 0.0509 | <0.0001 | |
| Lesion Complexity (integer) | 0.0994 | 0.3727 | 0.0262 | <0.0001 |
| Balloon-to-Vessel Ratio | 0.0519 | −0.4005 | −0.0356 | <0.0001 |
| Clinical presentation | 0.0508 | 0.0335 | <0.0001 | |
| History of CABG | 0.041 | 0.5709 | 0.048 | <0.0001 |
| Grade of Stenosis pre | 0.1083 | 0.0056 | 0.0283 | 0.0003 |
| Hypercholesterolemia | −0.0102 | - | - | |
| Lesion length | 0.0605 | - | - | |
| History of PCI | −0.0235 | −0.0166 | 0.037 | |
| Balloon Pressure | −0.0488 | 0.0141 | 0.0382 | |
| Vessel: LCA | −0.0364 | 0.1053 | - | - |
| TIMI Flow pre | −0.1066 | −0.0087 | 0.049 | |
| Age | −0.0136 | −0.0120 | 0.0619 | |
| BMI | 0.0041 | - | - | |
| Chronic occlusion | 0.0573 | 0.0308 | 0.0617 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Güldener, U.; Kessler, T.; von Scheidt, M.; Hawe, J.S.; Gerhard, B.; Maier, D.; Lachmann, M.; Laugwitz, K.-L.; Cassese, S.; Schömig, A.W.; et al. Machine Learning Identifies New Predictors on Restenosis Risk after Coronary Artery Stenting in 10,004 Patients with Surveillance Angiography. J. Clin. Med. 2023, 12, 2941. https://doi.org/10.3390/jcm12082941
Güldener U, Kessler T, von Scheidt M, Hawe JS, Gerhard B, Maier D, Lachmann M, Laugwitz K-L, Cassese S, Schömig AW, et al. Machine Learning Identifies New Predictors on Restenosis Risk after Coronary Artery Stenting in 10,004 Patients with Surveillance Angiography. Journal of Clinical Medicine. 2023; 12(8):2941. https://doi.org/10.3390/jcm12082941
Chicago/Turabian StyleGüldener, Ulrich, Thorsten Kessler, Moritz von Scheidt, Johann S. Hawe, Beatrix Gerhard, Dieter Maier, Mark Lachmann, Karl-Ludwig Laugwitz, Salvatore Cassese, Albert W. Schömig, and et al. 2023. "Machine Learning Identifies New Predictors on Restenosis Risk after Coronary Artery Stenting in 10,004 Patients with Surveillance Angiography" Journal of Clinical Medicine 12, no. 8: 2941. https://doi.org/10.3390/jcm12082941
APA StyleGüldener, U., Kessler, T., von Scheidt, M., Hawe, J. S., Gerhard, B., Maier, D., Lachmann, M., Laugwitz, K.-L., Cassese, S., Schömig, A. W., Kastrati, A., & Schunkert, H. (2023). Machine Learning Identifies New Predictors on Restenosis Risk after Coronary Artery Stenting in 10,004 Patients with Surveillance Angiography. Journal of Clinical Medicine, 12(8), 2941. https://doi.org/10.3390/jcm12082941





