Recent Insights in Noninvasive Diagnostic for the Assessment of Kidney and Cardiovascular Outcome in Kidney Transplant Recipients
Abstract
1. Introduction
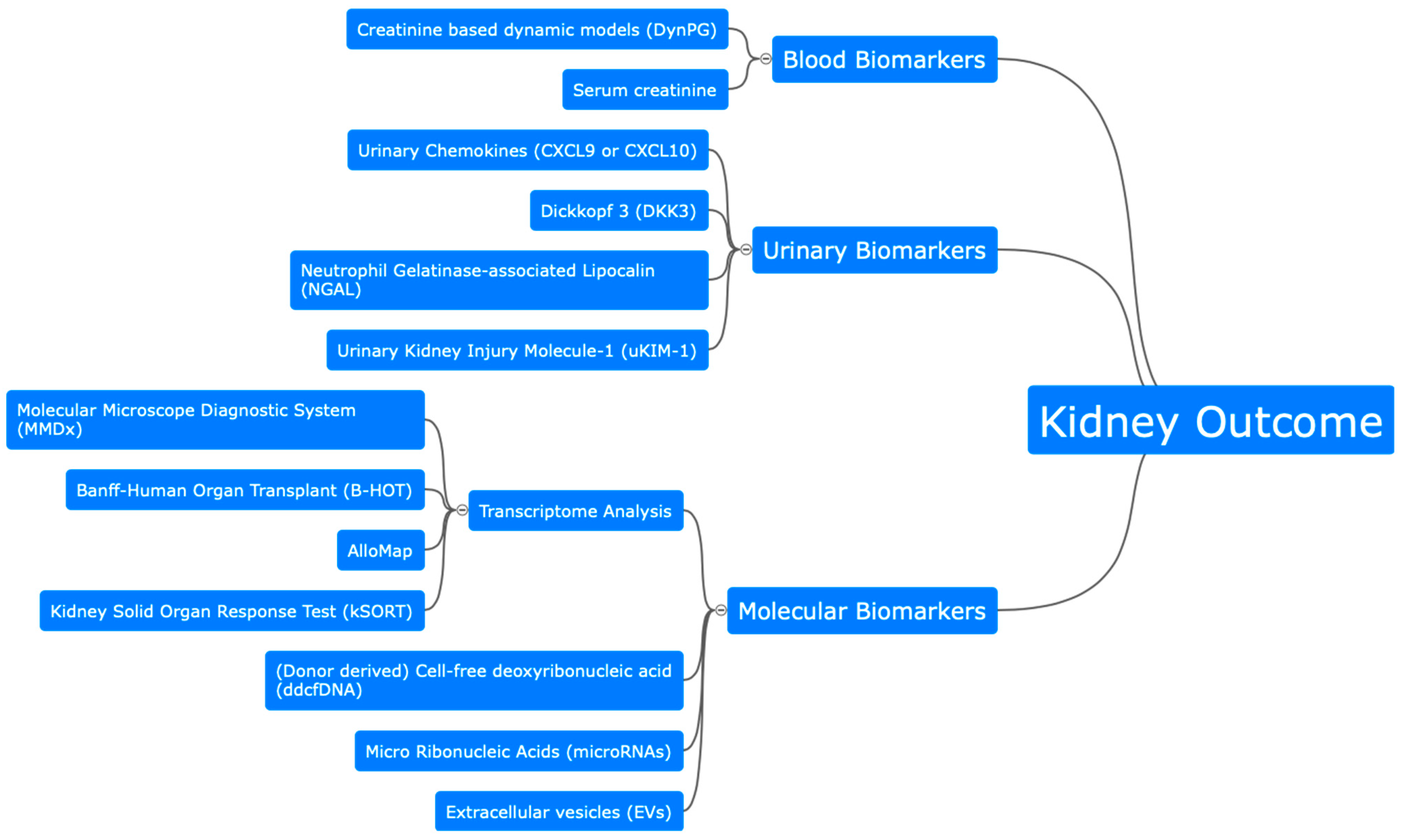
2. Assessment of Kidney Outcome after Kidney Transplantation
2.1. Biomarker for Glomerular and Tubular Function
2.2. Molecular Biomarkers
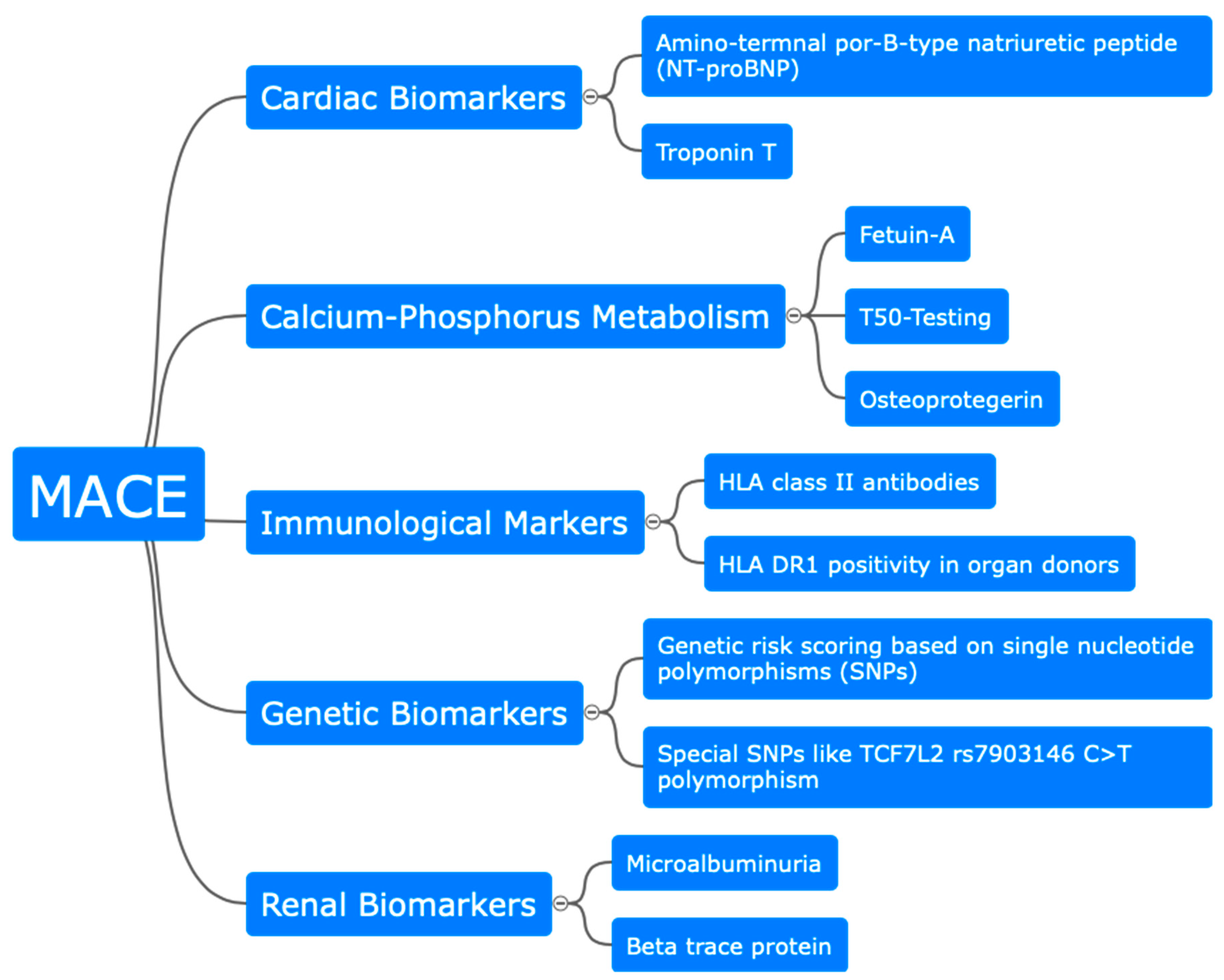
3. Biomarkers for Cardiovascular Risk in Kidney Transplant Recipients
3.1. Serum, Immunological, and Metabolic Biomarker
3.2. Insights from Genetic Analysis
4. Conclusions
Funding
Conflicts of Interest
References
- Loupy, A.; Aubert, O.; Orandi, B.J.; Naesens, M.; Bouatou, Y.; Raynaud, M.; Divard, G.; Jackson, A.M.; Viglietti, D.; Giral, M.; et al. Prediction system for risk of allograft loss in patients receiving kidney transplants: International derivation and validation study. BMJ 2019, 366, l4923. [Google Scholar] [CrossRef] [PubMed]
- Huang, E.; Mengel, M.; Clahsen-Van Groningen, M.C.; Jackson, A.M. Diagnostic Potential of Minimally Invasive Biomarkers: A Biopsy-centered Viewpoint From the Banff Minimally Invasive Diagnostics Working Group. Transplantation 2023, 107, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Loupy, A.; Haas, M.; Roufosse, C.; Naesens, M.; Adam, B.; Afrouzian, M.; Akalin, E.; Alachkar, N.; Bagnasco, S. The Banff 2019 Kidney Meeting Report (I): Updates on and clarification of criteria for T cell- and antibody-mediated rejection. Am. J. Transpl. 2020, 20, 2318–2331. [Google Scholar] [CrossRef]
- Loupy, A.; Mengel, M.; Haas, M. Thirty years of the International Banff Classification for Allograft Pathology: The past, present, and future of kidney transplant diagnostics. Kidney Int. 2022, 101, 678–691. [Google Scholar] [CrossRef] [PubMed]
- Lentine, K.L.; Brennan, D.C.; Schnitzler, M.A. Incidence and predictors of myocardial infarction after kidney transplantation. J. Am. Soc. Nephrol. 2005, 16, 496–506. [Google Scholar] [CrossRef] [PubMed]
- Lenain, R.M.; Dantan, E.; Giral, M.; Foucher, Y.; Asar, Ö.; Naesens, M.; Hazzan, M.; Fournier, M.C. External Validation of the DynPG for Kidney Transplant Recipients. Transplantation 2021, 105, 396–403. [Google Scholar] [CrossRef]
- Ostermann, M.; Zarbock, A.; Goldstein, S.; Kashani, K.; MacEdo, E.; Murugan, R.; Bell, M.; Forni, L.; Guzzi, L.; Joannidis, M.; et al. Recommendations on Acute Kidney Injury Biomarkers From the Acute Disease Quality Initiative Consensus Conference: A Consensus Statement. JAMA Netw. Open 2020, 3, e2019209. [Google Scholar] [CrossRef]
- Schuster, A.; Steines, L.; Müller, K.; Zeman, F.; Findeisen, P.; Banas, B.; Bergler, T. Dickkopf 3-A New Indicator for the Deterioration of Allograft Function After Kidney Transplantation. Front. Med. 2022, 9, 885018. [Google Scholar] [CrossRef]
- Rabant, M.; Amrouche, L.; Morin, L.; Bonifay, R.; Lebreton, X.; Aouni, L.; Benon, A.; Sauvaget, V.; Le Vaillant, L.; Aulagnon, F.; et al. Early Low Urinary CXCL9 and CXCL10 Might Predict Immunological Quiescence in Clinically and Histologically Stable Kidney Recipients. Am. J. Transplant. 2016, 16, 1868–1881. [Google Scholar] [CrossRef]
- Millán, O.; Budde, K.; Sommerer, C.; Aliart, I.; Rissling, O.; Bardaji, B.; Matz, M.; Zeier, M.; Silva, I.; Guirado, L. Urinary miR-155-5p and CXCL10 as prognostic and predictive biomarkers of rejection, graft outcome and treatment response in kidney transplantation. Br. J. Clin. Pharmacol. 2017, 83, 2636–2650. [Google Scholar] [CrossRef]
- Jackson, J.A.; Kim, E.J.; Begley, B.; Cheeseman, J.; Harden, T.; Perez, S.D.; Thomas, S.; Warshaw, B.; Kirk, A.D. Urinary chemokines CXCL9 and CXCL10 are noninvasive markers of renal allograft rejection and BK viral infection. Am. J. Transplant. 2011, 11, 2228–2234. [Google Scholar] [CrossRef] [PubMed]
- Lazzeri, E.; Rotondi, M.; Mazzinghi, B.; Lasagni, L.; Buonamano, A.; Rosati, A.; Pradella, F.; Fossombroni, V.; La Villa, G.; Gacci, M.; et al. High CXCL10 expression in rejected kidneys and predictive role of pretransplant serum CXCL10 for acute rejection and chronic allograft nephropathy. Transplantation 2005, 79, 1215–1220. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.M.; Li, Y.; Yan, L.; Wang, H.; Wu, X.J.; Tang, J.T.; Wang, L.L.; Shi, Y.Y. Comparison of urine and blood NGAL for early prediction of delayed graft function in adult kidney transplant recipients: A meta-analysis of observational studies. BMC Nephrol. 2019, 20, 291. [Google Scholar] [CrossRef]
- Kielar, M.; Dumnicka, P.; Gala-Błądzińska, A.; Będkowska-Prokop, A.; Ignacak, E.; Maziarz, B.; Ceranowicz, P.; Kusnierz-Cabala, B. Urinary NGAL Measured after the First Year Post Kidney Transplantation Predicts Changes in Glomerular Filtration over One-Year Follow-Up. J. Clin. Med. 2020, 10, 43. [Google Scholar] [CrossRef]
- Zhu, M.; Chen, Z.; Wei, Y.; Yuan, Y.; Ying, L.; Zhou, H.; Che, X.; Zhang, M.F.; Ni, Z.; Zhang, M.; et al. The predictive value of urinary kidney injury molecular-1 for long-term graft function in kidney transplant patients: A prospective study. Ann. Transl. Med. 2021, 9, 244. [Google Scholar] [CrossRef] [PubMed]
- Schröppel, B.; Krüger, B.; Walsh, L.; Yeung, M.; Harris, S.; Garrison, K.; Himmelfarb, J.; Lerner, S.M.; Bromberg, J.S.; Zhang, P.L.; et al. Tubular expression of KIM-1 does not predict delayed function after transplantation. J. Am. Soc. Nephrol. 2010, 21, 536–542. [Google Scholar] [CrossRef]
- Halloran, P.F.; Pereira, A.B.; Chang, J.; Matas, A.; Picton, M.; De Freitas, D.; Bromberg, J.; Seron, D.; Sellares, J.; Einecke, G.; et al. Potential impact of microarray diagnosis of T cell-mediated rejection in kidney transplants: The INTERCOM study. Am. J. Transplant. 2013, 13, 2352–2363. [Google Scholar] [CrossRef]
- Mengel, M.; Loupy, A.; Haas, M.; Roufosse, C.; Naesens, M.; Akalin, E.; Clahsen-van Groningen, M.; Dagobert, J.; Demetris, A.J.; Duong van Huyen, J.P.; et al. Banff 2019 Meeting Report: Molecular diagnostics in solid organ transplantation-Consensus for the Banff Human Organ Transplant (B-HOT) gene panel and open source multicenter validation. Am. J. Transpl. 2020, 20, 2305–2317. [Google Scholar] [CrossRef]
- Deng, M.C.; Eisen, H.J.; Mehra, M.R.; Billingham, M.; Marboe, C.C.; Berry, G.; Kobashigawa, J.; Johnson, F.L.; Starling, R.C.; Murali, S.; et al. Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling. Am. J. Transplant. 2006, 6, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, E.; Gazut, S.; Yazdani, S.; Lerut, E.; de Loor, H.; Coemans, M.; Noel, L.H.; Thorrez, L.; van Lommel, L.; Schuit, F. Development and validation of a peripheral blood mRNA assay for the assessment of antibody-mediated kidney allograft rejection: A multicentre, prospective study. EBioMedicine 2019, 46, 463–472. [Google Scholar] [CrossRef]
- Kurian, S.M.; Williams, A.N.; Gelbart, T.; Campbell, D.; Mondala, T.S.; Head, S.R.; Horvath, S.; Gaber, L.; Thompson, R.; Whisenant, T.; et al. Molecular classifiers for acute kidney transplant rejection in peripheral blood by whole genome gene expression profiling. Am. J. Transplant. 2014, 14, 1164–1172. [Google Scholar] [CrossRef]
- Roedder, S.; Sigdel, T.; Salomonis, N.; Hsieh, S.; Dai, H.; Bestard, O.; Metes, D.; Zeevi, A.; Gritsch, A.; Cheeseman, J.; et al. The kSORT Assay to Detect Renal Transplant Patients at High Risk for Acute Rejection: Results of the Multicenter AART Study. PLoS Med. 2014, 11, e1001759. [Google Scholar] [CrossRef] [PubMed]
- Bronkhorst, A.J.; Ungerer, V.; Holdenrieder, S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol. Detect. Quantif. 2019, 17, 100087. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.-K.; Lo, Y.D. Diagnostic developments involving cell-free (circulating) nucleic acids. Clin. Chim. Acta 2006, 363, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Martuszewski, A.; Paluszkiewicz, P.; Król, M.; Banasik, M.; Kepinska, M. Donor-Derived Cell-Free DNA in Kidney Transplantation as a Potential Rejection Biomarker: A Systematic Literature Review. J. Clin. Med. 2021, 10, 193. [Google Scholar] [CrossRef]
- Xiao, H.; Gao, F.; Pang, Q.; Xia, Q.; Zeng, X.; Peng, J.; Fan, L.; Liu, J.; Wang, Z.; Li, H. Diagnostic Accuracy of Donor-derived Cell-free DNA in Renal-allograft Rejection: A Meta-analysis. Transplantation 2021, 105, 1303–1310. [Google Scholar] [CrossRef]
- Aubert, O.; Ursule-Dufait, C.; Brousse, R.; Gueguen, J.; Racapé, M.; Raynaud, M.; van Loon, E.; Pagliazzi, A.; Huang, E. Cell-Free DNA for the detection of kidney allograft rejection. Nat. Med. 2024, 2024, 1. [Google Scholar] [CrossRef]
- Amrouche, L.; Desbuissons, G.; Rabant, M.; Sauvaget, V.; Nguyen, C.; Benon, A.; Barre, P.; Rabate, C.; Lebreton, X.; Gallazzini, M.; et al. MicroRNA-146a in Human and Experimental Ischemic AKI: CXCL8-Dependent Mechanism of Action. J. Am. Soc. Nephrol. 2017, 28, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Soltaninejad, E.; Nicknam, M.H.; Nafar, M.; Ahmadpoor, P.; Pourrezagholi, F.; Sharbafi, M.H. Differential expression of microRNAs in renal transplant patients with acute T-cell mediated rejection. Transpl. Immunol. 2015, 33, 1–6. [Google Scholar] [CrossRef]
- Betts, G.; Shankar, S.; Sherston, S.; Friend, P.; Wood, K.J. Examination of serum miRNA levels in kidney transplant recipients with acute rejection. Transplantation 2014, 97, e28–e30. [Google Scholar] [CrossRef]
- Tao, J.; Yang, X.; Han, Z.; Lu, P.; Wang, J.; Liu, X.; Wu, B.; Wang, Z.; Huang, Z.; Lu, Q.; et al. Serum MicroRNA-99a Helps Detect Acute Rejection in Renal Transplantation. Transplant. Proc. 2015, 47, 1683–1687. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, K.; Yamamoto, T.; Inanaga, Y.; Hiramitsu, T.; Miwa, Y.; Murotani, K.; Narumi, S.; Watarai, Y.; Katayama, A.; Uchida, K.; et al. MiR-142-5p and miR-486-5p as biomarkers for early detection of chronic antibody-mediated rejection in kidney transplantation. Biomarkers 2017, 22, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-J.; Wang, C. A review of the regulatory mechanisms of extracellular vesicles-mediated intercellular communication. Cell Commun. Signal 2023, 21, 77. [Google Scholar] [CrossRef] [PubMed]
- Buzas, E.I. The roles of extracellular vesicles in the immune system. Nat. Rev. Immunol. 2023, 23, 236–250. [Google Scholar] [CrossRef] [PubMed]
- Erdbrügger, U.; Blijdorp, C.J.; Bijnsdorp, I.V.; Borràs, F.E.; Burger, D.; Bussolati, B.; Byrd, J.B.; Clayton, A.; Dear, J.W.; Falcon-Perez, J.M.; et al. Urinary extracellular vesicles: A position paper by the Urine Task Force of the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2021, 10, e12093. [Google Scholar] [CrossRef]
- Ashcroft, J.; Leighton, P.; Elliott, T.R.; Hosgood, S.A.; Nicholson, M.L.; Kosmoliaptsis, V. Extracellular vesicles in kidney transplantation: A state-of-the-art review. Kidney Int. 2022, 101, 485–497. [Google Scholar] [CrossRef]
- Park, J.; Lin, H.-Y.; Assaker, J.P.; Jeong, S.; Huang, C.-H.; Kurdi, A.; Lee, K.; Fraser, K.; Min, C.; Eskandari, S.; et al. Integrated Kidney Exosome Analysis for the Detection of Kidney Transplant Rejection. ACS Nano 2017, 11, 11041–11046. [Google Scholar] [CrossRef]
- Woud, W.W.M.; Arykbaeva, A.S.B.; Alwayn, I.P.; Baan, C.C.; Minnee, R.C.; Hoogduijn, M.J.; Boer, K. Extracellular Vesicles Released During Normothermic Machine Perfusion Are Associated with Human Donor Kidney Characteristics. Transplantation 2022, 106, 2360–2369. [Google Scholar] [CrossRef] [PubMed]
- El Fekih, R.; Hurley, J.; Tadigotla, V.; Alghamdi, A.; Srivastava, A.; Coticchia, C.; Choi, J.; Allos, H.; Yatim, K.; Alhaddad, J.; et al. Discovery and Validation of a Urinary Exosome mRNA Signature for the Diagnosis of Human Kidney Transplant Rejection. J. Am. Soc. Nephrol. 2021, 32, 994–1004. [Google Scholar] [CrossRef]
- Mann, J.F.E.; Yi, Q.-L.; Gerstein, H.C. Albuminuria as a predictor of cardiovascular and renal outcomes in people with known atherosclerotic cardiovascular disease. Kidney Int. 2004, 66, S59–S62. [Google Scholar] [CrossRef]
- Halimi, J.M. Low-grade proteinuria and microalbuminuria in renal transplantation. Transplantation. 2013, 96, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Halimi, J.M.; Buchler, M.; Al-Najjar, A.; Laouad, I.; Chatelet, V.; Marlière, J.F.; Nivet, H.; Lebranchu, Y. Urinary albumin excretion and the risk of graft loss and death in proteinuric and non-proteinuric renal transplant recipients. Am. J. Transplant. 2007, 7, 618–625. [Google Scholar] [CrossRef] [PubMed]
- Bielopolski, D.; Rahamimov, R.; Zingerman, B.; Chagnac, A.; Azulay-Gitter, L.; Zvi, B.R. Microalbuminuria After Kidney Transplantation Predicts Cardiovascular Morbidity. Front. Med. 2021, 8, 635847. [Google Scholar] [CrossRef] [PubMed]
- Gerhardt, T.; Pöge, U.; Stoffel-Wagner, B.; Klein, B.; Klehr, H.-U.; Sauerbruch, T.; Woitas, R.P. Serum levels of beta-trace protein and its association to diuresis in haemodialysis patients. Nephrol. Dial. Transplant. 2008, 23, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Schwab, S.; Kleine, C.E.; Bös, D.; Bohmann, S.; Strassburg, C.P.; Lutz, P.; Woitas, R.P. Beta-trace protein as a potential biomarker of residual renal function in patients undergoing peritoneal dialysis. BMC Nephrol. 2021, 22, 87. [Google Scholar] [CrossRef] [PubMed]
- Schwab, S.; Pörner, D.; Kleine, C.; Werberich, R.; Werberich, L.; Reinhard, S.; Bös, D.; Strassburg, C.P.; von Vietinghoff, S.; Lutz, P. Pre-transplant serum Beta Trace Protein indicates risk for post-transplant major cardiac adverse events. Nephrology 2023, 28, 51–59. [Google Scholar] [CrossRef]
- Rørth, R.; Jhund, P.S.; Yilmaz, M.B.; Kristensen, S.L.; Welsh, P.; Desai, A.S.; Kober, L.; Prescott, M.F.; Rouleau, J.L.; Solomon, S.D.; et al. Comparison of BNP and NT-proBNP in Patients With Heart Failure and Reduced Ejection Fraction. Circ. Heart Fail. 2020, 13, E006541. [Google Scholar] [CrossRef] [PubMed]
- Mckie, P.M.; Burnett, J.C. NT-proBNP The Gold Standard Biomarker in Heart Failure. J. Am. Coll. Cardiol. 2016, 68, 2437–2439. [Google Scholar] [CrossRef]
- Pfister, R.; Scholz, M.; Wielckens, K.; Erdmann, E.; Schneider, C.A. Use of NT-proBNP in routine testing and comparison to BNP. Eur. J. Heart Fail. 2004, 6, 289–293. [Google Scholar] [CrossRef]
- Schwab, S.; Pörner, D.; Kleine, C.E.; Werberich, R.; Werberich, L.; Reinhard, S.; Bös, D.; Strassburg, C.P.; von Vietinghoff, S.; Lutz, P.; et al. NT-proBNP as predictor of major cardiac events after renal transplantation in patients with preserved left ventricular ejection fraction. BMC Nephrol. 2023, 24, 32. [Google Scholar] [CrossRef]
- Emrich, I.E.; Scheuer, A.L.; Rogacev, K.S.; Mahfoud, F.; Wagenpfeil, S.; Fliser, D.; Schirmer, S.H.; Böhm, M.; Heine, G.H. Plasma biomarkers outperform echocardiographic measurements for cardiovascular risk prediction in kidney transplant recipients: Results of the HOME ALONE study. Clin. Kidney, J. 2022, 15, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Connolly, G.M.; Cunningham, R.; McNamee, P.T.; Young, I.S.; Maxwell, A.P. Troponin T is an independent predictor of mortality in renal transplant recipients. Nephrol. Dial. Transplant. 2008, 23, 1019–1025. [Google Scholar] [CrossRef] [PubMed]
- Hansson, G.K. Inflammation, Atherosclerosis, and Coronary Artery Disease. N. Engl. J. Med. 2005, 352, 1685–1695. [Google Scholar] [CrossRef]
- Abedini, S.; Holme, I.; Marz, W. Inflammation in renal transplantation. Clin. J. Am. Soc. Nephrol. 2009, 4, 1246–1254. [Google Scholar] [CrossRef]
- Emerging Risk Factors Collaboration; Kaptoge, S.; Di Angelantonio, E.; Lowe, G.; Pepys, M.B.; Thompson, S.G.; Collins, R.; Danesh, J. C-reactive protein concentration and risk of coronary heart disease, stroke, and mortality: An individual participant meta-analysis. Lancet 2010, 375, 132–140. [Google Scholar]
- Ponticelli, C.; Campise, M.R. The inflammatory state is a risk factor for cardiovascular disease and graft fibrosis in kidney transplantation. Kidney Int. 2021, 100, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Hansson, G.K.; Robertson, A.K.L.; Söderberg-Nauclér, C. Inflammation and atherosclerosis. Annu. Rev. Pathol. 2006, 1, 297–329. [Google Scholar] [CrossRef] [PubMed]
- Söderlund, J.; Forsblom, C.; Ilonen, J.; Thorn, L.M.; Wadén, J.; Parkkonen, M.; Groop, P.H. HLA class II is a factor in cardiovascular morbidity and mortality rates in patients with type 1 diabetes. Diabetologia 2012, 55, 2963–2969. [Google Scholar] [CrossRef]
- Malfait, T.; Emonds, M.-P.; Daniëls, L.; Nagler, E.V.; Van Biesen, W.; Van Laecke, S. HLA Class II Antibodies at the Time of Kidney Transplantation and Cardiovascular Outcome: A Retrospective Cohort Study. Transplantation 2020, 104, 823–834. [Google Scholar] [CrossRef]
- Loupy, A.; Vernerey, D.; Viglietti, D.; Aubert, O.; Van Huyen, J.P.D.; Empana, J.P.; Bruneval, P.; Glotz, D.; Legendre, C.; Jouven, X.; et al. Determinants and Outcomes of Accelerated Arteriosclerosis: Major Impact of Circulating Antibodies. Circ. Res. 2015, 117, 470–482. [Google Scholar]
- Gupta, V.; Ekundayo, O.; Nemeth, Z.K.; Yang, Y.; Covic, A.; Mathe, Z.; Kovesdy, C.P.; Molnar, M.Z.; Mucsi, I. Association between serum osteoprotegerin level and mortality in kidney transplant recipients—A prospective observational cohort study. Transplant. Int. 2021, 34, 844–854. [Google Scholar] [CrossRef] [PubMed]
- Hjelmesæth, J.; Ueland, T.; Flyvbjerg, A.; Bollerslev, J.; Leivestad, T.; Jenssen, T.; Hansen, T.K.; Thiel, S.; Sagedal, S.; Roislien, J.; et al. Early posttransplant serum osteoprotegerin levels predict long-term (8-year) patient survival and cardiovascular death in renal transplant patients. J. Am. Soc. Nephrol. 2006, 17, 1746–1754. [Google Scholar] [CrossRef]
- Svensson, M.; Dahle, D.O.; Mjoen, G.; Weihrauch, G.; Scharnagl, H.; Dobnig, H.; März, W.; Jardine, A.; Fellström, B.; Holdaas, H. Osteoprotegerin as a predictor of renal and cardiovascular outcomes in renal transplant recipients: Follow-up data from the ALERT study. Nephrol. Dial. Transplant. 2012, 27, 2571–2575. [Google Scholar] [CrossRef]
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast differentiation and activation. Nature 2003, 423, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Tschiderer, L.; Willeit, J.; Schett, G.; Kiechl, S.; Willeit, P. Osteoprotegerin concentration and risk of cardiovascular outcomes in nine general population studies: Literature-based meta-analysis involving 26,442 participants. PLoS ONE 2017, 12, e0183910. [Google Scholar] [CrossRef] [PubMed]
- Holt, S.G.; Smith, E.R. Fetuin-A-containing calciprotein particles in mineral trafficking and vascular disease. Nephrol. Dial. Transplant. 2016, 31, 1583–1587. [Google Scholar] [CrossRef]
- Smith, E.R.; Ford, M.L.; Tomlinson, L.A.; Rajkumar, C.; McMahon, L.P.; Holt, S.G. Phosphorylated fetuin-A-containing calciprotein particles are associated with aortic stiffness and a procalcific milieu in patients with pre-dialysis CKD. Nephrol. Dial. Transplant. 2012, 27, 1957–1966. [Google Scholar] [CrossRef]
- Pasch, A.; Farese, S.; Gräber, S.; Wald, J.; Richtering, W.; Floege, J.; Jahnen-Dechent, W. Nanoparticle-based test measures overall propensity for calcification in serum. J. Am. Soc. Nephrol. 2012, 23, 1744–1752. [Google Scholar] [CrossRef] [PubMed]
- Pluquet, M.; Kamel, S.; Choukroun, G.; Liabeuf, S.; Laville, S.M. Serum Calcification Propensity Represents a Good Biomarker of Vascular Calcification: A Systematic Review. Toxins 2022, 14, 637. [Google Scholar] [CrossRef] [PubMed]
- Keyzer, C.A.; de Borst, M.H.; Berg, E.v.D.; Jahnen-Dechent, W.; Arampatzis, S.; Farese, S.; Bergmann, I.P.; Floege, J.; Navis, G.; Bakker, S.J.L.; et al. Calcification Propensity and Survival among Renal Transplant Recipients. J. Am. Soc. Nephrol. 2016, 27, 239–248. [Google Scholar] [CrossRef]
- Dahle, D.O.; Åsberg, A.; Hartmann, A.; Holdaas, H.; Bachtler, M.; Jenssen, T.G.; Dionisi, M.; Pasch, A. Serum calcification propensity is a strong and independent determinant of cardiac and all-cause mortality in kidney transplant recipients. Am. J. Transplant. 2016, 16, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Bostom, A.; Pasch, A.; Madsen, T.; Roberts, M.B.; Franceschini, N.; Steubl, D.; Garimella, P.S.; Ix, J.H.; Tuttle, K.R.; Ivanova, A.; et al. Serum Calcification Propensity and Fetuin-A: Biomarkers of Cardiovascular Disease in Kidney Transplant Recipients. Am. J. Nephrol. 2018, 48, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Pihlstrøm, H.K.; Mjøen, G.; Mucha, S.; Franke, A.; Jardine, A.; Fellström, B.; Dahle, D.O.; Holdaas, H.; Melum, E. Genetic markers associated with long-term cardiovascular outcome in kidney transplant recipients. Am. J. Transplant. 2019, 19, 1444–1451. [Google Scholar] [CrossRef]
- Mega, J.L.; O Stitziel, N.; Smith, J.G.; I Chasman, D.; Caulfield, M.J.; Devlin, J.J.; Nordio, F.; Hyde, C.; Cannon, C.P.; Sacks, F.; et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: An analysis of primary and secondary prevention trials. Lancet. 2015, 385, 2264–2271. [Google Scholar] [CrossRef] [PubMed]
- Quaglia, M.; Musetti, C.; Merlotti, G.; Genazzani, A.A.; Cargnin, S.; Cena, T.; Cantaluppi, V.; Terrazzino, S. Pilot cohort study on the potential role of TCF7L2 rs7903146 on ischemic heart disease among non-diabetic kidney transplant recipients. Clin. Transplant. 2017, 31, e12959. [Google Scholar] [CrossRef]
- Khwaja, A. KDIGO Clinical Practice Guideline for Acute Kidney Injury. Kidney Int. 2012, 2 (Suppl. S1), 1–138, Online Appendices A–F. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Falahat, P.; Scheidt, U.; Pörner, D.; Schwab, S. Recent Insights in Noninvasive Diagnostic for the Assessment of Kidney and Cardiovascular Outcome in Kidney Transplant Recipients. J. Clin. Med. 2024, 13, 3778. https://doi.org/10.3390/jcm13133778
Falahat P, Scheidt U, Pörner D, Schwab S. Recent Insights in Noninvasive Diagnostic for the Assessment of Kidney and Cardiovascular Outcome in Kidney Transplant Recipients. Journal of Clinical Medicine. 2024; 13(13):3778. https://doi.org/10.3390/jcm13133778
Chicago/Turabian StyleFalahat, Peyman, Uta Scheidt, Daniel Pörner, and Sebastian Schwab. 2024. "Recent Insights in Noninvasive Diagnostic for the Assessment of Kidney and Cardiovascular Outcome in Kidney Transplant Recipients" Journal of Clinical Medicine 13, no. 13: 3778. https://doi.org/10.3390/jcm13133778
APA StyleFalahat, P., Scheidt, U., Pörner, D., & Schwab, S. (2024). Recent Insights in Noninvasive Diagnostic for the Assessment of Kidney and Cardiovascular Outcome in Kidney Transplant Recipients. Journal of Clinical Medicine, 13(13), 3778. https://doi.org/10.3390/jcm13133778





