Abstract
Background/Objectives: In recent times, epigenetics alterations in Hidradenitis suppurativa (HS) have been explored and exploited translationally to guide investigation of new therapeutic approaches. On the other hand, long noncoding RNAs (LncRNAs), main regulators of the epigenetic status of the human genome, have been scarcely investigated, notwithstanding their potential relevance in broad pathogenesis comprehension. Here, we aim to explore the methylation pattern of lncRNAs in HS. Methods: In this case-control study, 24 HS patients and age-, sex- and BMI-matched controls were analyzed to characterize the methylome of lncRNA genes in peripheral blood cells. Gene ontology analysis (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, protein–protein interaction (PPI) network, and MCODE analysis were performed. Results: A set of fifteen lncRNA genes exhibited significantly differential methylation patterns, with ten of them showing hypomethylation and five displaying hypermethylation at specific CpG sites. The hypomethylated lncRNA genes were DLEU2, MESTIT1, CASC2, TUG1, KCNQ1DN, PSORS1C3, PCA3, DSCR8, RFPL1S, and PVT1, while the hypermethylated ones were HAR1A, FAM66B, SNHG9, HCG9, and HCP5. These lncRNA genes have been linked to various important biological processes, including cell proliferation, apoptosis, inflammation, chronic inflammatory skin diseases, and wound healing. Their altered methylation status suggests potential roles in regulating these processes, and may contribute to HS pathogenesis and healing mechanisms. Conclusions: This study revealed an interesting dysregulation pattern of definite lncRNAs in the methylome which is linked to both the development of HS and its comorbidities. Epigenetically altered lncRNAs genes could represent useful biomarkers, and could help in guiding innovative treatment strategies.
1. Introduction
Hidradenitis suppurativa (HS), also known as acne inversa or Verneuil’s disease, is a chronic and recurrent inflammatory dermatosis of the hair follicle with severe negative impact on quality of life and associated with important co-morbidities. HS lesions most commonly occur in intertriginous areas and areas rich in apocrine glands. Among the most common are the axillary, groin, perianal, perineal, and inframammary locations. Because of the associated pain, sensitive locations, drainage, odor, and scarring, this condition may have a negative psychosocial impact [1]. Numerous definite genetic changes [2,3] and epigenetic modifications [4,5,6] are associated with HS susceptibility, disease onset/progression and response to treatment. Current basic science research focusing on the etiology, pathophysiology, and treatment of HS is ever-increasing [7]; however, it remains elusive and poorly informative as regards targeted therapies [8,9,10]. Furthermore, while exposure to a number of environmental factors is capable of modulating disease severity, flares, and even drug response, the pathogenesis of disease outcomes modifications caused by ambient components remains unclear. The framework of disease mechanisms driving HS may involve long noncoding RNAs (lncRNAs), which are transcripts longer than 200 nucleotides in length that lack protein-coding capacity [11]. LncRNA biogenesis is related to specific subcellular localizations and functions; through definite interactions with DNA, RNA and proteins can control chromatin function, regulate the assembly and function of membraneless nuclear bodies, modify cytoplasmic mRNA stability and translation, and affect intracellular signalling pathways, in due course impacting gene expression in various biological and pathological settings [12].
Furthermore, lncRNAs fine-tune numerous cellular processes such as splicing, the cell cycle, apoptosis, pluripotency preservation, embryonic development, and cell differentiation [13,14]. lncRNAs are transcribed from different genomic regions, such as exons, promoters, and intergenic regions, and are important regulators of epigenetic status in the human genome, influencing gene expression [15]; according to Gencode Release v43, (https://www.gencodegenes.org, accessed on 23 March 2023), 19,928 lncRNA genes produce 25,407 lncRNA transcripts, and the number is growing steadily. LncRNAs are uniquely expressed in specific cell types to a greater degree than protein-coding RNAs (mRNAs), and are regarded as crucial in disease onset, showing specific expression in different cancer types. They are considered valuable biomarkers supportive of diagnosis. Furthermore, lncRNAs are enriched at numerous imprinted gene clusters and participate in genomic imprinting, a multifaceted and highly controlled process leading to the monoallelic silencing of definite genes based on the parent-of-origin of the allele imprinting processes [16].
Clinically, lncRNAs play a role in the pathogenesis of various diseases, including cardiovascular disease, atherosclerosis, Alzheimer’s disease [17], dyslipidemia, and metabolic syndrome [18]. lncRNAs represent valuable biomarkers and druggable targets depending on tissue-specific and condition-specific expression patterns.
Recently, lncRNAs have gained attention for their potential roles in modulating keratinocyte differentiation and inflammation [19]. Deregulated lncRNAs have been implicated in aberrant keratinocyte differentiation and disruption of epidermal homeostasis, and are crucially involved in the pathogenesis of several hyperproliferative skin diseases, including psoriasis, hypertrophic scars, cutaneous squamous cancer, melanoma and haemangiomas [20]. Recent studies have shown that lncRNAs play important roles in epidermal development, keratinocyte differentiation, and melanocyte function [21]. Notwithstanding this, the role played by lncRNAs in HS onset and progression is not adequately yet clear.
The aim of this study was to explore the outline of DNA methylation at lncRNA coding genes in the peripheral blood of HS patients and age-matched controls through genome-wide analysis of DNA methylation patterns.
2. Materials and Methods
2.1. Study Design
This study was approved by the Institutional Review Board of Beaumont Health System, Royal Oak, MI, USA (HIC#: 2015-172, 21 May 2015). The study followed ethical guidelines by obtaining written informed consent from all participating individuals, adhering to the principles outlined in the Helsinki Declaration. To ensure reliable and meaningful results, individuals diagnosed with HS were carefully paired with healthy controls matched based on similar characteristics such as age, gender, and body mass index (BMI). The demographic and clinical characteristics of patients and controls were previously reported [6].
2.2. Hidradenitis Suppurativa Sample Selection and Statistical Methods
Three independent board-certified dermatologists (RR, DGS, TM) at VS Hospital, Ahmedabad, India, employed a visual-aided questionnaire to conduct a thorough self-assessment of HS in patients. This questionnaire was specifically designed to aid in the evaluation of HS-related symptoms and their severity [22]. The diagnostic process and sample assessment were carried out according to the guidelines from the European Hidradenitis Suppurative Foundation (EHSF) [23].
The severity of the disease was assessed through grading system widely used to typify the extent of disease in HS patients and the impact of the condition on the affected individuals, including the Hurley score [24], HS Severity Score System (IHS4) [25], and Autoinflammatory Disease Damage index (ADDI) [26].
Inclusion criteria were (a) adult patients (>20 years of age) with a diagnosis of HS with a duration greater than 5 years, (b) at least a Hurley stage II severity, (c) IHS4 > 3 points, (d) ADDI < 3 points, (e) newly diagnosed HS (<3 months), and (f) untreated for at least 6 months.
Exclusion criteria were: (a) syndromic HS as defined by Van der Zee and Jemec clinical phenotypes [27], (b) smoking, (c) fasting regimens [28] and/or particular diet regimens different from omnivore, (d) alcohol abuse (Alcohol Use Disorders Identification Test (AUDIT) > 7 points), (e) drug addiction, (f) use of concomitant medications, including contraceptives and enzyme-inducing foods (i.e., grapefruit) (g) treated for HS, (h) chronic inflammatory/infectious diseases or history of cancer in the previous 5 years, and (m) persons unable to provide informed consent for any reason.
2.3. Statistics and Bioinformtic Analyes
Data (IDAT files) were normalized using Genome Studio 2.0 software (Illumina Inc., San Diego, CA, USA, accessed on 2 April 2024) functional normalization and determined Cytosine methylation levels (ß-value) for each CpG site. Before analysis, we removed all CpG-probes that had missing ß-values. Differential methylation was assessed by comparing the ß-values for cytosine at each CpG locus in HS versus controls. To avoid confounding factors, we removed probes associated with sex chromosomes, non-specific probes, and probes targeting CpG sites within 10 bp of SNPs (each of which listed dbSNP entries within 10 bp of the CpG site). Further, SNPs with a minor allele frequency (≤0.05) were only considered for forwarding analysis. Significantly differently methylated CpG sites between HS and controls were defined based on preset cutoff criteria (FDR p < 0.05). Multiple CpG sites within a gene were resolved by selecting the CpG with the highest AUC ROC ranking and the lowest p-value. The p-value for methylation differences between case and control groups at each locus was calculated as previously described [6]. Raw and FDR p-values corrected for multiple testing (Benjamini–Hochberg test) were calculated. The AUC for combinations of loci was calculated using the ‘R’ program ‘ROCR’ package (v3.5.0) (accessed on 2 April 2024).
2.4. DNA Preparation and Methylation Analysis
We collected whole blood samples from 24 individuals diagnosed with HS and 24 healthy individuals as controls. Genomic DNA was extracted from these blood samples using the Gentra Puregene® Blood Kit (Qiagen, Venlo, The Netherlands). To analyze DNA methylation patterns, the extracted DNA was subjected to a sodium bisulfite conversion process. This conversion process was carried out following the manufacturer’s protocol, which involved using the EZ 96-DNA methylation kit (Zymo Research, Irvine, CA, USA). A comprehensive description of the methodology used for the entire process has been previously published in [4].
2.5. Heatmap
The ComplexHeatmap module (v1.6.0) in the R package (v3.2.2) was utilized to create a heatmap. The heatmap displayed the distribution of methylated CpG sites within the CYP coding regions, with each site representing an individual data point. The purpose of this analysis was to visualize the methylation patterns across these regions. We utilized Ward’s method to perform hierarchical cluster analysis on the samples [29]. For comparison between HS and controls, CpG sites with FDR p-values ≤ 0.05 were considered significantly differently methylated. The area under the receiver operating characteristic (AUC-ROC) was calculated based on methylation levels at the most significantly differently methylated CpG loci.
2.6. Principal Component Analysis
Principal Component Analysis (PCA) is a powerful data transformation technique extensively employed for enhancing data visualization and performing feature extraction by reducing the dimensionality of the dataset. PCA serves as a valuable feature extraction tool. Used in various fields, including epigenetics, it allows the most important features that contribute the most to the overall variance between groups to be identified and selected. The R function “prcomp” was used to compute principal components (PCs), then PC1, PC2, and PC3 were used for the PCA distribution plot. The 3D PCA distribution plot was generated using the R package “ggplot2”.
2.7. Protein–Protein Interaction Network and MCODE Analysis
Based on a database search (http://bio-annotation.cn/lncrna2target/search.jsp, accessed on 2 April 2024) of all 15 lncRNAs, 38 mRNA transcripts were found to be their targets. These genes were further subjected to string protein–protein interaction network analysis to search for association between them. Protein–protein interaction mapping is the process of identifying the physical interactions between proteins in a biological system. The STRING database (https://string-db.org/, accessed on 2 April 2024) is a valuable resource for this purpose, as it provides information on known and predicted protein–protein interactions based on a variety of sources, including experimental data, co-expression patterns, and text mining.
In the present study, protein–protein interaction (PPI) mapping was performed using the STRING database for multi-protein options in Organism: Homo sapiens. A total of 38 target proteins retrieved from lncRNA targets were subjected to PPI. The interaction detection method was set to experimentally validated, co-expressed, and curated genes, with the confidence score threshold set to high (>0.7).
2.8. Protein–Protein Interaction Network and MCODE Analysis
During the initial phase of the study, a comprehensive list of genes of interest was meticulously compiled. After consolidating the data, the subsequent pivotal phase involved delving into the complexities of potential protein–protein interactions (PPIs) linked to these genes.
To fulfill this objective, the renowned STRING database (version 12.0 https://string-db.org/, accessed on 2 April 2024) played an indispensable role. Given the immense amount of data present in the realm of biological interactions, it was essential to filter and streamline the data to our specific needs. Thus, a set of interaction sources was activated to uphold the quality and pertinence of the gathered data. These sources encompassed: text mining, which derives interactions from the vast repository of scholarly literature; experiments, spotlighting interactions identified in tangible experimental scenarios; databases, integrating insights from various curated biological databases; and co-expression, shedding light on genes or proteins that express concurrently. To further refine the interaction data, a high-confidence interaction score threshold was set at 0.700. This strategic filtration ensured that only the most credible interactions were considered. Subsequently, to manifest these interactions in a more visually coherent manner, the PPI network extracted from STRING was channeled into Cytoscape software (https://cytoscape.org, accessed on 2 April 2024). This platform was instrumental in providing a more tangible representation, facilitating a deeper understanding and analysis of the interaction dynamics.
2.9. Ingenuity Pathway Analysis
We performed Ingenuity Pathway Analysis (IPA QIAGEN, Aarhus, Denmark) to implement wide-ranging data analysis to recognise investigational outcomes, envisage downstream influences, and recognize novel targets/biomarkers in the setting of HS.
3. Results
3.1. Identification of Dysregulated CpGs in HS
Epigenome-wide DNA methylation profiling with the Illumina Epic array (Illumina Inc., San Diego, USA) was performed on blood DNA samples from 24 HS patients and 24 age- and sex-matched controls to explore changes in DNA methylation at lncRNA coding genes. We identified fifteen significantly differentially methylated lncRNA coding genes in blood, of which ten were hypomethylated (DLEU2, MESTIT1, CASC2, TUG1, KCNQ1DN, PSORS1C3, PCA3, DSCR8, RFPL1S and PVT1) and five were hypermethylated HAR1A, FAM66B, SNHG9, HCG9, and HCP5) at CpG sites (FDR p-values ≤ 0.05) associated with the fifteen genes. All dysmethylated CPGs are listed in Table 1.

Table 1.
Significantly differentially methylated CpG sites of lncRNA associated with HS. Differentially methylated CpG sites and regions related to HS with Target ID, Gene ID, chromosome location, % methylation change, and FDR p-value for each CpG loci are provided. CpG sites with a significant FDR p-value indicating methylation status and ROC AUC > 0.75 appear to be potential diagnostic biomarkers for HS.
3.2. Validation
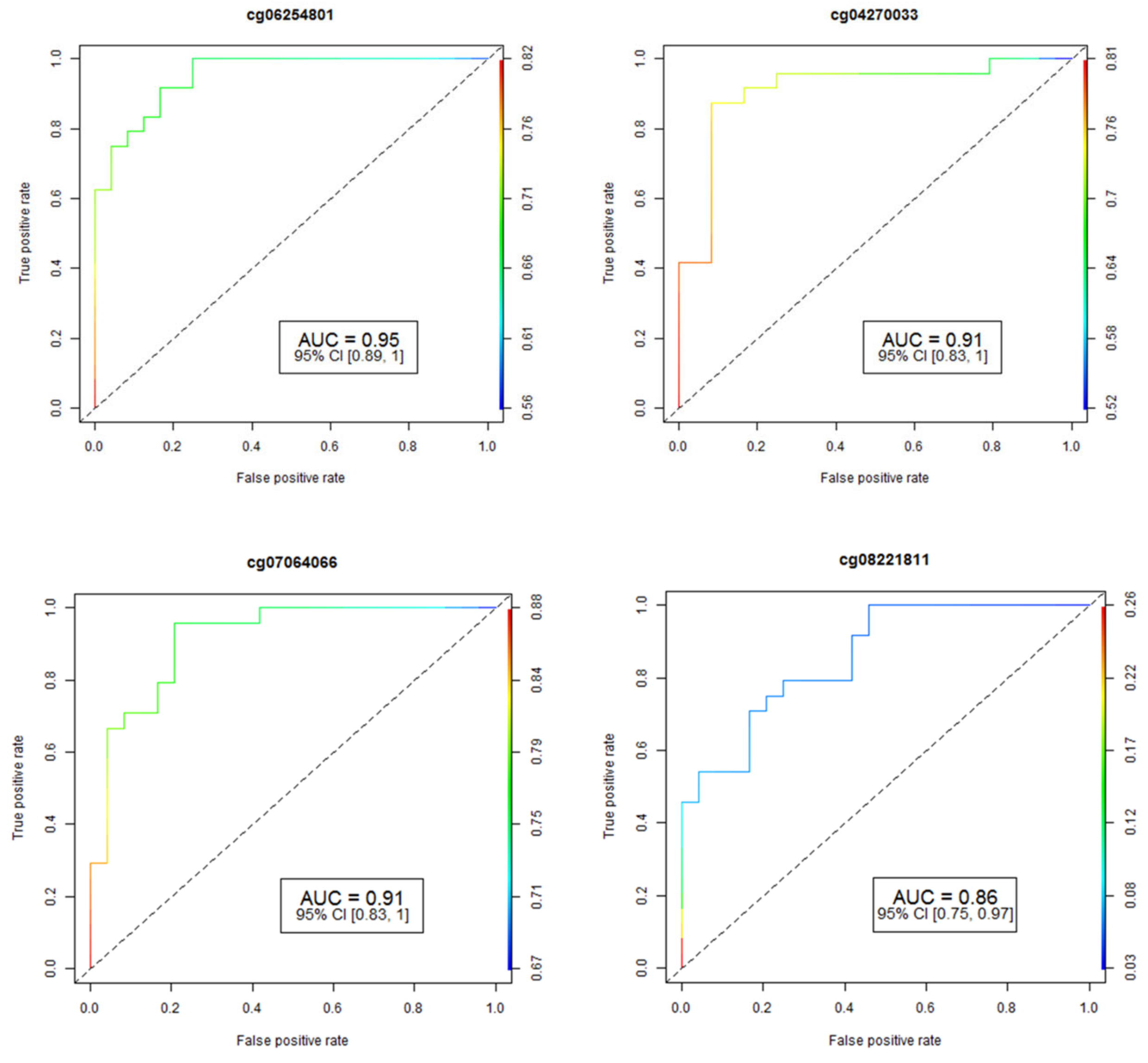
The accuracy and reliability of the data concerning CpG methylation changes were rigorously assessed through pyrosequencing, a well-established method for validation. To determine the performance of individual CpG loci, we employed the Area Under the Receiver Operating Curve (AUC-ROC), a widely accepted metric in this field. The results for the four most promising CpG loci are visually represented in Figure 1, providing valuable insights into their predictive capabilities.
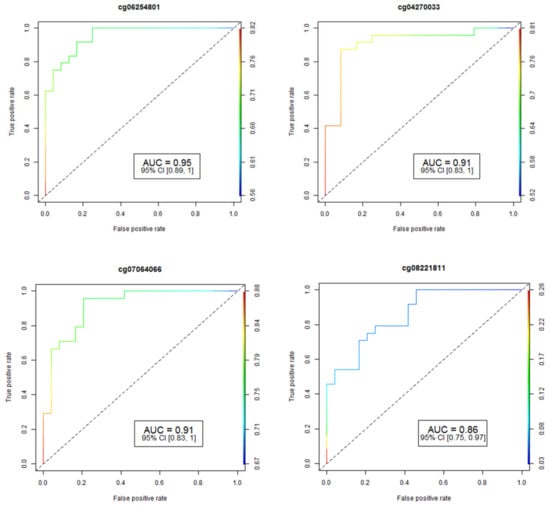
Figure 1.
Analysis of receiver operating characteristic curves (ROC) based on AUC ROC and FDR p-values for four of the most significant CpGs associated with HS. The study identified fifteen differentially-methylated CpG sites in fifteen genes that have an area under the ROC curve ≥ 0.75 (p-value ≤ 0.05) for HS prediction. AUC: area under the receiver operating characteristics curve; 95% CI: 95% confidence interval. Lower and upper confidence intervals are given in parentheses.
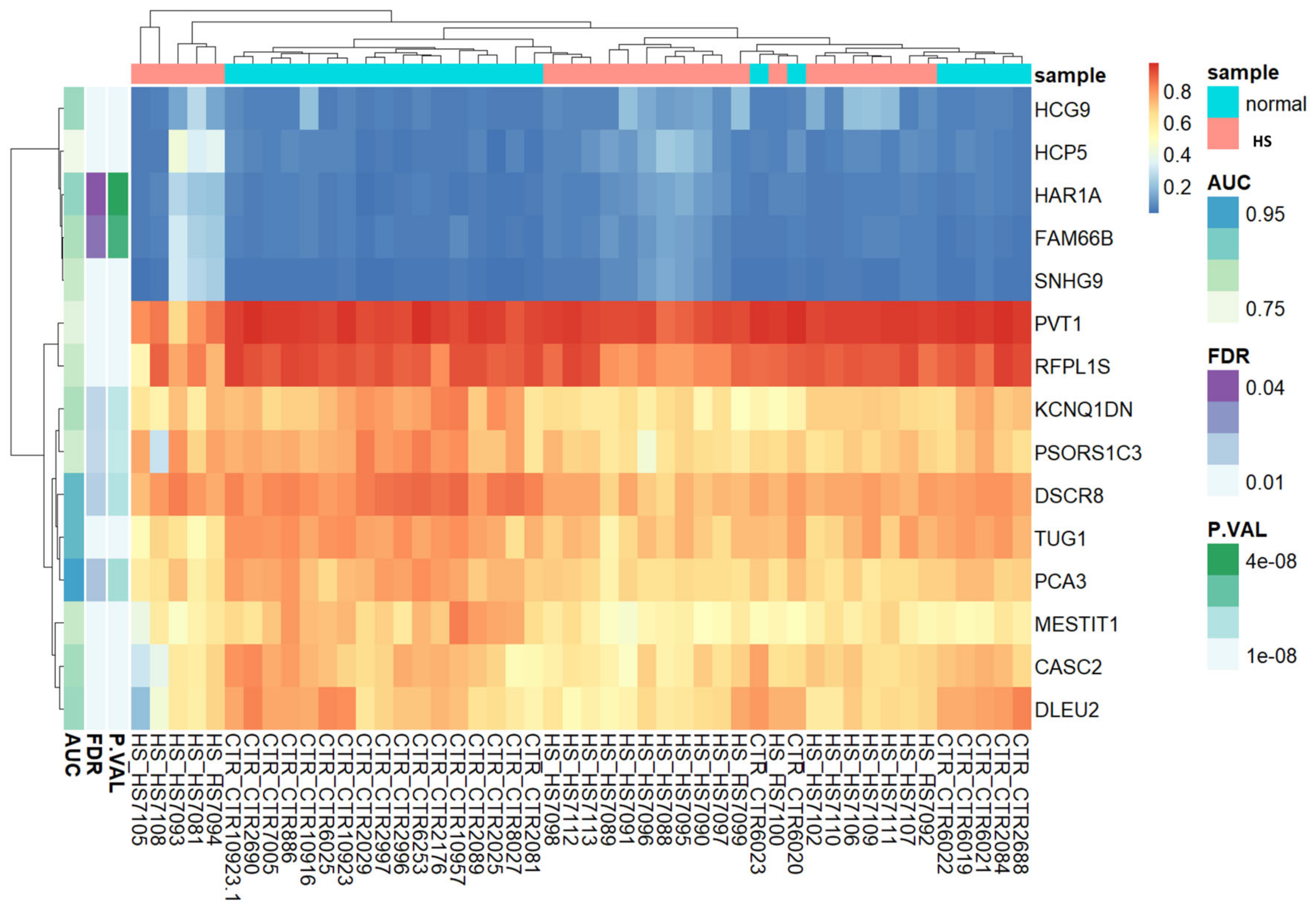
3.3. Evaluation of Heatmaps
Based on lncRNA-related CpG methylation markers, the heatmap unequivocally demonstrates the presence of two discernible clusters of CpGs: one corresponding to HS patients, and another representing the control group (Figure 2). This compelling evidence supports the notion that these methylation markers serve as highly reliable indicators for distinguishing between HS-affected patients and unaffected individuals. In essence, our findings corroborate the accuracy and efficacy of these methylation markers for accurately discriminating between the two study groups.
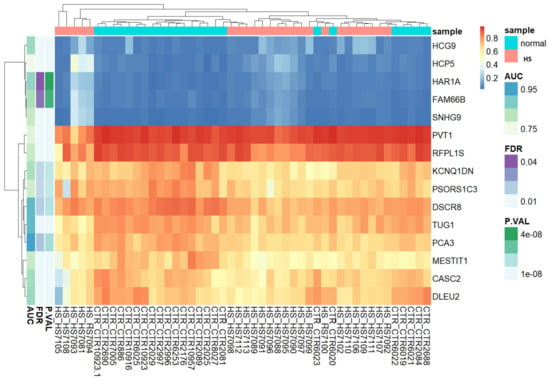
Figure 2.
Unsupervised hierarchical clustering and heat map of the methylation data of fifteen CpGs with (Δβ) > 0.2 and FDR p values < 0.05 between HS cases and controls. The heat map colors correspond to lncRNA expression, as indicated in the color legend. The cases with HS phenotype are shown in red boxes and normal controls in green boxes. Blue and yellow indicate 0 and 1 methylation, respectively. The probes showing “hypermethylated” cases are represented by yellow vertical bars, and those showing “hypomethylated” cases by blue ones.
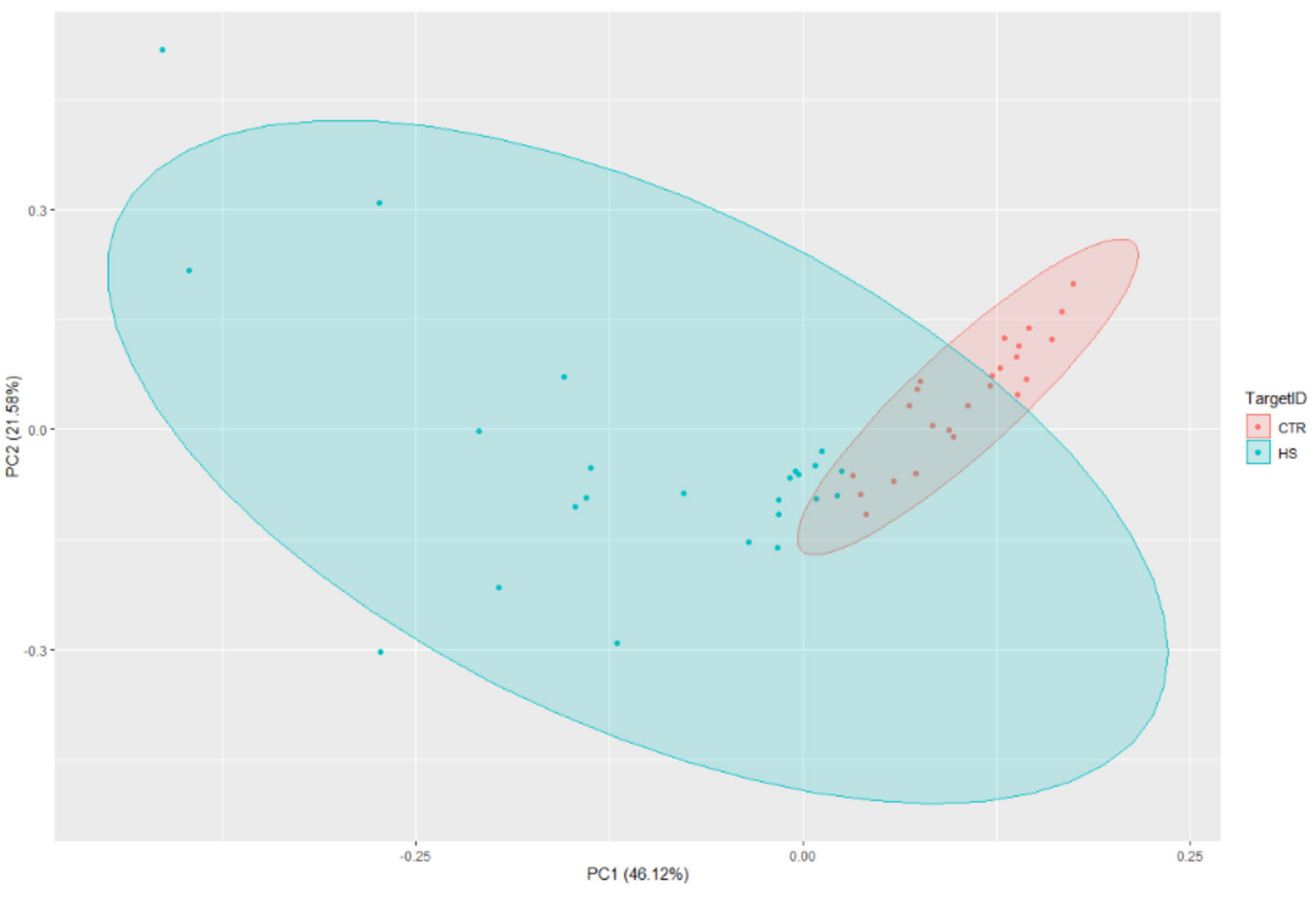
3.4. PCA
The PCA analysis provided strong evidence of distinct separation between the HS cases and controls (Figure 3).
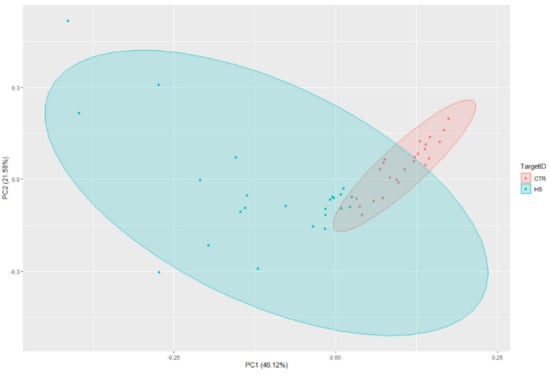
Figure 3.
Principal Component Analysis (PCA) with lncRNA associated gene markers.
3.5. Protein–Protein Interaction Network and Modular Analysis
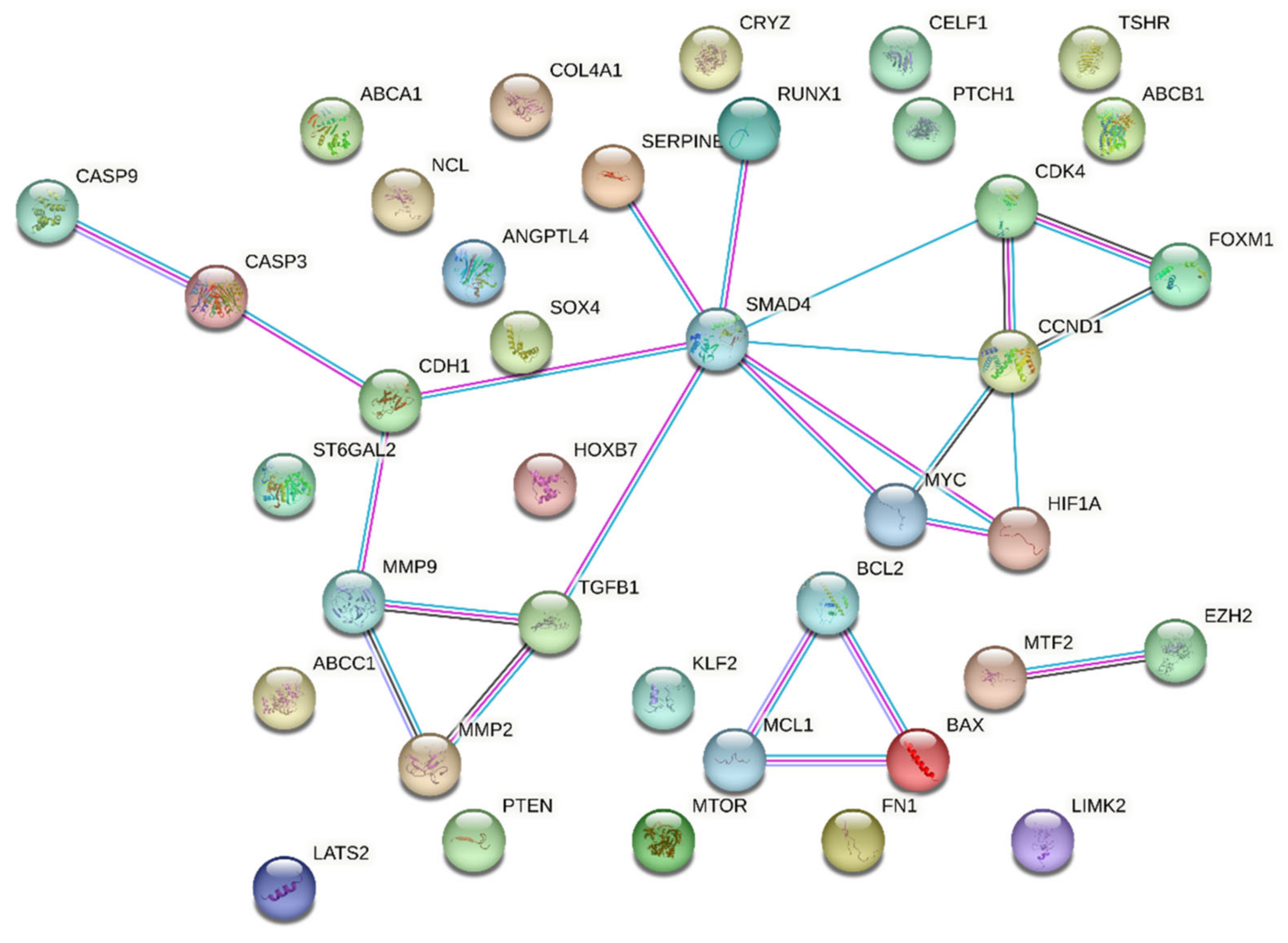
Upon utilizing the STRING database with the set parameters and interaction sources, the resulting protein–protein interaction network revealed some intriguing statistics. The network comprised a total of 38 nodes representing individual proteins or genes; these nodes were interconnected through 108 edges signifying potential interactions between the proteins. On average, each node in the network demonstrated a degree of 5.68, indicating that on average each protein or gene interacts with approximately five to six other entities in the network. SMAD4 and CCND1 were leading hub nodes, with node degrees of 8 and 5, respectively. Upon further examining the intricacy of these interactions, the average local clustering coefficient was found to be 0.535 (Figure 4).
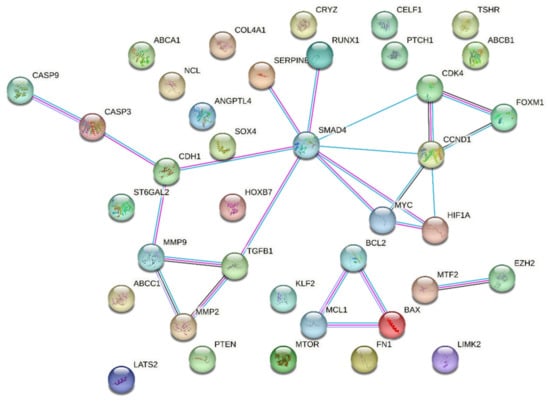
Figure 4.
String output showing protein–protein interactions of 38 queried proteins. Colors legend: Red: inactivated, Green: activated, Blue: Indifferent.
This metric suggests a moderate tendency for the proteins to cluster together, forming interconnected groups within the network. Interestingly, when contrasting the observed data against the expected norm, the expected number of edges for such a network was only 18. This significant deviation from the expected values indicates that the compiled list of genes showcases a denser interaction than would typically be predicted for a random set of proteins of a similar size. This observation was statistically fortified with a PPI enrichment p-value of less than 1.0 × 10−16, confirming that the network contained a significantly higher number of interactions than expected.
Moreover, delving deeper into the local network clusters from STRING, several crucial biological processes and pathways surfaced, showcasing the potential roles of our genes of interest. Among the four emergent clusters, the cluster associated with “Apoptosis—Multiple Species and TRAIL Signaling” sheds light on the roles of proteins such as BAX, CASP3, CASP9, MCL1, and BCL2. Next, our attention was drawn to the “Bcl-2 Family and BH3-only Proteins Associate with and Inactivate Anti-apoptotic BCL-2 Members”. This cluster underscores the intricate interactions and regulatory mechanisms orchestrated by proteins such as BAX, MCL1, and BCL2. A third significant cluster, titled “Extracellular Matrix Organization”, unveils insights into the operational dynamics of proteins pivotal for the ECM, including MMP2, TGFB1, FN1, MMP9, and COL4A1. Concluding the list of notable clusters, the “Activation of Caspases through Apoptosome-mediated Cleavage” cluster highlights the indispensable roles played by CASP3 and CASP9 in the intricate dance of apoptosis. A comprehensive overview of the False Discovery Rate (FDR) values and the strength associated with these clusters is available in Table 2.

Table 2.
Local Network Clusters from STRING.
3.6. Ingenuity Pathway Analysis
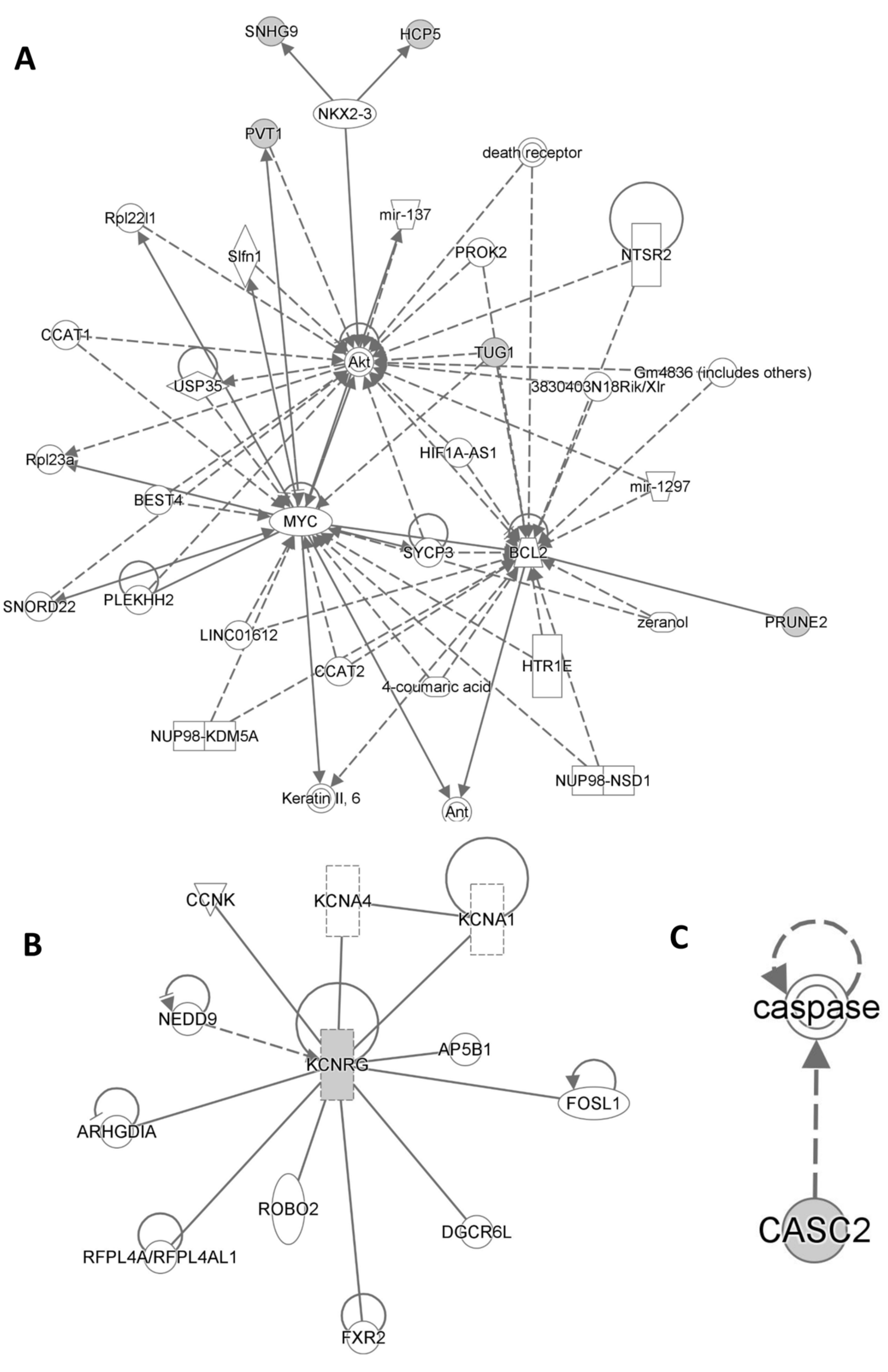
Ingenuity Pathway Analysis (IPA QIAGEN) evidenced that Cancer, Organismal Injury and Abnormalities, Cell Death and Survival, Organismal Injury and Abnormalities as well as Cellular Growth and Proliferation, Organ Development, Reproductive System Development and Function, and Tissue Development were included among statistically significant diseases and functions (Supplementary Tables S1 and S2). On the other hand, Cancer, Cell Death and Survival, Organismal Injury and Abnormalities, Cell Signaling, Cellular Development, Cellular Growth and Proliferation, Cardiovascular Disease, Neurological Disease, and Ophthalmic Disease were included among statistically significant networks (Figure 5).
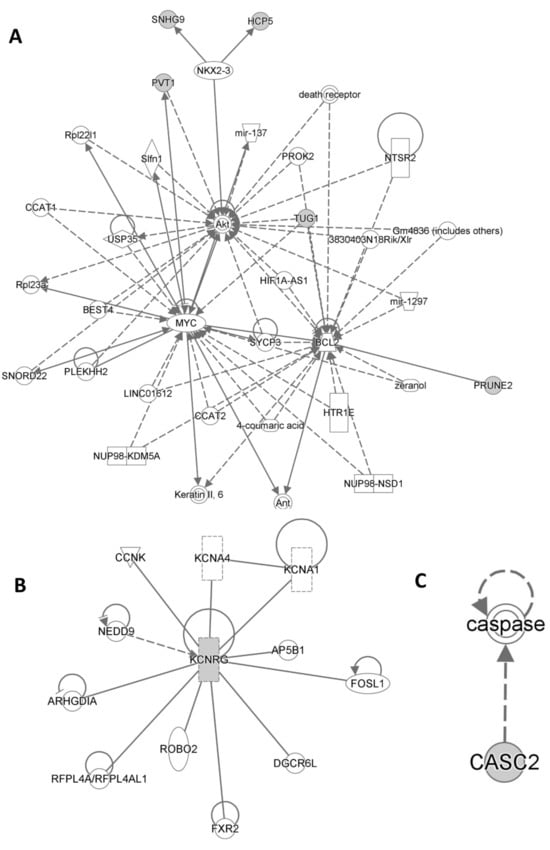
Figure 5.
Statistically significant networks and pathways related to the lncRNA typical of HS patients from Ingenuity Pathway Analysis are shown in (A), (B), (C) respectively.
4. Discussion
Several important stages are hallmarks in the course of HS development, during which distinctive lncRNAs play an essential role. Particularly when deregulated, they disrupt the delicate balance between damaging and reparative processes, which in turn can exacerbate chronic inflammation and hinder wound healing and tissue renovation. lncRNAs contribute significantly to HS pathogenesis and progression through regulation of gene expression and molecular signaling pathways [30]. Despite the fact that some lncRNAs have been linked to several biological processes, their precise roles have not yet been fully studied in the setting of HS pathogenetic mechanisms.
As reported in the Results section, we pinpointed statistically significant differential methylation at CpG sites of lncRNA coding genes; DLEU2, MESTIT1, CASC2, TUG1, KCNQ1DN, PSORS1C3, PCA3, DSCR8, RFPL1S, and PVT1 were hypomethylated, while HAR1A, FAM66B, SNHG9, HCG9, and HCP5 were hypermethylated.
4.1. PCA3
Prostate Cancer Antigen 3 (PCA3) is a gene that has primarily been associated with prostate cancer; its expression is used as a diagnostic marker for the disease [31]. PCA3 modulates prostate cancer (PCa) cell survival through modulating androgen receptor (AR) signaling [32]. AR is a nuclear hormone receptor that binds to androgens such as testosterone and dihydrotestosterone (DHT) and regulates the transcription of target genes involved in cell proliferation, differentiation, and survival. In HS, the overexpression and activation of AR have been implicated in the increased proliferation and abnormal differentiation of the hair follicles and apocrine glands in the affected areas, leading to the formation of inflammatory nodules and cysts [33].
4.2. DSCR8
DSCR8 gene (Down syndrome critical region 8), also known as MMA-1 (Malignant melanoma-associated protein 1), is highly expressed in uterine cancer and melanoma [34]. The prevalence of non-melanoma skin cancer risk is augmented in correlation with HS [35]. It may be possible that DSCR8 regulates immune responses and inflammation, which in turn contribute to HS development. Moreover, many studies have reported an increased prevalence of Down syndrome in the HS population [36,37,38].
4.3. TUG1
TUG1 gene has been implicated in various biological processes, including cell proliferation, apoptosis, and inflammation. Although the exact role of TUG1 in apocrine glands is not clear, some studies suggest that it may be involved in regulating the function of these glands. TUG1 is highly expressed in human apocrine sweat glands and its expression is regulated by androgen hormones. TUG1 is upregulated in the apocrine sweat glands of patients with HS. TUG1 is a regulator of the NLRP3 inflammasome, a key mediator of inflammation in HS. Earlier reports indicate that TUG1 is upregulated in HS lesional skin and that knockdown of TUG1 can reduce the production of IL-1β, a proinflammatory cytokine produced by the NLRP3 inflammasome.
4.4. HAR1A
HS and atopic dermatitis (AD) are both chronic inflammatory skin diseases [39]. AD, also known as eczema, produces dry, itchy, and red skin; the HAR1A gene is implicated in AD [40]. A strong clinical association between HS and AD has been previously reported [39], suggesting that HS may share a more common genetic landscape.
4.5. DLEU2
HS-related genes such as DLEU2 [41], which regulates Sirtuins and mitochondrial respiratory chain complex IV, are associated with the regulation of mitochondrial function, indicating that mitochondrial dysfunction could play a central role in HS pathogenesis. DLEU2 is involved in several types of cancer, including chronic lymphocytic leukemia and non-small-cell lung cancer. Loss of the DLEU2 gene is associated with an increased risk of squamous cell carcinoma of the skin. Furthermore, DLEU2 expression is significantly decreased in melanoma samples compared to normal skin.
4.6. HCG9
Several studies have found an association between HCG9 gene changes and bipolar disorder [42]. Although it is common for patients with HS to have psychiatric comorbidities, few studies have examined whether severe psychiatric disorders are associated with HS. However, a recent population-based study found that HS patients are more likely to suffer from bipolar disorder [43].
4.7. CASC2
CASC2 (Cancer susceptibility 2) is a tumor suppressor gene that has been associated with several types of cancers, including endometrial, lung, gastric, and colorectal cancers [44]; however, there is limited research on its possible involvement in HS. HS seems to cause an increased risk of developing several types of cancer, including lung, gastric, and colorectal cancers, as well as earlier onset [35]. Additionally, CASC2 may inhibit the development of malignant melanoma by regulating miR-18a-5p/RUNX1 axis [45]. Both CASC2 and RUNX1 were hypomethylated in the present study.
4.8. FAM66B
FAM66B gene changes have been reported to be involved in esophageal squamous cell carcinoma [46]. However, the association of such changes with HS has not been studied in depth yet.
4.9. KCNQ1DN
KCNQ1DN is an imprinted gene located between p57(KIP2) and KvLQT1 (KCNQ1) in chr11p15.5 within the Wilms’ tumorigenesis (WT2) critical region, and is considered a candidate for involvement in WT.
4.10. RFPL1S
The tumor suppressor gene RFPL1S may slow ovarian cancer progression by inhibiting IFN-β/STAT1 signaling [47].
4.11. SNHG9
The SNHG9 gene has been implicated in several cancer types, including glioblastoma [48], pancreatic cancer [49], and non-small cell lung cancer (SCLC) [50], although its function in HS remains unknown. The majority of HS patients are either active or passive smokers; smoking is a known risk factor for lung cancer, including SCLC, and smoking rates are higher among subjects suffering from HS compared to the general population.
4.12. MESTIT1
MESTIT1 is an imprinted gene preferentially expressed from the paternal allele [51]. MESTIT1 has been reported as a strong candidate gene for Silver–Russell syndrome (SRS) in chr7q31 [52]. Approximately 7–10% of SRS cases have been reported with maternal uniparental disomy (UPD) of chromosome 7, a segmental maternal UPD (7) restricted to 7q31-other. The involvement of MESTIT1 in HS pathophysiology is unknown at present.
4.13. PSORS1C3
PSORS1C3 is associated with the development of psoriasis, a common chronic immune-mediated inflammatory disease of the skin [53]. Psoriasis is one of the critical comorbidities associated with HS [54].
4.14. PVT1
PVT1 has been linked to a variety of cancers, including gastric cancer, breast cancer, ovarian cancer, and skin cancer [55]. PVT1 functions as an oncogene by inhibiting cancer cell apoptosis, promoting cell proliferation, and affecting tumor invasion and metastasis generation [56]. When downregulated, PVT1 is thought to be involved in the regulation of inflammation, as it reduces the expression of inflammatory cytokines such as TNF-α, IL-1β, IL-6, IL-10, IL-17, and IFN-γ [57], suggesting that PVT1 may contribute to the inflammatory response in HS.
4.15. HCP5
HCP5 (HLA complex P5) is a gene that encodes a protein involved in the immune response. Variants of the HCP5 gene are associated with an increased risk of developing Stevens–Johnson syndrome (SJS) and toxic epidermal necrolysis (TEN), which are severe and potentially life-threatening cutaneous drug reactions [58,59]. Sorafenib treatment has been found to cause adverse cutaneous reactions and side effects in HS patients [60]. There is evidence that hypermethylation of HCP5 relates to obesity and BMI in Africans [61]. Obesity and overweight are the most common comorbid conditions associated with HS.
4.16. Protein–Protein Interactions
PPIs and lncRNA expression are two important aspects of molecular biology that play crucial roles in cellular processes and gene regulation. LncRNAs regulate gene expression by interacting with intracellular molecules such as DNA, RNA, and proteins. They can bind DNA to influence protein recruitment, control mRNA stability and translation, and form ribonucleoprotein complexes for diverse cellular functions.
In the present study, we looked for lncRNA targets through database mining and retrieved 38 lncRNA target genes. These genes were further subjected to STRING protein–protein interaction network analysis to search for interactions among them. We found two hub nodes showing maximum interaction among the group components, namely, SMAD4 (target of lncRNA PVT1) and CCND1 (target of lncRNA TUG1). Increased risk for SCC or adenocarcinoma has been reported in HS patients with abnormal CCND1 expression, possibly due to chronic inflammation [62]. Results from a GWAS study from PIONEER I and II clinical trial participants have highlighted a pathway involving BCL2 in response to adalimumab, the only FDA approved drug for HS treatment [63]. In addition, among the 38 gene result lncRNA targets, SERPINE1, MTOR, and MMP9 have been reported to be suitably druggable [64].
Recent genome-wide association studies (GWASs) have been performed for HS, which have identified and replicated significant HS-associated risk loci. Of particular interest were the lead variants rs10512572 (p = 2.3 × 10−11) and rs17090189 (p = 2.1 × 10−8) near the SOX9 and KLF5 genes, respectively; variants at these loci resulted in enhanced regulatory elements detected in skin tissue [65].
Furthermore, the results of a recent cross-sectional genotype–phenotype study performed in a Maltese patient cohort propose that monogenic variation in NCSTN, one of the genes that code for proteins of the γ secretase complex, is associated with HS in a subset of patients with a different nonconforming phenotype. In addition, carriers of the NCSTN:c.671_682del variant were more likely to require adalimumab treatment [66].
4.17. Identifying Druggable Targets
Identifying druggable targets typically involves extensive research into the biological pathways, functions, and potential therapeutic applications of the studied molecules. LncRNAs presents unique challenges compared to protein-coding genes. Traditional small molecule drugs, which primarily target proteins, are not applicable for lncRNAs, as they lack a protein-coding function and instead function as regulators of gene expression. While several emerging strategies to potentially target lncRNAs for therapeutic purposes are currently being researched and developed in preclinical testing, these approaches are relatively new, albeit promising. They include antisense oligonucleotides (ASOs), small interfering RNAs (siRNAs), CRISPR-Cas9-based therapies, and RNA interference-based therapies.
4.18. Limitations
The current findings are limited by a lack of experimental in vivo and in vitro validation. Future experimental studies are needed in order to corroborate the expression and function of the identified genes at the protein level, determine whether these changes in pathway activation are associated with HS disease activity, and identify which molecules can be used to develop targeted therapies for HS. In addition, the validity of the lncRNAs needs to be verified in a large HS population study. While various lncRNAs are candidate central regulators in inflammatory signaling pathways, only a few of them have been pinpointed in HS. The shared mechanisms of HS and other inflammatory diseases imply a similar role of lncRNAs guiding the onset of HS. It would be informative to uncover the functions of these lncRNAs in the context of HS.
Furthermore, as evidenced by IPA, changes in genes encoding lncRNAs in the context of HS suggest enrichment of pathways linked to carcinogenesis or cellular processes essential for upholding tissue homeostasis, such as cell death/survival and cell growth/proliferation.
5. Conclusions
LncRNAs are excellent targets to (a) further understand HS pathogenesis, (b) decipher the molecular mechanisms involved in HS-driven inflammation, and (c) train the development of new effective targeted therapies. A recent systematic review aiming to classify all recognized HS biomarkers evaluated the results of randomized clinical trials, uncontrolled clinical trials, cohort studies, case-control studies, and other observational studies published up until 31 December 2020 without exclusion criteria related to patient age, sex, race or ethnicity, or language. Among the 48 acknowledged biomarkers, one diagnostic biomarkers (serum IL-2R), one monitoring biomarkers (dermal Doppler vascularity), and two predictive biomarkers (epithelialized tunnels and positive family history of HS) were found with high GRADE ratings. However, none of them could be recommended for routine use in the clinical setting [67].
HS ensues after puberty; thus, hormones could play an important role in its pathogenesis. However, randomized controlled trials and experimental studies investigating the influence of hormones in HS are lacking. Currently, no relationship among deregulated lncRNAs and hormone secretion has been found. On the other hand, the role of sex hormones such as androgens and estrogens, adipokines, thyroid hormones, insulin resistance, and alteration of the insulin-driven immune–metabolic axis is under intense evaluation in HS pathophysiology [68]. As proposed in our study, blood-based biomarkers could provide a less invasive and more efficient tool for detecting skin diseases, and specifically HS, particularly in the early stages when treatment may be more effective. Although more research is needed in order to fully understand the role of lncRNAs in skin diseases and develop reliable biomarkers, the increasing interest and investment in this area suggests that progress is being made. Understanding the role played by lncRNAs will likely lead to novel mechanistic insights into HS pathogenesis, leading to new therapeutic options.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jcm13103016/s1, Table S1: Diseases and Functions associated to HS-related lncRNAs; Table S2: Networks associated to HS-related lncRNAs.
Author Contributions
Conceptualization, methodology, software, validation, formal analysis, investigation, resources, data curation, writing—original draft preparation: U.R. (Uppala Radhakrishna) and G.D.; writing—review and editing: U.R. (Uppala Ratnamala), G.D., G.B.E.J., D.D.J., M.P., L.V.U., T.M., G.M., A.V., N.S., S.R.S. and R.M.R.; visualization: U.R. (Uppala Ratnamala), G.D. and G.B.E.J.; supervision: U.R. (Uppala Radhakrishna) and D.D.J.; project administration and funding acquisition: U.R. (Uppala Radhakrishna). All authors have read and agreed to the published version of the manuscript.
Funding
The study did not receive external fundings.
Institutional Review Board Statement
This study was approved by the Institutional Review Board of Beaumont Health System, Royal Oak, MI, USA (HIC#: 2015-172, 21 May 2015).
Informed Consent Statement
All patients signed an informed consent form.
Data Availability Statement
The published article and its Supplementary Materials contain all the data generated during this study.
Acknowledgments
We thank the patients who voluntarily participated to our research study. We thank Vix Kennedy of the HS-USA (Michigan, USA) forum for financial support and encouragement for the study. Our thanks go to dermatologists Ranjanben Rawal, Timir Mehta, and DG Saple for support in evaluating HS patients.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Nguyen, T.V.; Damiani, G.; Orenstein, L.A.V.; Hamzavi, I.; Jemec, G.B. Hidradenitis suppurativa: An update on epidemiology, phenotypes, diagnosis, pathogenesis, comorbidities and quality of life. J. Eur. Acad. Dermatol. Venereol. 2021, 35, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Duchatelet, S.; Miskinyte, S.; Delage, M.; Ungeheuer, M.N.; Lam, T.; Benhadou, F.; Del Marmol, V.; Vossen, A.R.V.; Prens, E.P.; Cogrel, O.; et al. Low Prevalence of GSC Gene Mutations in a Large Cohort of Predominantly Caucasian Patients with Hidradenitis Suppurativa. J. Investig. Dermatol. 2020, 140, 2085–2088.e14. [Google Scholar] [CrossRef] [PubMed]
- Ratnamala, U.; Jain, N.K.; Jhala, D.D.; Prasad, P.V.S.; Saiyed, N.; Nair, S.; Radhakrishna, U. An Updated Mutation Spectrum of the gamma-Secretase Complex: Novel NCSTN Gene Mutation in an Indian Family with Hidradenitis Suppurativa and Acne Conglobata. Indian J. Dermatol. 2023, 68, 141–147. [Google Scholar] [PubMed]
- Radhakrishna, U.; Ratnamala, U.; Jhala, D.D.; Uppala, L.V.; Vedangi, A.; Saiyed, N.; Patel, M.; Vadsaria, N.; Shah, S.R.; Rawal, R.M.; et al. Hidradenitis suppurativa associated telomere-methylome dysregulations in blood. J. Eur. Acad. Dermatol. Venereol. 2023, 38, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishna, U.; Ratnamala, U.; Jhala, D.D.; Vadsaria, N.; Patel, M.; Uppala, L.V.; Vedangi, A.; Saiyed, N.; Rawal, R.M.; Damiani, G.; et al. Cytochrome P450 Genes Mediated by DNA Methylation Are Involved in the Resistance to Hidradenitis Suppurativa. J. Investig. Dermatol. 2023, 143, 670–673.e19. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishna, U.; Ratnamala, U.; Jhala, D.D.; Uppala, L.V.; Vedangi, A.; Patel, M.; Vadsaria, N.; Shah, S.; Saiyed, N.; Rawal, R.M.; et al. Hidradenitis suppurativa presents a methylome dysregulation capable to explain the pro-inflammatory microenvironment. Are these DNA methylations potential therapeutic targets? J. Eur. Acad. Dermatol. Venereol. 2023, 37, 2109–2123. [Google Scholar] [CrossRef] [PubMed]
- Frew, J.W.; Hawkes, J.E.; Krueger, J.G. A systematic review and critical evaluation of immunohistochemical associations in hidradenitis suppurativa. F1000Research 2018, 7, 1923. [Google Scholar] [CrossRef] [PubMed]
- Kozera, E.K.; Lowes, M.A.; Hsiao, J.L.; Frew, J.W. Clinical considerations in the management of hidradenitis suppurativa in women. Int. J. Womens Dermatol. 2021, 7, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Frew, J.W.; Marzano, A.V.; Wolk, K.; Join-Lambert, O.; Alavi, A.; Lowes, M.A.; Piguet, V. A Systematic Review of Promising Therapeutic Targets in Hidradenitis Suppurativa: A Critical Evaluation of Mechanistic and Clinical Relevance. J. Investig. Dermatol. 2021, 141, 316–324.e2. [Google Scholar] [CrossRef]
- Orenstein, L.A.V.; Nguyen, T.V.; Damiani, G.; Sayed, C.; Jemec, G.B.E.; Hamzavi, I. Medical and Surgical Management of Hidradenitis Suppurativa: A Review of International Treatment Guidelines and Implementation in General Dermatology Practice. Dermatology 2020, 236, 393–412. [Google Scholar] [CrossRef]
- Gao, N.; Li, Y.; Li, J.; Gao, Z.; Yang, Z.; Li, Y.; Liu, H.; Fan, T. Long Non-Coding RNAs: The Regulatory Mechanisms, Research Strategies, and Future Directions in Cancers. Front. Oncol. 2020, 10, 598817. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Niu, F.; Humburg, B.A.; Liao, K.; Bendi, S.; Callen, S.; Fox, H.S.; Buch, S. Molecular mechanisms of long noncoding RNAs and their role in disease pathogenesis. Oncotarget 2018, 9, 18648–18663. [Google Scholar] [CrossRef]
- De Martino, M.; Esposito, F.; Pallante, P. Long non-coding RNAs regulating multiple proliferative pathways in cancer cell. Transl. Cancer Res. 2021, 10, 3140–3157. [Google Scholar] [CrossRef] [PubMed]
- Morlando, M.; Fatica, A. Alteration of Epigenetic Regulation by Long Noncoding RNAs in Cancer. Int. J. Mol. Sci. 2018, 19, 570. [Google Scholar] [CrossRef] [PubMed]
- Cao, J. The functional role of long non-coding RNAs and epigenetics. Biol. Proced. Online 2014, 16, 11. [Google Scholar] [CrossRef]
- Doxtater, K.; Tripathi, M.K.; Khan, M.M. Recent advances on the role of long non-coding RNAs in Alzheimer’s disease. Neural Regen. Res. 2020, 15, 2253–2254. [Google Scholar] [PubMed]
- Shefler, A.; Patrick, M.T.; Wasikowski, R.; Chen, J.; Sarkar, M.K.; Gudjonsson, J.E.; Tsoi, L.C. Skin-Expressing lncRNAs in Inflammatory Responses. Front. Genet. 2022, 13, 835740. [Google Scholar] [CrossRef] [PubMed]
- Shibasaki, K. Physiological significance of TRPV2 as a mechanosensor, thermosensor and lipid sensor. J. Physiol. Sci. 2016, 66, 359–365. [Google Scholar] [CrossRef]
- Tang, L.; Liang, Y.; Xie, H.; Yang, X.; Zheng, G. Long non-coding RNAs in cutaneous biology and proliferative skin diseases: Advances and perspectives. Cell Prolif. 2020, 53, e12698. [Google Scholar] [CrossRef]
- Wilusz, J.E.; Sunwoo, H.; Spector, D.L. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009, 23, 1494–1504. [Google Scholar] [CrossRef] [PubMed]
- Cazzaniga, S.; Naldi, L.; Damiani, G.; Atzori, L.; Patta, F.; Guidarelli, G.; Bettoli, V. Validation of a visual-aided questionnaire for the self-assessment of hidradenitits suppurativa. J. Eur. Acad. Dermatol. Venereol. 2018, 32, 1993–1998. [Google Scholar] [CrossRef] [PubMed]
- Lipsker, D.; Severac, F.; Freysz, M.; Sauleau, E.; Boer, J.; Emtestam, L.; Matusiak, Ł.; Prens, E.; Velter, C.; Lenormand, C.; et al. The ABC of Hidradenitis Suppurativa: A Validated Glossary on how to Name Lesions. Dermatology 2016, 232, 137–142. [Google Scholar] [CrossRef]
- Hurley, H.J. Axillary hyperhidrosis, apocrine bromhidrosis, hidradenitis suppurativa, and familial benign pemphigus: Surgical approach. In Roenigk and Roenigk’s Dermatologic Surgery: Principles and Practice, 2nd ed.; Roenigk, R.K., Roenigk, H.H., Jr., Eds.; Marcel Dekker: New York, NY, USA, 1996; pp. 623–645. [Google Scholar]
- Radhakrishna, U.; Ratnamala, U.; Jhala, D.D.; Vadsaria, N.; Patel, M.; Uppala, L.V.; Vishweswaraiah, S.; Vedangi, A.; Saiyed, N.; Damiani, G.; et al. Methylated miRNAs may serve as potential biomarkers and therapeutic targets for hidradenitis suppurativa. J. Eur. Acad. Dermatol. Venereol. 2022, 36, 2199–2213. [Google Scholar] [CrossRef] [PubMed]
- Damiani, G.; Della Valle, V.; Iannone, M.; Dini, V.; Marzano, A.V. Autoinflammatory Disease Damage Index (ADDI): A possible newborn also in hidradenitis suppurativa daily practice. Ann. Rheum. Dis. 2017, 76, e25. [Google Scholar] [CrossRef]
- van der Zee, H.H.; Jemec, G.B. New insights into the diagnosis of hidradenitis suppurativa: Clinical presentations and phenotypes. J. Am. Acad. Dermatol. 2015, 73, S23–S26. [Google Scholar] [CrossRef]
- Damiani, G.; Mahroum, N.; Pigatto, P.D.M.; Pacifico, A.; Malagoli, P.; Tiodorovic, D.; Conic, R.; Amital, H.; Bragazzi, N.L.; Watad, A.; et al. The Safety and Impact of a Model of Intermittent, Time-Restricted Circadian Fasting (“Ramadan Fasting”) on Hidradenitis Suppurativa: Insights from a Multicenter, Observational, Cross-Over, Pilot, Exploratory Study. Nutrients 2019, 11, 1781. [Google Scholar] [CrossRef]
- Gu, Z. ComplexHeatmap: Making Complex Heatmaps. R Package Version 1.6.0. 2015. Available online: https://githubcom/jokergoo/ComplexHeatmap (accessed on 2 April 2024).
- Herter, E.K.; Xu Landen, N. Non-Coding RNAs: New Players in Skin Wound Healing. Adv. Wound Care 2017, 6, 93–107. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.J.; Yao, X.D. Function of PCA3 in prostate tissue and clinical research progress on developing a PCA3 score. Chin. J. Cancer Res. 2014, 26, 493–500. [Google Scholar]
- Lemos, A.E.G.; Matos, A.D.R.; Ferreira, L.B.; Gimba, E.R.P. The long non-coding RNA PCA3: An update of its functions and clinical applications as a biomarker in prostate cancer. Oncotarget 2019, 10, 6589–6603. [Google Scholar] [CrossRef]
- Davey, R.A.; Grossmann, M. Androgen Receptor Structure, Function and Biology: From Bench to Bedside. Clin. Biochem. Rev. 2016, 37, 3–15. [Google Scholar] [PubMed]
- de Wit, N.J.; Weidle, U.H.; Ruiter, D.J.; van Muijen, G.N. Expression profiling of MMA-1a and splice variant MMA-1b: New cancer/testis antigens identified in human melanoma. Int. J. Cancer 2002, 98, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.M.; Lee, K.H.; Kim, Y.J.; Chang, S.E.; Lee, M.W.; Choi, J.H.; Won, C.H.; Lee, W.J. Assessment of Overall and Specific Cancer Risks in Patients With Hidradenitis Suppurativa. JAMA Dermatol. 2020, 156, 844–853. [Google Scholar] [CrossRef] [PubMed]
- Denny, G.; Anadkat, M.J. Hidradenitis suppurativa (HS) and Down syndrome (DS): Increased prevalence and a younger age of hidradenitis symptom onset. J. Am. Acad. Dermatol. 2016, 75, 632–634. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.; Strunk, A.; Midura, M.; Papagermanos, V.; Pomerantz, H. Prevalence of hidradenitis suppurativa among patients with Down syndrome: A population-based cross-sectional analysis. Br. J. Dermatol. 2018, 178, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Poizeau, F.; Sbidian, E.; Mircher, C.; Rebillat, A.S.; Chosidow, O.; Wolkenstein, P.; Ravel, A.; Hotz, C. Prevalence and Description of Hidradenitis Suppurativa in Down Syndrome: A Cross-sectional Study of 783 Subjects. Acta Derm. Venereol. 2019, 99, 351–352. [Google Scholar] [CrossRef] [PubMed]
- Kaakati, R.N.; Tanaka, J.; Liu, B.; Ward, R.; Macleod, A.S.; Green, C.L.; Jaleel, T. Atopic dermatitis is associated with hidradenitis suppurativa diagnosis: A single institution retrospective cohort study. JAAD Int. 2021, 4, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Nousbeck, J.; McAleer, M.A.; Irvine, A.D. Peripheral Blood Gene Expression Profile of Infants with Atopic Dermatitis. JID Innov. 2023, 3, 100165. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kay, M.K.; Park, M.H.; Meruvu, S.; Powell, C.; Choudhury, M. LncRNA DLEU2 regulates sirtuins and mitochondrial respiratory chain complex IV: A novel pathway in obesity and offspring’s health. Int. J. Obes. 2022, 46, 969–976. [Google Scholar] [CrossRef]
- Kaminsky, Z.; Tochigi, M.; Jia, P.; Pal, M.; Mill, J.; Kwan, A.; Ioshikhes, I.; Vincent, J.B.; Kennedy, J.L.; Strauss, J.; et al. A multi-tissue analysis identifies HLA complex group 9 gene methylation differences in bipolar disorder. Mol. Psychiatry 2012, 17, 728–740. [Google Scholar] [CrossRef]
- Tzur Bitan, D.; Berzin, D.; Cohen, A. Hidradenitis Suppurativa and Bipolar Disorders: A Population-Based Study. Dermatology 2020, 236, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zheng, H.; Tse, G.; Zhang, L.; Wu, W.K.K. CASC2: An emerging tumour-suppressing long noncoding RNA in human cancers and melanoma. Cell Prolif. 2018, 51, e12506. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qian, W.; Feng, F.; Cao, Q.; Li, Y.; Hou, Y.; Zhang, L.; Fan, J. Upregulated lncRNA CASC2 May Inhibit Malignant Melanoma Development Through Regulating miR-18a-5p/RUNX1. Oncol. Res. 2019, 27, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Takemoto, A.; Tanimoto, K.; Mori, S.; Inoue, J.; Fujiwara, N.; Noda, T.; Inazawa, J. Integrative genome-wide analyses reveal the transcriptional aberrations in Japanese esophageal squamous cell carcinoma. Cancer Sci. 2021, 112, 4377–4392. [Google Scholar] [CrossRef]
- Liu, S.; Chen, X.; Huang, K.; Xiong, X.; Shi, Y.; Wang, X.; Pan, X.; Cong, Y.; Sun, Y.; Ge, L.; et al. Long noncoding RNA RFPL1S-202 inhibits ovarian cancer progression by downregulating the IFN-beta/STAT1 signaling. Exp. Cell Res. 2023, 422, 113438. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Qin, D.; Jiang, Z.; Zhang, J. SNHG9/miR-199a-5p/Wnt2 Axis Regulates Cell Growth and Aerobic Glycolysis in Glioblastoma. J. Neuropathol. Exp. Neurol. 2019, 78, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, C.; Sun, Z. Long non-coding RNA LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 are novel prognostic markers for pancreatic cancer. Am. J. Transl. Res. 2018, 10, 2648–2658. [Google Scholar]
- Wang, R.; Chen, C.; Kang, W.; Meng, G. SNHG9 was upregulated in NSCLC and associated with DDP-resistance and poor prognosis of NSCLC patients. Am. J. Transl. Res. 2020, 12, 4456–4466. [Google Scholar] [PubMed]
- Nakabayashi, K.; Bentley, L.; Hitchins, M.P.; Mitsuya, K.; Meguro, M.; Minagawa, S.; Bamforth, J.S.; Stanier, P.; Preece, M.; Weksberg, R.; et al. Identification and characterization of an imprinted antisense RNA (MESTIT1) in the human MEST locus on chromosome 7q32. Hum. Mol. Genet. 2002, 11, 1743–1756. [Google Scholar] [CrossRef]
- Meyer, E.; Wollmann, H.A.; Eggermann, T. Searching for genomic variants in the MESTIT1 transcript in Silver-Russell syndrome patients. J. Med. Genet. 2003, 40, e65. [Google Scholar] [CrossRef]
- Linh, N.T.T.; Giang, N.H.; Lien, N.T.K.; Trang, B.K.; Trang, D.T.; Ngoc, N.T.; Nghia, V.X.; My, L.T.; Van Mao, C.; Hoang, N.H.; et al. Association of PSORS1C3, CARD14 and TLR4 genotypes and haplotypes with psoriasis susceptibility. Genet. Mol. Biol. 2022, 45, e20220099. [Google Scholar] [CrossRef] [PubMed]
- Gau, S.Y.; Preclaro, I.A.C.; Wei, J.C.; Lee, C.Y.; Kuan, Y.H.; Hsiao, Y.P.; Juang, S.-E.; Ma, K.S.-K. Risk of psoriasis in people with hidradenitis suppurativa: A systematic review and meta-analysis. Front. Immunol. 2022, 13, 1033844. [Google Scholar] [CrossRef] [PubMed]
- Onagoruwa, O.T.; Pal, G.; Ochu, C.; Ogunwobi, O.O. Oncogenic Role of PVT1 and Therapeutic Implications. Front. Oncol. 2020, 10, 17. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wei, M.; Jiang, X.; Tan, J.; Xu, W.; Fan, X.; Zhang, R.; Ding, C.; Zhao, F.; Shao, X.; et al. lncRNA PVT1 Promotes Tumorigenesis of Colorectal Cancer by Stabilizing miR-16-5p and Interacting with the VEGFA/VEGFR1/AKT Axis. Mol. Ther. Nucleic Acids 2020, 20, 438–450. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.; Qi, Y.; Dong, C.; Yang, C. PVT1 regulates inflammation and cardiac function via the MAPK/NF-kappaB pathway in a sepsis model. Exp. Ther. Med. 2018, 16, 4471–4478. [Google Scholar] [PubMed]
- Tohkin, M.; Kaniwa, N.; Saito, Y.; Sugiyama, E.; Kurose, K.; Nishikawa, J.; Hasegawa, R.; Aihara, M.; Matsunaga, K.; Abe, M.; et al. A whole-genome association study of major determinants for allopurinol-related Stevens-Johnson syndrome and toxic epidermal necrolysis in Japanese patients. Pharmacogenom. J. 2013, 13, 60–69. [Google Scholar] [CrossRef]
- Stern, R.S.; Divito, S.J. Stevens-Johnson Syndrome and Toxic Epidermal Necrolysis: Associations, Outcomes, and Pathobiology-Thirty Years of Progress but Still Much to Be Done. J. Investig. Dermatol. 2017, 137, 1004–1008. [Google Scholar] [CrossRef]
- Morse, D.C.; Chockalingam, R.; Pye, A.; Huen, A. Hidradenitis suppurativa associated with sorafenib initiation. Dermatol. Online J. 2019, 25, AB219. [Google Scholar] [CrossRef]
- Meeks, K.A.C.; Henneman, P.; Venema, A.; Burr, T.; Galbete, C.; Danquah, I.; Schulze, M.B.; Mockenhaupt, F.P.; Owusu-Dabo, E.; Rotimi, C.N.; et al. An epigenome-wide association study in whole blood of measures of adiposity among Ghanaians: The RODAM study. Clin. Epigenet. 2017, 9, 103. [Google Scholar] [CrossRef]
- Fimmel, S.; Zouboulis, C.C. Comorbidities of hidradenitis suppurativa (acne inversa). Dermatoendocrinol 2010, 2, 9–16. [Google Scholar] [CrossRef]
- Zouboulis, C.C.; Desai, N.; Emtestam, L.; Hunger, R.E.; Ioannides, D.; Juhasz, I.; Lapins, J.; Matusiak, L.; Prens, E.P.; Revuz, J.; et al. European S1 guideline for the treatment of hidradenitis suppurativa/acne inversa. J. Eur. Acad. Dermatol. Venereol. 2015, 29, 619–644. [Google Scholar] [CrossRef] [PubMed]
- Zouboulis, V.A.; Zouboulis, K.C.; Zouboulis, C.C. Hidradenitis Suppurativa and Comorbid Disorder Biomarkers, Druggable Genes, New Drugs and Drug Repurposing-A Molecular Meta-Analysis. Pharmaceutics 2021, 14, 44. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Broadaway, K.A.; Edmiston, S.N.; Fajgenbaum, K.; Miller-Fleming, T.; Westerkam, L.L.; Melendez-Gonzalez, M.; Bui, H.; Blum, F.R.; Levitt, B.; et al. Genetic Variants Associated With Hidradenitis Suppurativa. JAMA Dermatol. 2023, 159, 930–938. [Google Scholar] [CrossRef] [PubMed]
- Mintoff, D.; Pace, N.P.; Borg, I. NCSTN In-Frame Deletion in Maltese Patients with Hidradenitis Suppurativa. JAMA Dermatol. 2023, 159, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Der Sarkissian, S.; Hessam, S.; Kirby, J.S.; Lowes, M.A.; Mintoff, D.; Naik, H.B.; Ring, H.C.; Chandran, N.S.; Frew, J.W. Identification of Biomarkers and Critical Evaluation of Biomarker Validation in Hidradenitis Suppurativa: A Systematic Review. JAMA Dermatol. 2022, 158, 300–313. [Google Scholar] [CrossRef]
- Abu Rached, N.; Gambichler, T.; Dietrich, J.W.; Ocker, L.; Seifert, C.; Stockfleth, E.; Bechara, F.G. The Role of Hormones in Hidradenitis Suppurativa: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 15250. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).