Advancements in Artificial Intelligence for Kidney Transplantology: A Comprehensive Review of Current Applications and Predictive Models
Abstract
:1. Introduction
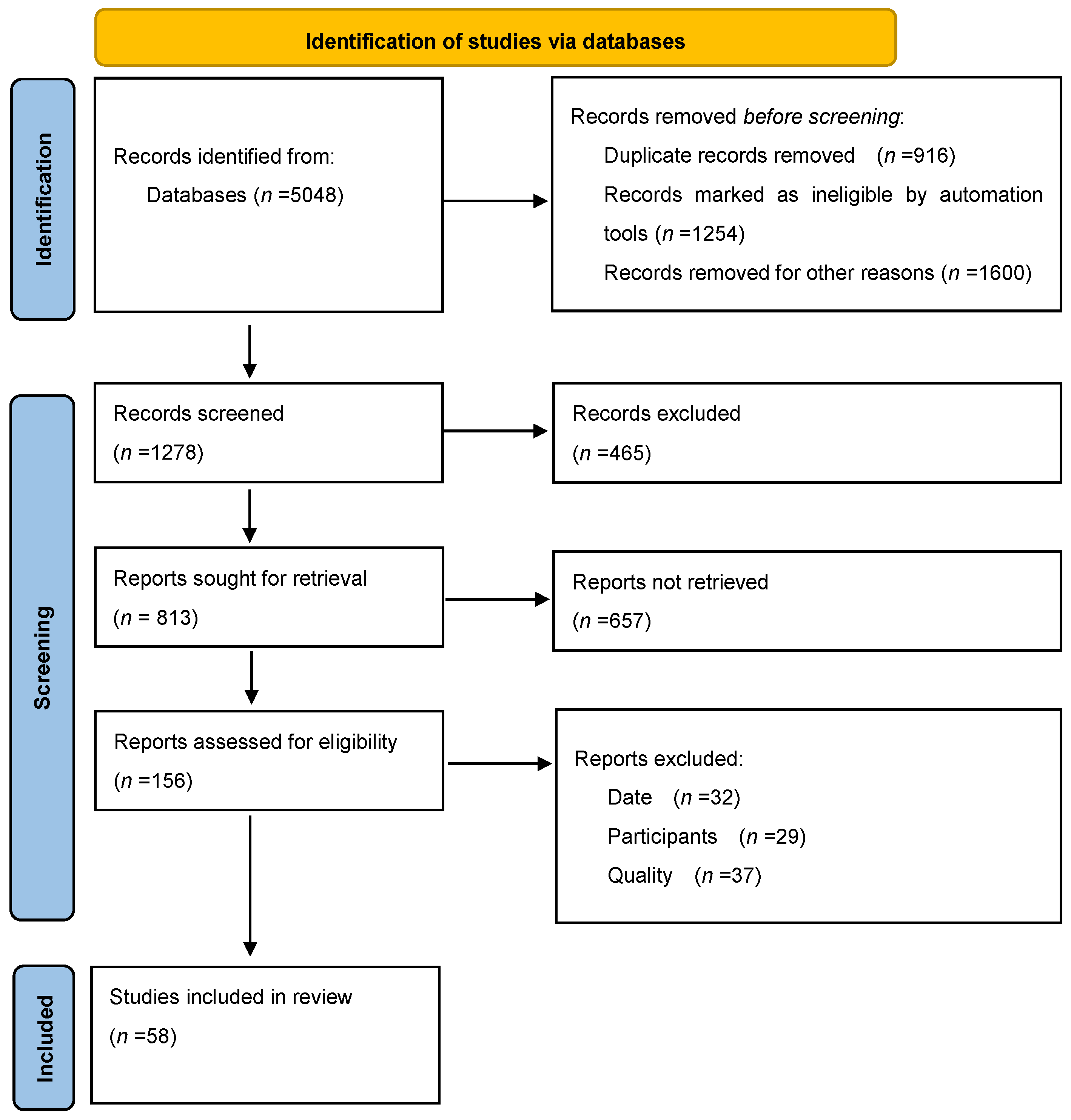
2. Materials and Methods
2.1. Literature Review—Methodology
- RQ1: What machine learning (ML) algorithms and techniques have been applied in the context of kidney transplantation as reported in the literature?
- RQ2: What specific application areas of machine learning in kidney transplantation are identified in the literature?
- RQ3: Which method should be selected to achieve the most accurate results for the intended analysis?
- RQ4: What variables (features) are commonly used in predictive models related to kidney transplantation in the existing body of research?
- (A)
- Defining the Sources for Literature Analysis
- (B)
- Establishing the Search Strategy and Scope
- The time period;
- The type of publications (peer-reviewed journals and conference papers);
- The language.
- (C)
- Selection Criteria for Inclusion
- Relevance to the research questions (RQ1, RQ2, and RQ3);
- Methodological rigor and quality of the studies;
- Theoretical or empirical focus;
- Availability of full-text versions for detailed analysis. Papers that meet these criteria will be retained for further analysis.
- (D)
- Screening and Extraction of Relevant Papers
- Initial screening: Title and abstract review to eliminate papers that clearly do not meet the selection criteria.
- Detailed screening: Full-text analysis to ensure alignment with the research questions. Relevant papers will be extracted and compiled into a dataset for further analysis.
- (E)
- Addressing Research Questions (RQ1, RQ2, and RQ3)
- (F)
- Synthesis of Results and Conclusion Formulation
2.2. Literature Review—Findings
- (A)
- Defining the Sources for Literature Analysis
- (B)
- Establishing the Search Strategy and Scope
- (C)
- Selection Criteria for Inclusion
- (D)
- Screening and Extraction of Relevant Papers
- (E)
- Addressing Research Questions (RQ1, RQ2, and RQ3)
- (F)
- Synthesis of Results and Conclusion Formulation
3. Selection of AI Techniques and Machine Learning Algorithms
- (A)
- Similarity Metrics: Clustering algorithms typically utilize mathematical measures of similarity or dissimilarity (e.g., Euclidean distance and cosine similarity) to quantify the relationship between data points. Points within a cluster are expected to exhibit higher similarity to one another compared to those in different clusters.
- (B)
- Prominent Algorithms Selection: This allows for final feature combination selection, which maximizes the intercluster variability (e.g., Self-Organizing Map (SOM), Consensus Clustering, and Agglomerative Clustering). SOMs, also known as Kohonen maps, help to visualize and analyze high-dimensional data by creating a low-dimensional representation that preserves the data’s structure. Consensus Clustering improves the clustering accuracy by combining the results from multiple algorithms to produce a single, reliable solution. Agglomerative Clustering, a type of hierarchical clustering, merges the clusters based on their similarity, forming a hierarchy that can be visualized as a dendrogram.
4. AI Techniques in Kidney Transplantation—A Range of Applications
4.1. Machine Learning
4.1.1. Predicting the Graft Survival
4.1.2. Immunosuppressive Agent Dosage Estimation
4.1.3. Donor–Recipient Pairing
4.1.4. Virtual Biopsy
4.2. Deep Learning
5. AI Model Summary—Variable Selection
6. Discussion—Challenges, Limitations, and Future Directions
6.1. Challenges and Limitations
6.2. Possible Future Directions
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Woderska-Jasińska, A.; Hermanowicz, M.; Włodarczyk, Z. Transplantacja Nerki Jako Metoda Leczenia Nerkozastępczego Pacjenta z Przewlekłą Chorobą Nerek. Innow. Pielęgniarstwie Nauk. Zdrowiu 2021, 6, 73–83. [Google Scholar] [CrossRef]
- Stites, E.; Wiseman, A.C. Multiorgan Transplantation. Transplant. Rev. 2016, 30, 253–260. [Google Scholar] [CrossRef]
- Mizera, J.; Pilch, J.; Giordano, U.; Krajewska, M.; Banasik, M. Therapy in the Course of Kidney Graft Rejection—Implications for the Cardiovascular System—A Systematic Review. Life 2023, 13, 1458. [Google Scholar] [CrossRef]
- Rebillard, X.; Mourad, G.; Cristol, J.P.; Daures, J.P.; Iborra, F.; Guiter, J.; Mion, C.; Grasset, D. [Prognostic Factors of Success in Renal Transplantation]. Presse Med. 1991, 20, 1985–1988. [Google Scholar] [PubMed]
- Weissenbacher, A.; Jara, M.; Ulmer, H.; Biebl, M.; Bösmüller, C.; Schneeberger, S.; Mayer, G.; Pratschke, J.; Öllinger, R. Recipient and Donor Body Mass Index as Important Risk Factors for Delayed Kidney Graft Function. Transplantation 2012, 93, 524–529. [Google Scholar] [CrossRef]
- Lee, C.Y.; Yang, C.Y.; Lin, W.C.; Chen, C.C.; Tsai, M.K. Prognostic Factors for Renal Transplant Graft Survival in a Retrospective Cohort of 1000 Cases: The Role of Desensitization Therapy. J. Formos. Med. Assoc. 2020, 119, 829–837. [Google Scholar] [CrossRef]
- Koukoulaki, M.; Kitsiou, V.; Balaska, A.; Pistolas, D.; Loukopoulos, I.; Drakopoulos, V.; Athanasiadis, T.; Tarassi, K.; Vougas, V.; Metaxatos, G.; et al. Immunologic Prognostic Factors of Renal Allograft Survival. Transplant. Proc. 2014, 46, 3175–3178. [Google Scholar] [CrossRef]
- Fang, F.; Liu, P.; Song, L.; Wagner, P.; Bartlett, D.; Ma, L.; Li, X.; Rahimian, M.A.; Tseng, G.; Randhawa, P.; et al. Diagnosis of T-Cell-Mediated Kidney Rejection by Biopsy-Based Proteomic Biomarkers and Machine Learning. Front. Immunol. 2023, 14, 1090373. [Google Scholar] [CrossRef] [PubMed]
- Saatchi, M.; Poorolajal, J.; Ali Amirzargar, M.; Mahjub, H.; Esmailnasabs, N. Long-Term Survival Rate of Kidney Graft and Associated Prognostic Factors: A Retrospective Cohort Study, 1994–2011. Ann. Transplant. 2013, 18, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Tapak, L.; Hamidi, O.; Amini, P.; Poorolajal, J. Prediction of Kidney Graft Rejection Using Artificial Neural Network. Healthc. Inform. Res. 2017, 23, 277. [Google Scholar] [CrossRef]
- Amisha; Malik, P.; Pathania, M.; Rathaur, V.K. Overview of Artificial Intelligence in Medicine. J. Fam. Med. Prim. Care 2019, 8, 2328. [Google Scholar] [CrossRef]
- Rahman, M.A.; Yilmaz, I.; Albadri, S.T.; Salem, F.E.; Dangott, B.J.; Taner, C.B.; Nassar, A.; Akkus, Z. Artificial Intelligence Advances in Transplant Pathology. Bioengineering 2023, 10, 1041. [Google Scholar] [CrossRef] [PubMed]
- Salvagno, M.; De Cassai, A.; Zorzi, S.; Zaccarelli, M.; Pasetto, M.; Sterchele, E.D.; Chumachenko, D.; Gerli, A.G.; Azamfirei, R.; Taccone, F.S. The State of Artificial Intelligence in Medical Research: A Survey of Corresponding Authors from Top Medical Journals. PLoS ONE 2024, 19, e0309208. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, 71. [Google Scholar] [CrossRef]
- Schardt, C.; Adams, M.B.; Owens, T.; Keitz, S.; Fontelo, P. Utilization of the PICO Framework to Improve Searching PubMed for Clinical Questions. BMC Med. Inform. Decis. Mak. 2007, 7, 16. [Google Scholar] [CrossRef] [PubMed]
- Qiao, N. A Systematic Review on Machine Learning in Sellar Region Diseases: Quality and Reporting Items. Endocr. Connect. 2019, 8, 952. [Google Scholar] [CrossRef] [PubMed]
- Ravindhran, B.; Chandak, P.; Schafer, N.; Kundalia, K.; Hwang, W.; Antoniadis, S.; Haroon, U.; Zakri, R.H. Machine Learning Models in Predicting Graft Survival in Kidney Transplantation: Meta-Analysis. BJS Open 2023, 7, zrad011. [Google Scholar] [CrossRef]
- Díez-Sanmartín, C.; Sarasa Cabezuelo, A.; Andrés Belmonte, A. Ensemble of Machine Learning Techniques to Predict Survival in Kidney Transplant Recipients. Comput. Biol. Med. 2024, 180, 108982. [Google Scholar] [CrossRef] [PubMed]
- Ishwaran, H.; Kogalur, U.B.; Blackstone, E.H.; Lauer, M.S. Random Survival Forests. Ann. Appl. Stat. 2008, 2, 841–860. [Google Scholar] [CrossRef]
- Zou, H.; Hastie, T. Regularization and Variable Selection Via the Elastic Net. J. R. Stat. Soc. Series B Stat. Methodol. 2005, 67, 301–320. [Google Scholar] [CrossRef]
- Kursa, M.B.; Rudnicki, W.R. Feature Selection with the Boruta Package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar] [CrossRef]
- Yang, Z. FCM Clustering on Interaction Pattern Analysis of Chinese Language Learner Behavior. Comput. Intell. Neurosci. 2022, 2022, 8256646. [Google Scholar] [CrossRef]
- Barah, M.; Mehrotra, S. Predicting Kidney Discard Using Machine Learning. Transplantation 2021, 105, 2054. [Google Scholar] [CrossRef] [PubMed]
- Scholkopf, B.; Smola, A. Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond; MIT Press: Cambridge, MA, USA, 2018. [Google Scholar]
- Soffer, S.; Ben-Cohen, A.; Shimon, O.; Amitai, M.M.; Greenspan, H.; Klang, E. Convolutional Neural Networks for Radiologic Images: A Radiologist’s Guide. Radiology 2019, 290, 590–606. [Google Scholar] [CrossRef]
- Haykin, S. Neural Networks: A Comprehensive Foundation; Prentice Hall PTR: Hoboken, NJ, USA, 1998. [Google Scholar]
- Paquette, F.X.; Ghassemi, A.; Bukhtiyarova, O.; Cisse, M.; Gagnon, N.; Della Vecchia, A.; Rabearivelo, H.A.; Loudiyi, Y. Machine Learning Support for Decision-Making in Kidney Transplantation: Step-by-Step Development of a Technological Solution. JMIR Med. Inform. 2022, 10, e34554. [Google Scholar] [CrossRef] [PubMed]
- Mark, E.; Goldsman, D.; Gurbaxani, B.; Keskinocak, P.; Sokol, J. Using Machine Learning and an Ensemble of Methods to Predict Kidney Transplant Survival. PLoS ONE 2019, 14, e0209068. [Google Scholar] [CrossRef]
- Aksoy, G.K.; Akçay, H.G.; Arı, Ç.; Adar, M.; Koyun, M.; Çomak, E.; Akman, S. Predicting Graft Survival in Paediatric Kidney Transplant Recipients Using Machine Learning. Pediatr. Nephrol. 2024, 40, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Jing, Y.; Liu Mr, G.; Jiang Mr, X.; Liu, H.; Kong, Y.; Hou, X.; Cao, L.; Deng, P.; Xiao, P.; et al. Machine Learning-Based Method for Tacrolimus Dose Predictions in Chinese Kidney Transplant Perioperative Patients. J. Clin. Pharm. Ther. 2022, 47, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Ramalhete, L.; Almeida, P.; Ferreira, R.; Abade, O.; Teixeira, C.; Araújo, R. Revolutionizing Kidney Transplantation: Connecting Machine Learning and Artificial Intelligence with Next-Generation Healthcare—From Algorithms to Allografts. BioMedInformatics 2024, 4, 673–689. [Google Scholar] [CrossRef]
- IntHout, J.; Ioannidis, J.P.A.; Rovers, M.M.; Goeman, J.J. Plea for Routinely Presenting Prediction Intervals in Meta-Analysis. BMJ Open 2016, 6, e010247. [Google Scholar] [CrossRef]
- Rao, P.S.; Schaubel, D.E.; Guidinger, M.K.; Andreoni, K.A.; Wolfe, R.A.; Merion, R.M.; Port, F.K.; Sung, R.S. A Comprehensive Risk Quantification Score for Deceased Donor Kidneys: The Kidney Donor Risk Index. Transplantation 2009, 88, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Parsons, R.F.; Locke, J.E.; Redfield, R.R.; Roll, G.R.; Levine, M.H. Kidney Transplantation of Highly Sensitized Recipients Under the New Kidney Allocation System: A Reflection from Five Different Transplant Centers Across the United States. Hum. Immunol. 2016, 78, 30. [Google Scholar] [CrossRef]
- Tolstyak, Y.; Zhuk, R.; Yakovlev, I.; Shakhovska, N.; Gregus Ml, M.; Chopyak, V.; Melnykova, N. The Ensembles of Machine Learning Methods for Survival Predicting after Kidney Transplantation. Appl. Sci. 2021, 11, 10380. [Google Scholar] [CrossRef]
- Naqvi, S.A.A.; Tennankore, K.; Vinson, A.; Roy, P.C.; Abidi, S.S.R. Predicting Kidney Graft Survival Using Machine Learning Methods: Prediction Model Development and Feature Significance Analysis Study. J. Med. Internet Res. 2021, 23, e26843. [Google Scholar] [CrossRef] [PubMed]
- Collins, G.S.; De Groot, J.A.; Dutton, S.; Omar, O.; Shanyinde, M.; Tajar, A.; Voysey, M.; Wharton, R.; Yu, L.M.; Moons, K.G.; et al. External Validation of Multivariable Prediction Models: A Systematic Review of Methodological Conduct and Reporting. BMC Med. Res. Methodol. 2014, 14, 40. [Google Scholar] [CrossRef] [PubMed]
- Steyerberg, E.W.; Harrell, F.E. Prediction Models Need Appropriate Internal, Internal-External, and External Validation. J. Clin. Epidemiol. 2015, 69, 245. [Google Scholar] [CrossRef]
- Zhang, Q.; Tian, X.; Chen, G.; Yu, Z.; Zhang, X.; Lu, J.; Zhang, J.; Wang, P.; Hao, X.; Huang, Y.; et al. A Prediction Model for Tacrolimus Daily Dose in Kidney Transplant Recipients with Machine Learning and Deep Learning Techniques. Front. Med. 2022, 9, 813117. [Google Scholar] [CrossRef]
- Basuli, D.; Roy, S. Beyond Human Limits: Harnessing Artificial Intelligence to Optimize Immunosuppression in Kidney Transplantation. J. Clin. Med. Res. 2023, 15, 391–398. [Google Scholar] [CrossRef]
- Størset, E.; Åsberg, A.; Skauby, M.; Neely, M.; Bergan, S.; Bremer, S.; Midtvedt, K. Improved Tacrolimus Target Concentration Achievement Using Computerized Dosing in Renal Transplant Recipients—A Prospective, Randomized Study. Transplantation 2015, 99, 2158. [Google Scholar] [CrossRef] [PubMed]
- Gören, S.; Karahoca, A.; Onat, F.Y.; Gören, M.Z. Prediction of Cyclosporine A Blood Levels: An Application of the Adaptive-Network-Based Fuzzy Inference System (ANFIS) in Assisting Drug Therapy. Eur. J. Clin. Pharmacol. 2008, 64, 807–814. [Google Scholar] [CrossRef]
- Yoo, D.; Divard, G.; Raynaud, M.; Cohen, A.; Mone, T.D.; Rosenthal, J.T.; Bentall, A.J.; Stegall, M.D.; Naesens, M.; Zhang, H.; et al. A Machine Learning-Driven Virtual Biopsy System For Kidney Transplant Patients. Nat. Commun. 2024, 15, 554. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Coursera—Deep Learning vs. Machine Learning: A Beginner’s Guide. Available online: https://www.coursera.org/articles/ai-vs-deep-learning-vs-machine-learning-beginners-guide (accessed on 28 January 2025).
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Yi, Z.; Xi, C.; Menon, M.C.; Cravedi, P.; Tedla, F.; Soto, A.; Sun, Z.; Liu, K.; Zhang, J.; Wei, C.; et al. A Large-Scale Retrospective Study Enabled Deep-Learning Based Pathological Assessment of Frozen Procurement Kidney Biopsies to Predict Graft Loss and Guide Organ Utilization. Kidney Int. 2024, 105, 281–292. [Google Scholar] [CrossRef]
- Kers, J.; Bülow, R.D.; Klinkhammer, B.M.; Breimer, G.E.; Fontana, F.; Abiola, A.A.; Hofstraat, R.; Corthals, G.L.; Peters-Sengers, H.; Djudjaj, S.; et al. Deep Learning-Based Classification of Kidney Transplant Pathology: A Retrospective, Multicentre, Proof-of-Concept Study. Lancet Digit. Health 2022, 4, e18–e26. [Google Scholar] [CrossRef]
- Senanayake, S.; Kularatna, S.; Healy, H.; Graves, N.; Baboolal, K.; Sypek, M.P.; Barnett, A. Development and Validation of a Risk Index to Predict Kidney Graft Survival: The Kidney Transplant Risk Index. BMC Med. Res. Methodol. 2021, 21, 127. [Google Scholar] [CrossRef]
- Weinrauch, L.A.; Anis, K.H.; D’Elia, J.A. Diabetes and the Solid Organ Transplant Recipient. Diabetes Res. Clin. Pract. 2018, 146, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Hart, A.; Schladt, D.P.; Matas, A.J.; Itzler, R.; Israni, A.K.; Kasiske, B.L. Incidence, Risk Factors, and Long-Term Outcomes Associated with Antibody-Mediated Rejection—The Long-Term Deterioration of Kidney Allograft Function (DeKAF) Prospective Cohort Study. Clin. Transplant. 2021, 35, e14337. [Google Scholar] [CrossRef]
- Malik, M.S.; Akoh, J.; Houlberg, K. Exploring the Risk Factors and Preventable Measures for Early Graft Loss in Kidney Transplantation: A Single-Center Analysis in the Southwest of the United Kingdom. Exp. Clin. Transplant. 2023, 21, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Zulkhash, N.; Shanazarov, N.; Kissikova, S.; Kamelova, G.; Ospanova, G. Review of Prognostic Factors for Kidney Transplant Survival. Urologia 2023, 90, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Oomen, L.; de Wall, L.L.; Cornelissen, E.A.M.; Feitz, W.F.J.; Bootsma-Robroeks, C.M.H.H.T. Prognostic Factors on Graft Function in Pediatric Kidney Recipients. Transplant. Proc. 2021, 53, 889–896. [Google Scholar] [CrossRef]
- Truchot, A.; Raynaud, M.; Kamar, N.; Naesens, M.; Legendre, C.; Delahousse, M.; Thaunat, O.; Buchler, M.; Crespo, M.; Linhares, K.; et al. Machine Learning Does Not Outperform Traditional Statistical Modelling for Kidney Allograft Failure Prediction. Kidney Int. 2023, 103, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Vagliano, I.; Chesnaye, N.C.; Leopold, J.H.; Jager, K.J.; Abu-Hanna, A.; Schut, M.C. Machine Learning Models for Predicting Acute Kidney Injury: A Systematic Review and Critical Appraisal. Clin. Kidney J. 2022, 15, 2266–2280. [Google Scholar] [CrossRef] [PubMed]
- Solez, K.; Eknoyan, G. Transplant Nephropathology: Wherefrom, Wherein, and Whereto. Clin. Transplant. 2024, 38, e15309. [Google Scholar] [CrossRef] [PubMed]
- Tanveer, Y.; Arif, A.; Tsenteradze, T.; Anika, N.N.; Bakht, D.; Masood, Q.F.; Affaf, M.; Batool, W.; Yadav, I.; Gasim, R.W.; et al. Revolutionizing Heart Transplantation: A Multidisciplinary Approach to Xenotransplantation, Immunosuppression, Regenerative Medicine, Artificial Intelligence, and Economic Sustainability. Cureus 2023, 15, e46176. [Google Scholar] [CrossRef]

| AIM, STUDY DESIGN | VARIABLES | OUTCOMES | AUTHORS, YEAR, COUNTRY |
|---|---|---|---|
| Prediction of kidney graft survival—Original retrospective research | Model for ages 50 and under: CDD, CIT, CREA, DDM, DHT, ETH, HCV, KFR, PS, RA, REG, RDM, RFS, RMC. | The proposed models outperformed the US kidney allocation system’s Estimated Post-Transplant Survival (EPTS) model and other recent models, achieving a five-year concordance index of 0.724 compared to 0.697 for the EPTS. Key variables (per the Breiman–Cutler permutation importance) included the RA, RDM and KFR | Mark et al., 2019, USA, [28] |
| Model for ages 51 and older: CDD, CIT, COPD, CREA, DA, DHT, DIA, ETH, HCV, KFR, RA, RDM, RFS. | |||
| Facilitation and improved prediction of kidney transplant survival for individual donor–recipient pairs—Original retrospective research | CDD, COPD, DA, DBMI, DBT, DCREA, DDM, DETH, DHT, DHCV, DIA, DG, DSM, ETH, HLA, KFR, RA, RAI, RBMI, RBT, RDM, RG, RFS, RMC, RWT | The model enabled the accurate prediction of kidney transplant survival, offering critical support to medical professionals and transplant candidates in identifying the optimal donor–recipient pairing | Paquette et al., 2022, USA, [27] |
| To estimate the risk of kidney graft failure across three distinct temporal cohorts (within 1 year, within 5 years, and beyond 5 years post-transplant), based on the clinical and demographic characteristics of both donor and recipient—Original retrospective research | CDD, CIT, DA, DBMI, DCREA, DDM, DETH, DG, DHCV, DHT, DIA, DRC, ETH, HLA, KFR, PKT, PRA, RA, RBMI, RDM, RFS, RG, RHT, RMA, RVD, TT | ML algorithms can effectively predict graft survival by leveraging donor and recipient factors that are routinely collected during standard clinical care. Most important features included: HLA, DIA, KFR, and RDM | Naqvi et al., 2021, USA, [36] |
| To develop a risk index for application in pre-transplant decision-making processes—Original retrospective research | Cox model: DA, DHT, DIA, KFR, RA, RVD. | The methods demonstrate a moderate ability to differentiate patients at higher risk of graft failure and provide predictions of graft failure with a moderate level of accuracy | Senanayake et al., 2021, Australia, [49] |
| RSF model: CDD, DA, DCREA, DBMI, DDM, DETH, DIA, DHT, HLA, KFR, OG, PKT, PTX, RA, RSM, RVD | |||
| To develop a predictive model capable of accurately identifying kidneys at risk of being discarded—Original retrospective research | CDD, DA, DBMI, DBT, DCREA, DDM, DETH, DGC, DHT, DHCV, DMIN, DMA, DSM, DTTA, HBCA, HBSA, HBSAB, HTLV | Machine learning techniques, such as random forest, can enhance the accuracy of identifying kidneys at risk of discard | Barah et al., 2021, USA, [23] |
| Variable | Mark et al. 2019 (Ages 50 and Under) [28] | Mark et al. 2019 (Ages 51 and Above) [28] | Paquette et al. 2022 [27] | Naqvi et al. 2021 [36] | Senanayake et al. (Cox) 2021 [49] | Senanayake et al. (RSF) 2021 [49] | Barah et al. 2021 [23] |
|---|---|---|---|---|---|---|---|
| CDD | + | + | + | + | + | + | |
| COPD | + | + | |||||
| CREA | + | + | |||||
| CIT | + | + | + | ||||
| DA | + | + | + | + | + | + | |
| DBMI | + | + (weight and height analyzed separately) | + (only height) | + (weight and height analyzed separately) | |||
| DBT | + | + | |||||
| DDM | + | + | + | + | + (type) | ||
| DETH | + | + | + | + | |||
| DCREA | + | + | + | + | |||
| DHCV | + | + | + | ||||
| DG | + | + | + | ||||
| DHT | + | + | + | + | + | + | + |
| DIA | + | + | + | + | + | ||
| DMA | + | ||||||
| DMIN | + | ||||||
| DRC | + | + (only donor) | |||||
| DSM | + | + | |||||
| DTTA | + | ||||||
| ETH | + | + | + | + | |||
| HBCA | + | ||||||
| HBSA | + | ||||||
| HBSAB | + | ||||||
| HCV | + | + | |||||
| HLA | + | + | + | ||||
| HTLV | + | ||||||
| KDPI | + | ||||||
| KFR | + | + | + | + | + | + | |
| OG | + | ||||||
| PKT | + | + | |||||
| PRA | + | ||||||
| PS | + | ||||||
| PTX | + | ||||||
| RA | + | + | + | + | + | + | |
| RAI | + | ||||||
| RBMI | + | + (weight and height analyzed separately) | |||||
| RBT | + | ||||||
| REG | + | ||||||
| RG | + | + | |||||
| RFS | + | + | + | + | |||
| RHT | + | ||||||
| RMA | + | ||||||
| RMC | + | + | |||||
| RSM | + | ||||||
| RVD | + | + | + | ||||
| RDM | + | + | + (type) | + | |||
| RWT | + | ||||||
| TT | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mizera, J.; Pondel, M.; Kepinska, M.; Jerzak, P.; Banasik, M. Advancements in Artificial Intelligence for Kidney Transplantology: A Comprehensive Review of Current Applications and Predictive Models. J. Clin. Med. 2025, 14, 975. https://doi.org/10.3390/jcm14030975
Mizera J, Pondel M, Kepinska M, Jerzak P, Banasik M. Advancements in Artificial Intelligence for Kidney Transplantology: A Comprehensive Review of Current Applications and Predictive Models. Journal of Clinical Medicine. 2025; 14(3):975. https://doi.org/10.3390/jcm14030975
Chicago/Turabian StyleMizera, Jakub, Maciej Pondel, Marta Kepinska, Patryk Jerzak, and Mirosław Banasik. 2025. "Advancements in Artificial Intelligence for Kidney Transplantology: A Comprehensive Review of Current Applications and Predictive Models" Journal of Clinical Medicine 14, no. 3: 975. https://doi.org/10.3390/jcm14030975
APA StyleMizera, J., Pondel, M., Kepinska, M., Jerzak, P., & Banasik, M. (2025). Advancements in Artificial Intelligence for Kidney Transplantology: A Comprehensive Review of Current Applications and Predictive Models. Journal of Clinical Medicine, 14(3), 975. https://doi.org/10.3390/jcm14030975







