Enhancing Diagnostic Precision: Evaluation of Preprocessing Filters in Simple Diffusion Kurtosis Imaging for Head and Neck Tumors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Phantom
2.2. Patients
2.3. MRI Devices and Sequences
2.3.1. Phantom Imaging Conditions
2.3.2. Clinical Imaging Conditions
2.4. Creation of Phantom DW Images
2.5. Preprocessing of DW Images with Filters and Parameter Setting
2.6. DK Image Creation
2.7. Setting the ROI for the Evaluation
2.7.1. Setting the ROI of the Phantom Image
2.7.2. Setting the ROI for the Clinical Study
2.8. Image Analysis
2.9. Statistical Analysis
3. Results
3.1. Changes in Median MK Values in the Phantoms
3.2. Changes in Variance and AUC in the Phantoms
3.3. Clinical Case Information
3.4. MK Values for Tumor and Normal ROIs in Clinical Practice
3.5. Distinguishability between Tumor and Normal Tissues in Clinical Practice
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lutsep, H.L.; Albers, G.W.; DeCrespigny, A.; Kamat, G.N.; Marks, M.P.; Moseley, M.E. Clinical utility of diffusion-weighted magnetic resonance imaging in the assessment of ischemic stroke. Ann. Neurol. 1997, 41, 574–580. [Google Scholar] [CrossRef] [PubMed]
- van Everdingen, K.J.; van der Grond, J.; Kappelle, L.J.; Ramos, L.M.; Mali, W.P. Diffusion-weighted magnetic resonance imaging in acute stroke. Stroke 1998, 29, 1783–1790. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Cai, Y.Q.; Cai, Z.L.; Gao, Y.G.; An, N.Y.; Ma, L.; Mahankali, S.; Gao, J.H. Differentiation of clinically benign and malignant breast lesions using diffusion-weighted imaging. J. Magn. Reson. Imaging 2002, 16, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Tamada, T.; Sone, T.; Jo, Y.; Toshimitsu, S.; Yamashita, T.; Yamamoto, A.; Tanimoto, D.; Ito, K. Apparent diffusion coefficient values in peripheral and transition zones of the prostate. Comparison between normal and malignant prostatic tissues and correlation with histologic grade. J. Magn. Reson. Imaging 2008, 28, 720–726. [Google Scholar] [CrossRef] [PubMed]
- Hui, E.S.; Fieremans, E.; Jensen, J.H.; Tabesh, A.; Feng, W.; Bonilha, L.; Spampinato, M.V.; Adams, R.; Helpern, J.A. Stroke assessment with diffusional kurtosis imaging. Stroke 2012, 43, 2968–2973. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Lin, W.Y.; Lu, C.S.; Weng, Y.H.; Ng, S.H.; Wang, C.H.; Liu, H.L.; Hsieh, R.H.; Wan, Y.L.; Wai, Y.Y. Parkinson disease: Diagnostic utility of diffusion kurtosis imaging. Radiology 2011, 261, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Arab, A.; Wojna-Pelczar, A.; Khairnar, A.; Szabó, N.; Ruda-Kucerova, J. Principles of diffusion kurtosis imaging and its role in early diagnosis of neurodegenerative disorders. Brain Res. Bull. 2018, 139, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Tamura, C.; Shinmoto, H.; Soga, S.; Okamura, T.; Sato, H.; Okuaki, T.; Pang, Y.; Kosuda, S.; Kaji, T. Diffusion kurtosis imaging study of prostate cancer: Preliminary findings. J. Magn. Reson. Imaging 2014, 40, 723–729. [Google Scholar] [CrossRef]
- Liu, C.; Xing, Y.; Wei, D.; Jiao, Q.; Yang, Q.; Lei, D.; Tao, X.; Yao, W. Diffusion kurtosis imaging as a prognostic marker in osteosarcoma patients with preoperative chemotherapy. BioMed Res. Int. 2020, 2020, 3268138. [Google Scholar] [CrossRef]
- Hempel, J.M.; Brendle, C.; Adib, S.D.; Behling, F.; Tabatabai, G.; Vega, S.C.; Schittenhelm, J.; Ernemann, U.; Klose, U. Glioma-specific diffusion signature in diffusion kurtosis imaging. J. Clin. Med. 2021, 10, 2325. [Google Scholar] [CrossRef]
- Jiang, L.; Zhou, L.; Ai, Z.; Xiao, C.; Liu, W.; Geng, W.; Chen, H.; Xiong, Z.; Yin, X.; Chen, Y.C. Machine learning based on diffusion kurtosis imaging histogram parameters for glioma grading. J. Clin. Med. 2022, 11, 2310. [Google Scholar] [CrossRef] [PubMed]
- Tramontano, L.; Cavaliere, C.; Salvatore, M.; Brancato, V. The role of non-Gaussian models of diffusion weighted MRI in hepatocellular carcinoma: A systematic review. J. Clin. Med. 2021, 10, 2641. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Song, Q.; Wang, Y.; Lin, L.; Tian, S.; Wang, N.; Wang, J.; Liu, A. Amide proton transfer weighted combined with diffusion kurtosis imaging for predicting lymph node metastasis in cervical cancer. Magn. Reson. Imaging 2024, 106, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Ren, A.; Xu, X.; Meng, Z.; Feng, X.; Pylypenko, D.; Dou, W.; Yu, D. Application of DKI and IVIM imaging in evaluating histologic grades and clinical stages of clear cell renal cell carcinoma. Front. Oncol. 2023, 13, 1203922. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Li, L.; Shi, J.; Gong, X.; Cheng, Y.; Wang, W.; Cao, Y.; Cao, M.; Jiang, F.; Wang, L.; et al. Predicting histopathological types and molecular subtype of breast tumors: A comparative study using amide proton transfer-weighted imaging, intravoxel incoherent motion and diffusion kurtosis imaging. Magn. Reson. Imaging 2024, 105, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Yu, C.; Pan, J.; Xu, Y.; Luo, Y.; Yang, T.; Yang, X.; Xie, L.; Zhang, J.; Zhuo, R. Prediction of the Nottingham prognostic index and molecular subtypes of breast cancer through multimodal magnetic resonance imaging. Magn. Reson. Imaging 2024, 108, 168–175. [Google Scholar] [CrossRef]
- Hamada, K.; Kuroda, M.; Yoshimura, Y.; Khasawneh, A.; Barham, M.; Tekiki, N.; Sugianto, I.; Bamgbose, B.O.; Konishi, K.; Sugimoto, K.; et al. Evaluation of the imaging process for a novel subtraction method using apparent diffusion coefficient values. Acta Med. Okayama 2021, 75, 139–145. [Google Scholar]
- Kuroda, M.; Konishi, K.; Sugimoto, K.; Yoshimura, Y.; Hamada, K.; Khasawnehc, A.; Barham, M.; Tekiki, N.; Sugianto, I.; Bamgbose, O.B.; et al. Evaluation of fast diffusion kurtosis imaging using new software designed for widespread clinical use. Acta Med. Okayama 2022, 76, 297–305. [Google Scholar]
- Shimizu, Y.; Kuroda, M.; Nakamitsu, Y.; Al-Hammad, W.E.; Yoshida, S.; Fukumura, Y.; Nakamura, Y.; Kuroda, K.; Kamizaki, R.; Imajoh, S.; et al. Usefulness of simple diffusion kurtosis imaging for head and neck tumors: An early clinical study. Acta Med Okayama 2023, 77, 273–280. [Google Scholar]
- Fukumura, Y.; Kuroda, M.; Yoshida, S.; Nakamura, Y.; Nakamitsu, Y.; Al-Hammad, W.E.; Kuroda, K.; Kamizaki, R.; Shimizu, Y.; Tanabe, Y.; et al. Characteristic mean kurtosis values in simple diffusion kurtosis imaging of dentigerous cysts. Diagnostics 2023, 13, 3619. [Google Scholar] [CrossRef]
- Usman, O.L.; Muniyandi, R.C.; Omar, K.; Mohamad, M. Gaussian smoothing and modified histogram normalization methods to improve neural biomarker interpretations for dyslexia classification mechanism. PLoS ONE 2021, 16, e0245579. [Google Scholar] [CrossRef] [PubMed]
- Falangola, M.F.; Jensen, J.H.; Babb, J.S.; Hu, C.; Castellanos, F.X.; Di Martino, A.; Ferris, S.H.; Helpern, J.A. Age-related non-Gaussian diffusion patterns in the prefrontal brain. J. Magn. Reson. Imaging 2008, 28, 1345–1350. [Google Scholar] [CrossRef] [PubMed]
- Wahid, F.; Ghazali, R.; Fayaz, M.; Shah, A.S. Using probabilistic classification technique and statistical features for brain magnetic resonance imaging (MRI) classification: An application of AI technique in bio-science. Int. J. Bio-Sci. Bio-Technol. 2016, 8, 93–106. [Google Scholar] [CrossRef]
- Zhang, Z.P.; Vernekar, D.; Qian, W.S.; Kim, M.N. Non-local means based Rician noise filtering for diffusion tensor and kurtosis imaging in human brain and spinal cord. BMC Med. Imaging 2021, 21, 16. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.X.; Yan, X.; Xie, H.B.; Zheng, H.; Xu, D.R.; Yang, G. Evaluation of non-local means based denoising filters for diffusion kurtosis imaging using a new phantom. PLoS ONE 2015, 10, e0116986. [Google Scholar] [CrossRef] [PubMed]
- Khasawneh, A.; Kuroda, M.; Yoshimura, Y.; Sugianto, I.; Bamgbose, B.O.; Hamada, K.; Barham, M.; Tekiki, N.; Konishi, K.; Sugimoto, K.; et al. Development of a novel phantom using polyethylene glycol for the visualization of restricted diffusion in diffusion kurtosis imaging and apparent diffusion coefficient subtraction method. Biomed. Rep. 2020, 13, 52. [Google Scholar] [CrossRef] [PubMed]
- Matsuya, R.; Kuroda, M.; Matsumoto, Y.; Kato, H.; Matsuzaki, H.; Asaumi, J.; Murakami, J.; Katashima, K.; Ashida, M.; Sasaki, T.; et al. A new phantom using polyethylene glycol as an apparent diffusion coefficient standard for MR imaging. Int. J. Oncol. 2009, 35, 893–900. [Google Scholar]
- Wilson, B.; Dhas, J.P.M. Denoising in magnetic resonance images using improved Gaussian smoothing technique. Int. J. Eng. Technol. 2019, 8, 2. [Google Scholar] [CrossRef]
- Antoni, B.; Bartomeu, C.; Michel, M.J. Non-local means denoising. Image Process. Line 2011, 1, 208–212. [Google Scholar]
- Zhu, Q.Q.; Xu, Q.; Dou, W.Q.; Zhu, W.R.; Wu, J.T.; Chen, W.X.; Ye, J. Diffusion kurtosis imaging features of renal cell carcinoma: A preliminary study. Br. J. Radiol. 2021, 94, 1122. [Google Scholar] [CrossRef]
- Delgado, A.; Fahlstrom, M.; Nilsson, M.; Berntsson, S.G.; Zetterling, M.; Libard, S.; Alafuzoff, I.; Van Westen, D.; Latt, J.; Smits, A.; et al. Diffusion kurtosis imaging of gliomas grades II and III—A study of perilesional tumor infiltration, tumor grades and subtypes at clinical presentation. Radiol. Oncol. 2017, 51, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Minosse, S.; Marzi, S.; Piludu, F.; Vidiri, A. Correlation study between DKI and conventional DWI in brain and head and neck tumors. Magn. Reson. Imaging 2017, 42, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Tang, L.; Li, Z.Y.; Li, X.T.; Zhu, H.F.; Sun, Y.S.; Ji, J.F. Diffusion kurtosis imaging in the prediction of poor responses of locally advanced gastric cancer to neoadjuvant chemotherapy. Eur. J. Radiol. 2020, 128, 108974. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.L.; Jia, F.; Li, X.K.; Zhang, M.; Han, D.M.; Jia, Z.Y. Amide proton transfer imaging vs diffusion kurtosis imaging for predicting histological grade of hepatocellular carcinoma. J. Hepatocell. Carcinoma 2020, 7, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.Q.; Xiao, Q.; Hu, F.X.; Fu, C.X.; Jia, H.X.; Yan, X.; Xin, C.; Cai, S.J.; Peng, W.J.; Wang, X.L.; et al. Diffusion kurtosis imaging in the characterisation of rectal cancer: Utilizing the most repeatable region-of-interest strategy for diffusion parameters on a 3T scanner. Eur. J. Radiol. 2018, 28, 5211–5220. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.B.; Tang, Z.H.; Qiang, J.W.; Qian, W.; Zhong, Y.F.; Wang, R.; Wang, J.; Wu, L.J.; Tang, W.L. Differentiation of olfactory neuroblastomas from nasal squamous cell carcinomas using MR diffusion kurtosis imaging and dynamic contrast-enhanced MRI. J. Magn. Reson. Imaging 2018, 47, 354–361. [Google Scholar] [CrossRef]
- Cao, L.K.; Chen, J.; Duan, T.; Wang, M.; Jiang, H.Y.; Wei, Y.; Xia, C.C.; Zhou, X.Y.; Yan, X.; Song, B. Diffusion kurtosis imaging (DKI) of hepatocellular carcinoma: Correlation with microvascular invasion and histologic grade. Quant. Imaging Med. Surg. 2019, 9, 590–602. [Google Scholar] [CrossRef]
- Pasicz, K.; Podgórska, J.; Jasieniak, J.; Fabiszewska, E.; Skrzyński, W.; Anysz-Grodzicka, A.; Cieszanowski, A.; Kukołowicz, P.; Grabska, I. Optimal b-values for diffusion kurtosis imaging of the liver and pancreas in MR examinations. Phys. Medica 2019, 66, 119–123. [Google Scholar] [CrossRef]
- Rosenkrantz, A.B.; Padhani, A.R.; Chenevert, T.L.; Koh, D.; Keyzer, F.D.; Taouli, B.; Bihan, D.L. Body diffusion kurtosis imaging: Basic principles, applications, and considerations for clinical practice. J. Magn. Reson. Imaging 2015, 42, 1190–1202. [Google Scholar] [CrossRef]
- Fusco, R.; Sansone, M.; Granata, V.; Grimm, R.; Pace, U.; Delrio, P.; Tatangelo, F.; Botti, G.; Avallone, A.; Pecori, B.; et al. Diffusion and perfusion MR parameters to assess preoperative short-course radiotherapy response in locally advanced rectal cancer: A comparative explorative study among Standardized Index of Shape by DCE-MRI, intravoxel incoherent motion- and diffusion kurtosis imaging-derived parameters. Abdom. Radiol. 2019, 44, 3683–3700. [Google Scholar]
- Quentin, M.; Pentang, G.; Schimmöller, L.; Kott, O.; Müller-Lutz, A.; Blondin, D.; Arsov, C.; Hiester, A.; Rabenalt, R.; Wittsack, H.J. Feasibility of diffusional kurtosis tensor imaging in prostate MRI for the assessment of prostate cancer: Preliminary results. Magn. Reson. Imaging 2014, 32, 880–885. [Google Scholar] [CrossRef]



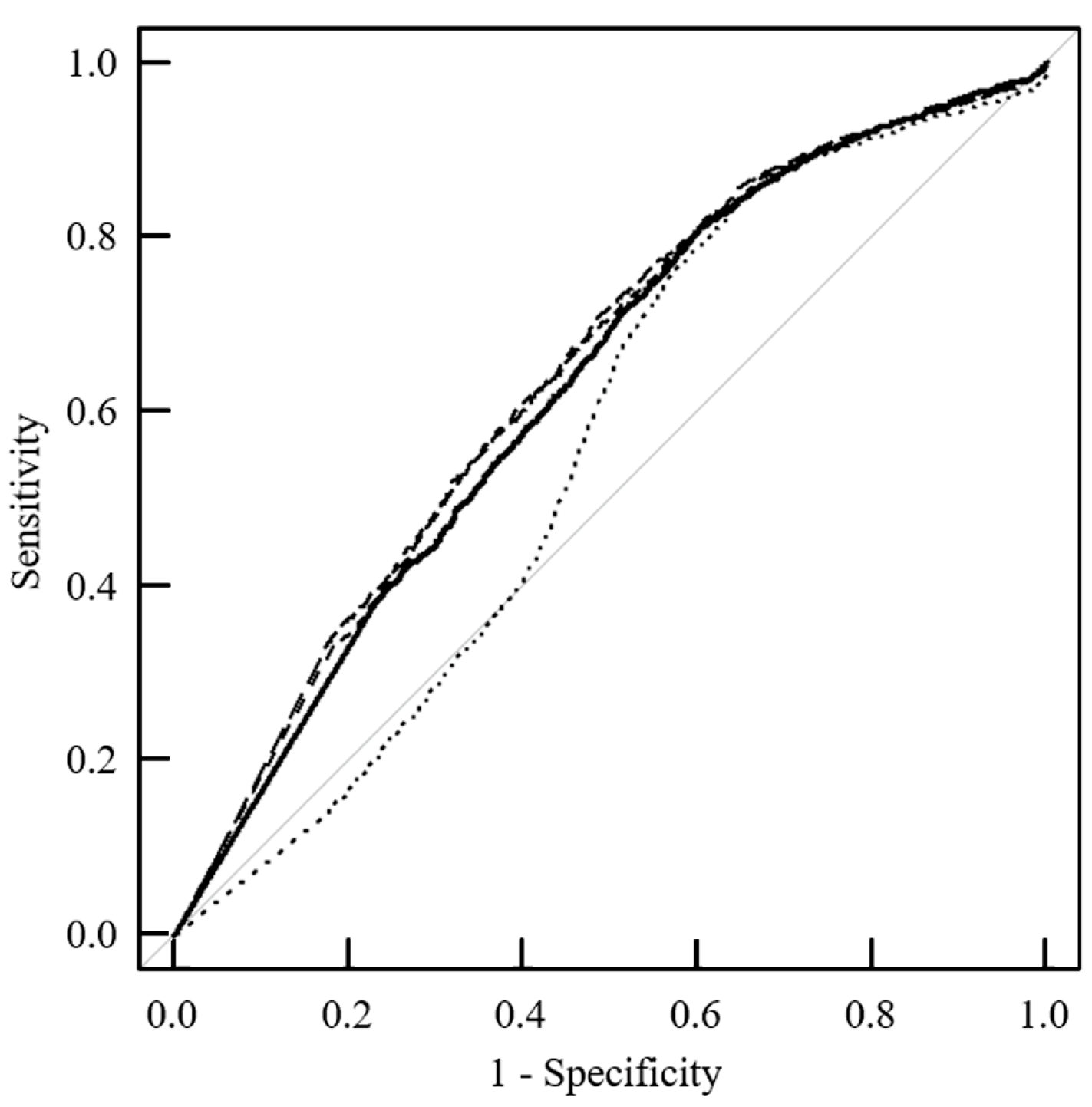
| Tumor ROI | p-Value | Normal ROI | p-Value | ||
|---|---|---|---|---|---|
| Filter Type | Parameter | Median (Q1, Q3) | Median (Q1, Q3) | ||
| Gaussian | σ = 0 | 0.715 (0.327, 1.014) | 0.055 (0.000, 0.346) | ||
| σ = 0.1 | 0.715 (0.327, 1.014) | 1.00 | 0.055 (0.000, 0.346) | 1.00 | |
| σ = 0.2 | 0.715 (0.327, 1.014) | 1.00 | 0.055 (0.000, 0.346) | 1.00 | |
| σ = 0.3 | 0.717 (0.334, 1.011) | 1.00 | 0.058 (0.000, 0.347) | 1.00 | |
| σ = 0.4 | 0.710 (0.406, 0.981) | 1.00 | 0.075 (0.000, 0.349) | 1.00 | |
| σ = 0.5 | 0.712 (0.504, 0.920) | 1.00 | 0.104 (0.000, 0.358) | 1.00 | |
| σ = 0.6 | 0.718 (0.556, 0.879) | 1.00 | 0.153 (0.000, 0.358) | 1.00 | |
| σ = 0.7 | 0.728 (0.597, 0.869) | 1.00 | 0.179 (0.000, 0.356) | 1.00 | |
| σ = 0.8 | 0.731 (0.618, 0.856) | 1.00 | 0.206 (0.000, 0.356) | 1.00 | |
| σ = 0.9 | 0.730 (0.632, 0.854) | 0.76 | 0.225 (0.000, 0.353) | 1.00 | |
| σ = 1.0 | 0.733 (0.645, 0.845) | 0.47 | 0.232 (0.000, 0.353) | 1.00 | |
| Median | Radius = 0 | 0.715 (0.327, 1.014) | 0.055 (0.000, 0.346) | ||
| Radius = 0.5 | 0.713 (0.525, 0.905) | 1.00 | 0.027 (0.000, 0.351) | 1.00 | |
| Radius = 1.0 | 0.692 (0.562, 0.874) | 1.00 | 0.066 (0.000, 0.352) | 1.00 | |
| Radius = 1.5 | 0.701 (0.558, 0.855) | 1.00 | 0.066 (0.000, 0.346) | 0.74 | |
| NLM | σ = 0 | 0.715 (0.327, 1.014) | 0.055 (0.000, 0.346) | ||
| σ = 1 | 0.723 (0.317, 1.013) | 1.00 | 0.051 (0.000, 0.344) | 1.00 | |
| σ = 2 | 0.723 (0.393, 0.982) | 1.00 | 0.031 (0.000, 0.344) | 1.00 | |
| σ = 3 | 0.707 (0.430, 0.938) | 1.00 | 0.041 (0.000, 0.351) | 1.00 | |
| σ = 4 | 0.705 (0.451, 0.951) | 1.00 | 0.067 (0.000, 0.367) | 1.00 | |
| σ = 5 | 0.703 (0.458, 0.951) | 1.00 | 0.061 (0.000, 0.354) | 1.00 | |
| σ = 6 | 0.714 (0.447, 0.952) | 1.00 | 0.077 (0.000, 0.360) | 1.00 | |
| σ = 9 | 0.718 (0.457, 0.962) | 1.00 | 0.049 (0.000, 0.358) | 1.00 | |
| σ = 10 | 0.718 (0.454, 0.946) | 1.00 | 0.030 (0.000, 0.353) | 1.00 | |
| σ = 15 | 0.732 (0.514, 0.927) | 1.00 | 0.024 (0.000, 0.351) | 1.00 | |
| σ = 20 | 0.741 (0.555, 0.904) | 1.00 | 0.067 (0.000, 0.354) | 1.00 | |
| σ = 25 | 0.727 (0.578, 0.892) | 1.00 | 0.092 (0.000, 0.364) | 1.00 | |
| σ = 30 | 0.718 (0.583, 0.893) | 1.00 | 0.057 (0.000, 0.360) | 1.00 |
| Filter | Homogeneity Evaluated by Fligner–Killeen Test | Discernment Ability | ||
|---|---|---|---|---|
| Filter Type | Filter Parameter | p-Value for Tumor ROI | p-Value for Normal ROI | AUC Value |
| Gaussian | σ = 0.0 | 0.835 | ||
| σ = 0.1 | 1.000 | 1.000 | 0.835 | |
| σ = 0.2 | 0.999 | 1.000 | 0.835 | |
| σ = 0.3 | 0.637 | 0.000 | 0.837 * | |
| σ = 0.4 | 0.000 | 0.000 | 0.865 * | |
| σ = 0.5 | 0.000 | 0.003 | 0.912 * | |
| σ = 0.6 | 0.000 | 0.567 | 0.948 * | |
| σ = 0.7 | 0.000 | 0.962 | 0.967 * | |
| σ = 0.8 | 0.000 | 0.502 | 0.978 * | |
| σ = 0.9 | 0.000 | 0.945 | 0.984 * | |
| σ = 1.0 | 0.000 | 0.854 | 0.988 * | |
| Median | Radius = 0 | 0.835 | ||
| Radius = 0.5 | 0.000 | 0.000 | 0.919 * | |
| Radius = 1.0 | 0.000 | 0.000 | 0.956 * | |
| Radius = 1.5 | 0.000 | 0.001 | 0.965 * | |
| NLM | σ = 0 | 0.835 | ||
| σ = 1 | 0.445 | 0.000 | 0.836 | |
| σ = 2 | 0.041 | 0.000 | 0.858 * | |
| σ = 3 | 0.000 | 0.000 | 0.871 * | |
| σ = 4 | 0.000 | 0.000 | 0.881 * | |
| σ = 5 | 0.000 | 0.000 | 0.884 * | |
| σ = 6 | 0.000 | 0.000 | 0.880 * | |
| σ = 9 | 0.000 | 0.000 | 0.889 * | |
| σ = 10 | 0.000 | 0.000 | 0.896 * | |
| σ = 15 | 0.000 | 0.000 | 0.924 * | |
| σ = 20 | 0.000 | 0.000 | 0.957 * | |
| σ = 25 | 0.000 | 0.002 | 0.959 * | |
| σ = 30 | 0.000 | 0.000 | 0.965 * | |
| ROI Setting | |||||
|---|---|---|---|---|---|
| Case | Histological Classification | Tumor ROI * | Normal ROI ** | ||
| Position | Number of Pixels | Position | Number of Pixels | ||
| 1 | Squamous cell carcinoma | Maxilla | 434 | Erector spinae muscle | 40 |
| 2 | 334 | Masseter muscle | 119 | ||
| 3 | 219 | Masseter muscle | 214 | ||
| 4 | 132 | Lateral pterygoid muscle | 100 | ||
| 5 | Mandible | 63 | Masseter muscle | 87 | |
| 6 | Tongue | 289 | Masseter muscle | 322 | |
| 7 | 245 | Masseter muscle | 65 | ||
| 8 | 21 | Erector spinae muscle | 300 | ||
| 9 | Adenoid cystic carcinoma | Palate | 412 | Temporal muscle | 200 |
| 10 | 59 | Masseter muscle | 177 | ||
| 11 | Acinic cell carcinoma | Parotid gland | 223 | Masseter muscle | 110 |
| 12 | Malignant lymphoma | Maxilla | 154 | Masseter muscle | 186 |
| 13 | Osteosarcoma | Mandible | 117 | Erector spinae muscle | 222 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakamitsu, Y.; Kuroda, M.; Shimizu, Y.; Kuroda, K.; Yoshimura, Y.; Yoshida, S.; Nakamura, Y.; Fukumura, Y.; Kamizaki, R.; Al-Hammad, W.E.; et al. Enhancing Diagnostic Precision: Evaluation of Preprocessing Filters in Simple Diffusion Kurtosis Imaging for Head and Neck Tumors. J. Clin. Med. 2024, 13, 1783. https://doi.org/10.3390/jcm13061783
Nakamitsu Y, Kuroda M, Shimizu Y, Kuroda K, Yoshimura Y, Yoshida S, Nakamura Y, Fukumura Y, Kamizaki R, Al-Hammad WE, et al. Enhancing Diagnostic Precision: Evaluation of Preprocessing Filters in Simple Diffusion Kurtosis Imaging for Head and Neck Tumors. Journal of Clinical Medicine. 2024; 13(6):1783. https://doi.org/10.3390/jcm13061783
Chicago/Turabian StyleNakamitsu, Yuki, Masahiro Kuroda, Yudai Shimizu, Kazuhiro Kuroda, Yuuki Yoshimura, Suzuka Yoshida, Yoshihide Nakamura, Yuka Fukumura, Ryo Kamizaki, Wlla E. Al-Hammad, and et al. 2024. "Enhancing Diagnostic Precision: Evaluation of Preprocessing Filters in Simple Diffusion Kurtosis Imaging for Head and Neck Tumors" Journal of Clinical Medicine 13, no. 6: 1783. https://doi.org/10.3390/jcm13061783





