Mortality Prediction of Septic Patients in the Emergency Department Based on Machine Learning
Abstract
:1. Introduction
2. Methods
2.1. Study Design
2.2. Study Setting and Population
2.3. Dataset Creation and Definition
2.4. Data Processing
2.5. Outcome
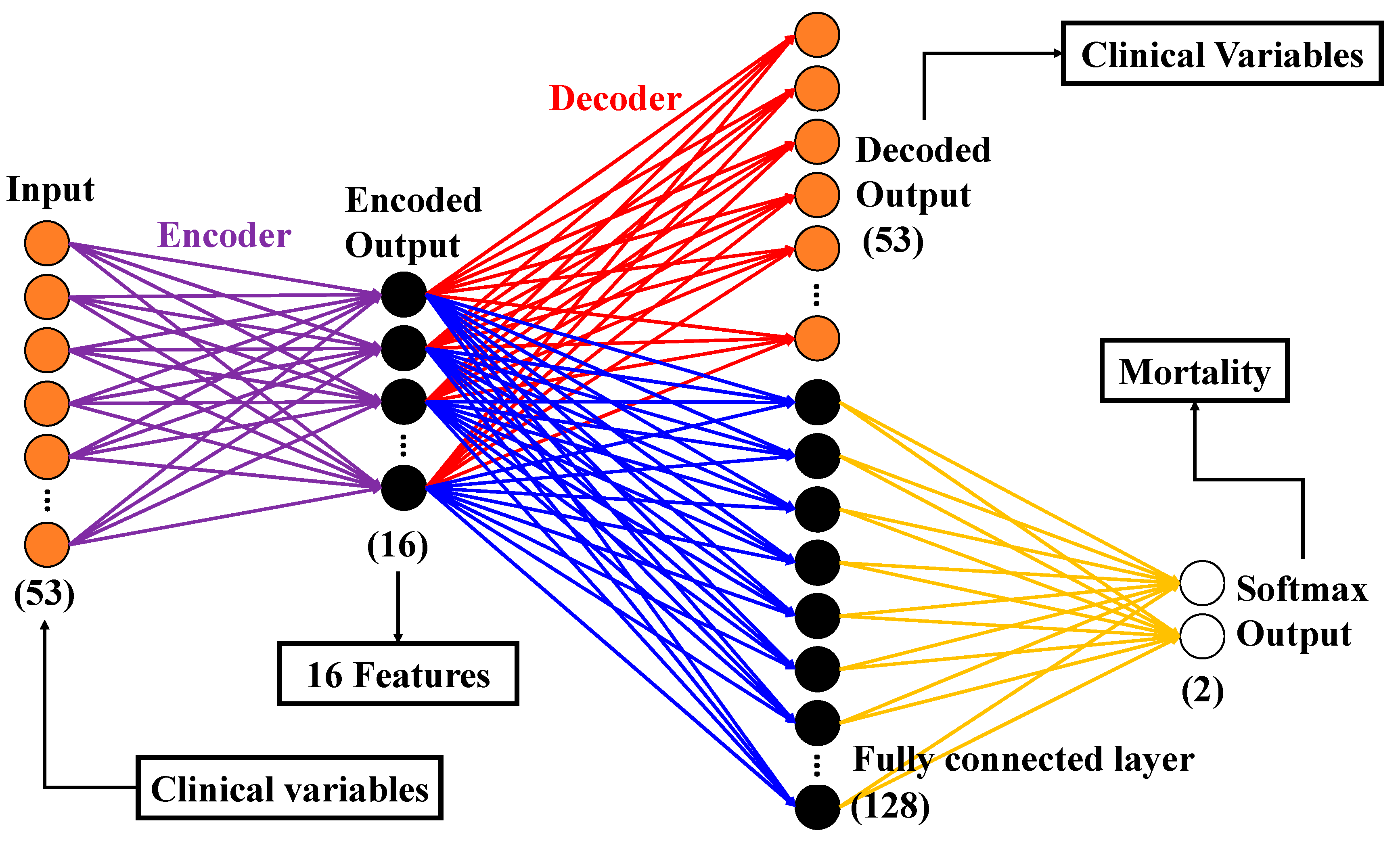
2.6. Machine Learning Model
2.6.1. Autoencoder (AE)
2.6.2. Convolutional Neural Network (CNN)
2.6.3. PCA
2.6.4. Classification Models
2.7. Statistics
3. Results
3.1. Patient Management Results
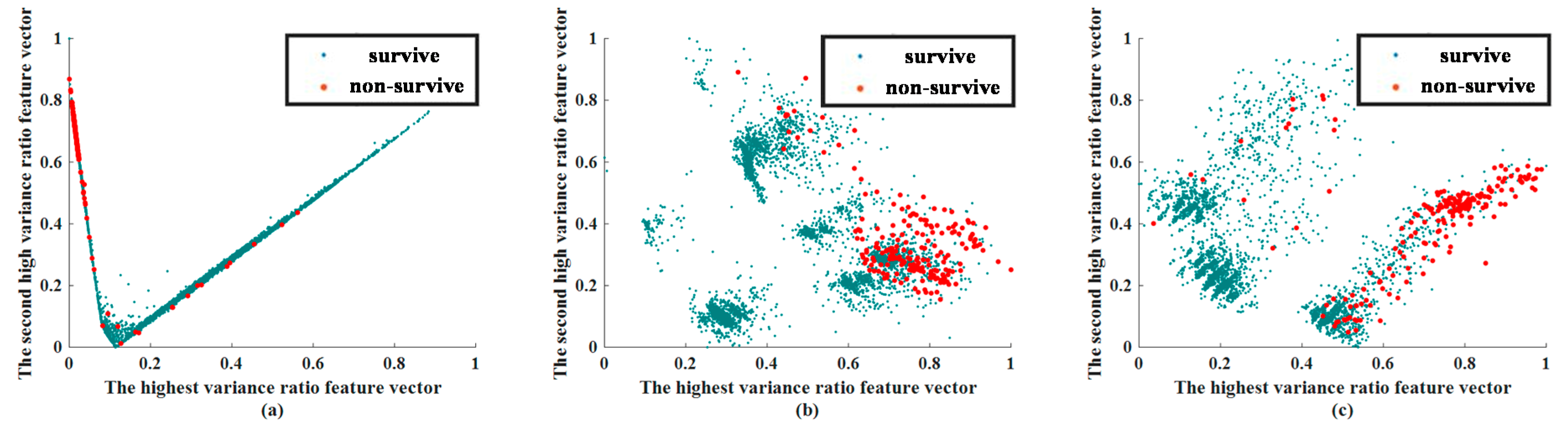
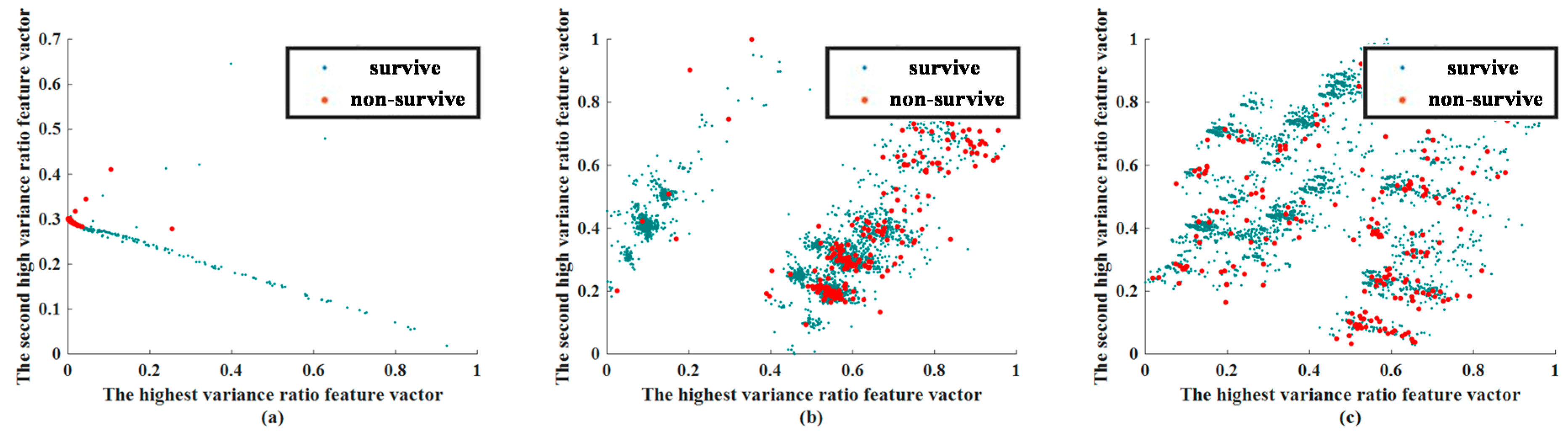
3.2. Feature Extraction Results
3.3. Classification Results
3.4. Importance of Feature
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
| No. | Clinical Variables | Survived | Non-Surviving | p | Miss (%) |
|---|---|---|---|---|---|
| 1 | Blood Pressure | 139.76 ± 32.48 | 121.17 ± 43.80 | <0.001 | 0 |
| 2 | Triage | <0.001 | 0 | ||
| 1 | 6.3% | 27.3% | |||
| 2 | 33.4% | 40.7% | |||
| 3 | 57.0% | 29.9% | |||
| 4 | 3.1% | 2.0% | |||
| 5 | 0.2% | 0.1% | |||
| 3 | GCS (E) | <0.001 | 0 | ||
| 1 | 2.1% | 14.2% | |||
| 2 | 2.5% | 8.1% | |||
| 3 | 3.4% | 7.9% | |||
| 4 | 92.0% | 69.8% | |||
| 4 | GCS (V) | <0.001 | 0 | ||
| 1 | 8.4% | 27.5% | |||
| 2 | 3.5% | 9.3% | |||
| 3 | 1.4% | 3.0% | |||
| 4 | 3.0% | 5.7% | |||
| 5 | 83.7% | 54.5% | |||
| 5 | GCS (M) | <0.001 | 0 | ||
| 1 | 0.9% | 10.2% | |||
| 2 | 1.1% | 4.4% | |||
| 3 | 2.8% | 6.8% | |||
| 4 | 4.8% | 10.9% | |||
| 5 | 5.5% | 11.6% | |||
| 6 | 84.9% | 56.2% | |||
| 6 | WBC | 11.32 ± 8.36 | 14.29 ± 16.95 | <0.001 | 0.017 |
| 7 | Hb | 12.01 ± 2.39 | 10.74 ± 2.61 | <0.001 | 0.008 |
| Seg | 76.98 ± 13.37 | 77.96 ± 16.84 | <0.001 | 0.064 | |
| 9 | Lymph | 14.81 ± 10.67 | 12.02 ± 12.57 | <0.001 | 0.067 |
| 10 | PT-INR | 1.21 ± 0.55 | 1.58 ± 0.93 | <0.001 | 21.94 |
| 11 | BUN | 24.76 ± 24.33 | 45.21 ± 37.56 | <0.001 | 0 |
| 12 | Cr | 1.47 ± 1.79 | 2.29 ± 2.30 | <0.001 | 0 |
| 13 | Bil | 2.42 ± 3.74 | 6.13 ± 8.51 | <0.001 | 20.16 |
| 14 | AST | 73.29 ± 258.18 | 258.10 ± 1099.87 | <0.001 | 16.46 |
| 15 | ALT | 47.24 ± 129.98 | 105.73 ± 364.09 | <0.001 | 0 |
| 16 | Troponin I | 0.33 ± 3.20 | 1.43 ± 8.52 | <0.001 | 24.21 |
| 17 | pH | 7.40 ± 0.11 | 7.33 ± 0.18 | <0.001 | 22.79 |
| 18 | HCO3 | 23.41 ± 6.49 | 20.49 ± 8.01 | <0.001 | 22.79 |
| 19 | Atypical lymphocyte | 0.078 ± 0.51 | 0.18 ± 0.66 | <0.001 | 0.067 |
| 20 | Promyelocyte | 0.0071 ± 0.45 | 0.044 ± 1.23 | <0.001 | 0.067 |
| 21 | Metamyelocyte | 0.11 ± 0.51 | 0.55 ± 1.60 | <0.001 | 0.067 |
| 22 | Myelocyte | 0.15 ± 0.71 | 0.61 ± 1.53 | <0.001 | 0.001 |
| 23 | Sodium ion | 135.48 ± 5.60 | 134.51 ± 8.67 | <0.001 | 0.067 |
| 24 | Potassium ion | 3.91 ± 0.71 | 4.30 ± 1.14 | <0.001 | 0 |
| 25 | Albumin | 2.99 ± 0.72 | 2.55 ± 0.63 | <0.001 | 25.79 |
| 26 | Sugar | 163.16 ± 106.24 | 192.04 ± 167.08 | <0.001 | 15.10 |
| 27 | RDW-SD | 46.46 ± 7.54 | 53.69 ± 11.54 | <0.001 | 0.012 |
| 28 | MCV | 88.44 ± 8.14 | 90.35 ± 9.26 | <0.001 | 0.011 |
| 29 | RDW-CV | 14.49 ± 22.24 | 16.50 ± 3.27 | <0.001 | 0.012 |
| 30 | Base excess | −1.12 ± 6.44 | −5.08 ± 9.15 | <0.001 | 22.79 |
| 31 | MCH | 29.53 ± 3.16 | 29.91 ± 3.35 | <0.001 | 0.010 |
| 32 | MCHC | 33.36 ± 1.39 | 33.11 ± 1.71 | <0.001 | 0.011 |
| 33 | MAP | 101.14 ± 26.83 | 88.25 ± 32.54 | <0.001 | 0.011 |
| 34 | RR | 19.58 ± 2.77 | 20.25 ± 6.04 | <0.001 | 0 |
| 35 | Temperature | 37.37 ± 1.26 | 36.46 ± 4.21 | <0.001 | 0.001 |
| 36 | Heart rate | 99.90 ± 23.00 | 102.66 ± 33.95 | <0.001 | 0 |
| 37 | Age | 61.05 ± 18.11 | 68.52 ± 15.02 | <0.001 | 0 |
| 38 | Sex (male%) | 52.6% | 61.3% | <0.001 | 0 |
| 39 | qSOFA Score | <0.001 | 0 | ||
| 0 | 69.7% | 33.1% | 0 | ||
| 1 | 23.7% | 38.1% | 0 | ||
| 2 | 6.0% | 23.9% | 0 | ||
| 3 | 0.6% | 4.8% | 0 | ||
| 40 | Shock episode | 2.5% | 26.8% | <0.001 | 0 |
| 41 | Liver cirrhosis | 6.9% | 17.6% | <0.001 | 0 |
| 42 | DM | 25.2% | 26.4% | 0.029 | 0 |
| 43 | CRF | 10.3% | 28.1% | <0.001 | 0 |
| 44 | CHF | 4.5% | 9.1% | <0.001 | 0 |
| 45 | CVA | 8.6% | 12.5% | <0.001 | 0 |
| 46 | Solid tumor | 18.0% | 43.6% | <0.001 | 0 |
| 47 | RI | 66.0% | 48.9% | <0.001 | 0 |
| 48 | UTI | 21.1% | 15.7% | <0.001 | 0 |
| 49 | Soft tissue infection | 13.7% | 4.7% | <0.001 | 0 |
| 50 | Intra-abdominal infection | 11.2% | 10.6% | 0.141 | 0 |
| 51 | Other infection | 35.7% | 33.4% | <0.001 | 0 |
| 52 | Bacteremia | 8.1% | 16.5% | <0.001 | 0 |
| 53 | Antibiotic used within 24 h | 77.9% | 85.5% | <0.001 | 0 |
References
- Chen, F.-C.; Kung, C.T.; Cheng, H.H.; Cheng, C.Y.; Tsai, T.C.; Hsiao, S.Y.; Su, C.M. Quick Sepsis-related Organ Failure Assessment predicts 72-h mortality in patients with suspected infection. Eur. J. Emerg. Med. 2019, 26, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Askim, A.; Moser, F.; Gustad, L.T.; Stene, H.; Gundersen, M.; Åsvold, B.O.; Dale, J.; Bjørnsen, L.P.; Damås, J.K.; Solligård, E. Poor performance of quick-SOFA (qSOFA) score in predicting severe sepsis and mortality—A prospective study of patients admitted with infection to the emergency department. Scand. J. Trauma Resusc. Emerg. Med. 2017, 25, 56. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.M.; Greenslade, J.H.; McKenzie, J.V.; Chu, K.; Brown, A.F.T.; Lipman, J. Systemic Inflammatory Response Syndrome, Quick Sequential Organ Function Assessment, and Organ Dysfunction: Insights from a Prospective Database of ED Patients with Infection. Chest 2017, 151, 586–596. [Google Scholar] [CrossRef]
- Award, A.; Bader-El-Den, M.; McNicholas, J.; Briggs, J. Early hospital mortality prediction of intensive care unit patients using an ensemble learning approach. Int. J. Med. Inf. 2017, 108, 185–195. [Google Scholar] [Green Version]
- Gupta, A.; Liu, T.; Shepherd, S.; Paiva, W. Using Statistical and Machine Learning Methods to Evaluate the Prognostic Accuracy of SIRS and qSOFA. Healthc. Inform. Res. 2018, 24, 139–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Layeghian, J.S.; Sepehri, M.M.; Layeghian Javan, M.; Khatibi, T. An intelligent warning model for early prediction of cardiac arrest in sepsis patients. Comput. Methods Programs Biomed. 2019, 178, 47–58. [Google Scholar] [CrossRef]
- Ribas, V.J. Severe Sepsis Mortality Prediction with Relevance Vector Machines. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, 30 August 2011; pp. 100–103. [Google Scholar]
- Zhou, W.; Yeh, C.; Jin, R.; Li, Z.; Song, S.; Yang, J. ISAR imaging of targets with rotating parts based on robust principal component analysis. IET Radar Sonar Navig. 2017, 11, 563–569. [Google Scholar] [CrossRef]
- Maligo, A.; Lacroix, S. Classification of Outdoor 3D Lidar Data Based on Unsupervised Gaussian Mixture Models. IEEE Trans. Autom. Sci. Eng. 2017, 14, 5–16. [Google Scholar] [CrossRef]
- Ahmad, A.; Dey, L. A k-mean clustering algorithm for mixed numeric and categorical data. Data Knowl. Eng. 2007, 63, 503–527. [Google Scholar] [CrossRef]
- Bawane, M.N.; Bhurchandi, K.M. Classification of Mental Task Based on EEG Processing Using Self Organising Feature Map. In Proceedings of the 2014 Sixth International Conference on Intelligent Human-Machine Systems and Cybernetics, Hangzhou, China, 26–27 August 2014; pp. 240–244. [Google Scholar]
- Jan, S.U.; Lee, Y.; Shin, J.; Koo, I. Sensor Fault Classification Based on Support Vector Machine and Statistical Time-Domain Features. IEEE Access 2017, 5, 8682–8690. [Google Scholar] [CrossRef]
- Jiang, M.; Jiang, L.; Jiang, D.; Xiong, J.; Shen, J.; Ahmed, S.H.; Luo, J.; Song, H. Dynamic measurement errors prediction for sensors based on firefly algorithm optimize support vector machine. Sustain. Cities Soc. 2017, 35, 250–256. [Google Scholar] [CrossRef]
- Szadkowski, Z.; Głas, D.; Pytel, K.; Wiedeński, M. Optimization of an FPGA Trigger Based on an Artificial Neural Network for the Detection of Neutrino-Induced Air Showers. IEEE Trans. Nucl. Sci. 2017, 64, 1271–1281. [Google Scholar] [CrossRef]
- Wu, F.Y.; Asada, H.H. Implicit and Intuitive Grasp Posture Control for Wearable Robotic Fingers: A Data-Driven Method Using Partial Least Squares. IEEE Trans. Robot. 2016, 32, 176–186. [Google Scholar] [CrossRef]
- Quan, H.; Sundararajan, V.; Halfon, P.; Fong, A.; Burnand, B.; Luthi, J.C.; Saunders, L.D.; Beck, C.A.; Feasby, T.E.; Ghali, W.A. Coding Algorithms for Defining Comorbidities in ICD-9-CM and ICD-10 Administrative Data. Med. Care 2005, 43, 1130–1139. [Google Scholar] [CrossRef]
- Bone, R.C.; Balk, R.A.; Cerra, F.B.; Dellinger, R.P.; Fein, A.M.; Knaus, W.A.; Schein, R.M.; Sibbald, W.J. Definitions for Sepsis and Organ Failure and Guidelines for the Use of Innovative Therapies in Sepsis. Chest 1992, 101, 1644–1655. [Google Scholar] [CrossRef] [Green Version]
- Seymour, C.W.; Liu, V.X.; Iwashyna, T.J.; Brunkhorst, F.M.; Rea, T.D.; Scherag, A.; Rubenfeld, G.; Kahn, J.M.; Shankar-Hari, M.; Singer, M.; et al. Assessment of clinical criteria for sepsis: For the third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA 2016, 315, 762–774. [Google Scholar] [CrossRef]
- Yu, Q.; Miche, Y.; Eirola, E.; van Heeswijk, M.; Severin, E.; Lendasse, A. Regularized extreme learning machine for regression with missing data. Neurocomputing 2013, 102, 45–51. [Google Scholar] [CrossRef]
- Jerez, J.M.; Molina, I.; García-Laencina, P.J.; Alba, E.; Ribelles, N.; Martín, M.; Franco, L. Missing data imputation using statistical and machine learning methods in a real breast cancer problem. Artif. Intell. Med. 2010, 50, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Seltzer, M.L.; Yu, D.; Acero, A.; Mohamed, A.; Hinton, G. Binary Coding of Speech Spectrograms Using a Deep Auto-Encoder. In Proceedings of the 11th Annual Conference of the International Speech Communication Association, Chiba, Japan, 26–30 September 2010. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv 2018, arXiv:1502.03167. [Google Scholar]
- Srivastava, N.; Hinton, G.; Kriszhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Kingma, D.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the 3rd International Conference for Learning Representations, San Diego, CA, USA, 7 May 2015. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Proceedings of the Advances in Neural Information Processing Systems, Lake Tahoe, NV, USA, 3 December 2012. [Google Scholar]
- Liu, G.; Yin, Z.; Jia, Y.; Xie, Y. Passenger flow estimation based on convolutional neural network in public transportation system. Knowl. Based Syst. 2017, 123, 102–115. [Google Scholar] [CrossRef]
- Kao, I.; Wang, W.; Lai, Y.; Perng, J. Analysis of Permanent Magnet Synchronous Motor Fault Diagnosis Based on Learning. IEEE Trans. Instrum. Meas. 2018, 68, 310–324. [Google Scholar] [CrossRef]
- Suykens, J.A.K.; Chua, L.O. n-Double Scroll Hypercubes in 1-D CNNs. Int. J. Bifurc. Chaos 1997, 07, 1873–1885. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Zhao, R.; Yan, R.; Shao, S.; Chen, X. Convolutional Discriminative Feature Learning for Induction Motor Fault Diagnosis. IEEE Trans. Ind. Inf. 2017, 13, 1350–1359. [Google Scholar] [CrossRef]
- Keller, J.M.; Gary, M.R.; Givens, J.A. A fuzzy K-nearest neighbor algorithm. IEEE Trans. Syst. Man Cybern. 1985, SMC-15, 580–585. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Liu, B.; Chen, Y.X.; Yin, Q.; Zhao, Y.Z.; Li, C.S. Diagnostic value and prognostic evaluation of Presepsin for sepsis in an emergency department. Crit. Care 2013, 17, R244. [Google Scholar] [CrossRef]
- Lee, C.; Chen, S.Y.; Tsai, C.L.; Wu, S.C.; Chiang, W.C.; Wang, J.L.; Sun, H.Y.; Chen, S.C.; Chen, W.J.; Hsueh, P.R. Prognostic value of mortality in emergency department sepsis score, procalcitonin, and C-reactive protein in patients with sepsis at the emergency department. Shock 2008, 29, 322–327. [Google Scholar] [CrossRef]
- Lichtenstern, C.; Brenner, T.; Bardenheuer, H.J.; Weigand, M.A. Predictors of survival in sepsis: What is the best inflammatory marker to measure? Curr. Opin. Infect. Dis. 2012, 25, 328–336. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, C.; Jia, Y. Evaluation of the Mortality in Emergency Department Sepsis score combined with procalcitonin in septic patients. Am. J. Emerg. Med. 2013, 31, 1086–1091. [Google Scholar] [CrossRef]
- Hermans, M.A.; Leffers, P.; Jansen, L.M.; Keulemans, Y.C.; Stassen, P.M. The value of the Mortality in Emergency Department Sepsis (MEDS) score, C reactive protein and lactate in predicting 28-day mortality of sepsis in a Dutch emergency department. Emerg. Med. J. 2012, 29, 295–300. [Google Scholar] [CrossRef]
- Goulden, R.; Hoyle, M.C.; Monis, J.; Railton, D.; Riley, V.; Martin, P.; Martina, R.; Nsutebu, E. qSOFA, SIRS and NEWS for predicting inhospital mortality and ICU admission in emergency admissions treated as sepsis. Emerg. Med. J. 2018, 35, 345–349. [Google Scholar] [CrossRef]
- Macdonald, S.P.; Arendts, G.; Fatovich, D.M.; Brown, S.G. Comparison of PIRO, SOFA, and MEDS scores for predicting mortality in emergency department patients with severe sepsis and septic shock. Acad. Emerg. Med. 2014, 21, 1257–1263. [Google Scholar] [CrossRef] [PubMed]
- Nathan, S.; Wolfe, R.E.; Moore, R.B.; Smith, E.; Burdick, E.; Bates, D.W. Mortality in Emergency Department Sepsis (MEDS) score: A prospectively derived and validated clinical prediction rule. Crit. Care Med. 2003, 31, 670–675. [Google Scholar]
- Taylor, R.A.; Pare, J.R.; Venkatesh, A.K.; Mowafi, H.; Melnick, E.R.; Fleischman, W.; Hall, M.K. Prediction of In-hospital Mortality in Emergency Department Patients with Sepsis: A Local Big Data-Driven, Machine Learning Approach. Acad. Emerg. Med. 2016, 23, 269–278. [Google Scholar] [CrossRef]
- Kao, I.H.; Hsu, Y.-W.; Yang, Y.-Z.; Chen, Y.-L.; Lai, Y.-H.; Perng, J.-W. Determination of Lycopersicon maturity using convolutional autoencoders. Sci. Hortic. 2019, 256, 108538. [Google Scholar] [CrossRef]
- Kao, I.H.; Hsu, Y.-W.; Lai, Y.-H.; Perng, J.-W. Laser Cladding Quality Monitoring Using Coaxial Image Based on Machine Learning. IEEE Trans. Instrum. Meas. Early Access 2019, 256, 1–11. [Google Scholar] [CrossRef]
- Gattinoni, L.; Vasques, F.; Camporota, L.; Meessen, J.; Romitti, F.; Pasticci, I.; Duscio, E.; Vassalli, F.; Forni, L.G.; Payen, D.; et al. Understanding Lactatemia in Human Sepsis. Potential Impact for Early Management. Am. J. Respir. Crit. Care Med. 2019, 200, 582–589. [Google Scholar] [CrossRef]
- Arayici, S.; Şimşek, G.K.; Canpolat, F.E.; Oncel, M.Y.; Uras, N.; Oguz, S.S. Can Base Excess be Used for Prediction to Early Diagnosis of Neonatal Sepsis in Preterm Newborns? Mediterr. J. Hematol. Infect. Dis. 2019, 11, 1–5. [Google Scholar] [CrossRef]
- Han, Y.Q.; Zhang, L.; Yan, L.; Li, P.; Ouyang, P.H.; Lippi, G.; Hu, Z.D. Red blood cell distribution width predicts long-term outcomes in sepsis patients admitted to the intensive care unit. Clin. Chim. Acta 2018, 487, 112–116. [Google Scholar] [CrossRef]


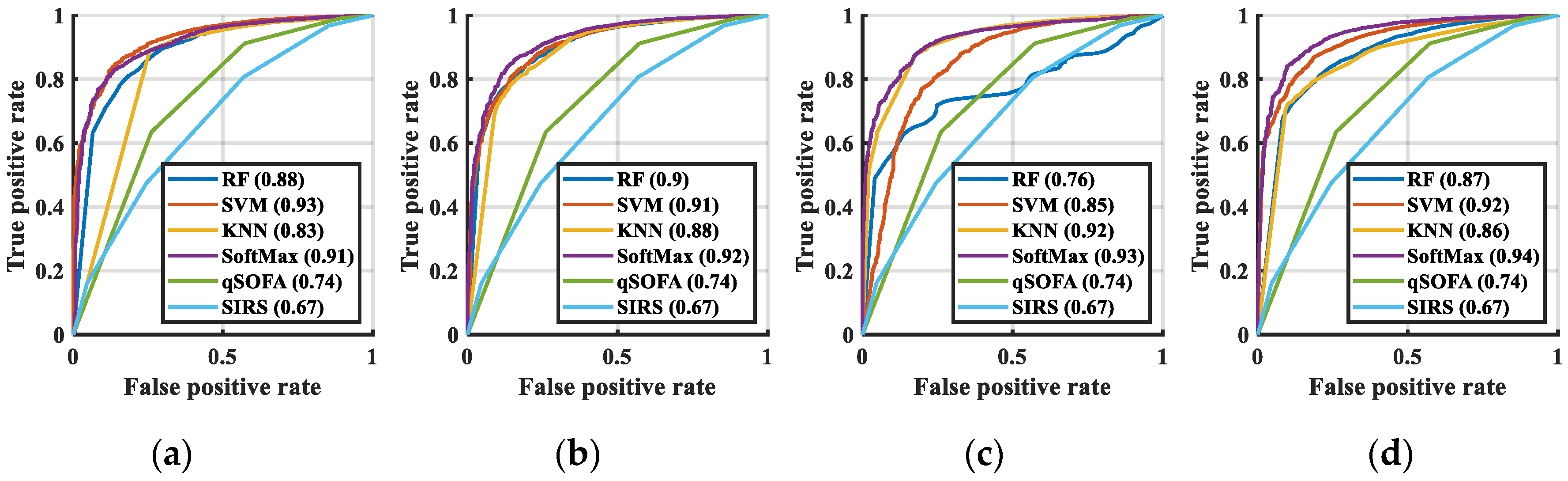

| Algorithms | AUC | SE | 95%CI | Compared with CNN + SoftMax | Acc (%) |
|---|---|---|---|---|---|
| SIRS | 0.67 | 0.0101 | 0.67–0.68 | p < 0.0001 | 59.43 |
| qSOFA | 0.74 | 0.0101 | 0.73–0.74 | p < 0.0001 | 67.27 |
| RF | 0.89 | 0.0067 | 0.88–0.89 | p < 0.0001 | 62.56 |
| KNN | 0.83 | 0.0087 | 0.83–0.84 | p < 0.0001 | 77.31 |
| SVM | 0.93 | 0.0044 | 0.92–0.93 | p < 0.0001 | 74.33 |
| SoftMax | 0.91 | 0.0052 | 0.91–0.92 | p < 0.0001 | 82.73 |
| PCA + RF | 0.90 | 0.0059 | 0.90–0.91 | p < 0.0001 | 62.62 |
| PCA + KNN | 0.88 | 0.0071 | 0.88–0.89 | p < 0.0001 | 81.67 |
| PCA + SVM | 0.91 | 0.0055 | 0.90–0.91 | p < 0.0001 | 78.91 |
| PCA + SoftMax | 0.92 | 0.0050 | 0.92–0.93 | p < 0.0001 | 83.48 |
| AE + RF | 0.77 | 0.0064 | 0.76–0.77 | p < 0.0001 | 63.52 |
| AE + KNN | 0.92 | 0.0053 | 0.91–0.92 | p < 0.0001 | 80.64 |
| AE + SVM | 0.85 | 0.0086 | 0.85–0.85 | p < 0.0001 | 78.76 |
| AE + SoftMax | 0.93 | 0.0042 | 0.92–0.93 | p < 0.0001 | 84.17 |
| CNN + RF | 0.87 | 0.0069 | 0.87–0.88 | p < 0.0001 | 61.03 |
| CNN + KNN | 0.86 | 0.0069 | 0.85–0.86 | p < 0.0001 | 81.73 |
| CNN + SVM | 0.92 | 0.0047 | 0.92–0.92 | p < 0.0001 | 84.96 |
| CNN + SoftMax | 0.94 | 0.0043 | 0.94–0.94 | None | 87.01 |
| Algorithms | AUC | SE | 95%CI | Compared with CNN + SoftMax | Acc (%) |
|---|---|---|---|---|---|
| SIRS | 0.59 | 0.0063 | 0.59–0.60 | p < 0.0001 | 59.43 |
| qSOFA | 0.68 | 0.0061 | 0.67–0.69 | p < 0.0001 | 67.27 |
| RF | 0.89 | 0.0032 | 0.89–0.89 | p < 0.0001 | 62.56 |
| KNN | 0.84 | 0.0047 | 0.83–0.84 | p < 0.0001 | 77.31 |
| SVM | 0.90 | 0.0031 | 0.89–0.90 | p < 0.0001 | 74.33 |
| SoftMax | 0.88 | 0.0034 | 0.90–0.89 | p < 0.0001 | 82.73 |
| PCA + RF | 0.89 | 0.0034 | 0.89–0.89 | p < 0.0001 | 62.62 |
| PCA + KNN | 0.84 | 0.0050 | 0.84–0.85 | p < 0.0001 | 81.67 |
| PCA + SVM | 0.89 | 0.0033 | 0.88–0.89 | p < 0.0001 | 78.91 |
| PCA + SoftMax | 0.91 | 0.0031 | 0.90–0.91 | p < 0.0001 | 83.48 |
| AE + RF | 0.84 | 0.0037 | 0.83–0.84 | p < 0.0001 | 63.52 |
| AE + KNN | 0.81 | 0.0042 | 0.81–0.82 | p < 0.0001 | 80.64 |
| AE + SVM | 0.89 | 0.0033 | 0.89–0.90 | p < 0.0001 | 78.76 |
| AE + SoftMax | 0.90 | 0.0032 | 0.89–0.90 | p < 0.0001 | 84.17 |
| CNN + RF | 0.90 | 0.0032 | 0.90–0.91 | p < 0.0001 | 61.03 |
| CNN + KNN | 0.86 | 0.0040 | 0.85–0.86 | p < 0.0001 | 81.73 |
| CNN + SVM | 0.92 | 0.0027 | 0.91–0.92 | p < 0.0001 | 84.96 |
| CNN + SoftMax | 0.92 | 0.0027 | 0.92–0.92 | None | 87.01 |
| Test 1 | Test 2 | Test 3 | Test 4 | ||||
|---|---|---|---|---|---|---|---|
| Feature | Importance | Feature | Importance | Feature | Importance | Feature | Importance |
| BE | 35.60 | BE | 39.50 | BE | 33.59 | BE | 36.50 |
| Shock episode | 12.89 | Shock episode | 11.86 | Shock episode | 13.89 | Shock episode | 13.00 |
| GCS (V) | 7.62 | ||||||
| ~ Lower than 5% ignored ~ | |||||||
| Test 1 | Test 2 | Test 3 | Test 4 | ||||
|---|---|---|---|---|---|---|---|
| Feature | Importance | Feature | Importance | Feature | Importance | Feature | Importance |
| BE | 20.39 | BE | 23.38 | BE | 19.88 | BE | 20.29 |
| RDW-SD | 9.07 | Solid tumor | 6.00 | RDW-SD | 10.11 | RDW-CV | 8.55 |
| RDW-CV | 5.53 | RDW-CV | 5.80 | Solid tumor | 5.55 | ||
| Solid tumor | 5.35 | RDW-SD | 5.43 | RDW-SD | 5.54 | ||
| ~ Lower than 5% ignored ~ | |||||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perng, J.-W.; Kao, I.-H.; Kung, C.-T.; Hung, S.-C.; Lai, Y.-H.; Su, C.-M. Mortality Prediction of Septic Patients in the Emergency Department Based on Machine Learning. J. Clin. Med. 2019, 8, 1906. https://doi.org/10.3390/jcm8111906
Perng J-W, Kao I-H, Kung C-T, Hung S-C, Lai Y-H, Su C-M. Mortality Prediction of Septic Patients in the Emergency Department Based on Machine Learning. Journal of Clinical Medicine. 2019; 8(11):1906. https://doi.org/10.3390/jcm8111906
Chicago/Turabian StylePerng, Jau-Woei, I-Hsi Kao, Chia-Te Kung, Shih-Chiang Hung, Yi-Horng Lai, and Chih-Min Su. 2019. "Mortality Prediction of Septic Patients in the Emergency Department Based on Machine Learning" Journal of Clinical Medicine 8, no. 11: 1906. https://doi.org/10.3390/jcm8111906
APA StylePerng, J.-W., Kao, I.-H., Kung, C.-T., Hung, S.-C., Lai, Y.-H., & Su, C.-M. (2019). Mortality Prediction of Septic Patients in the Emergency Department Based on Machine Learning. Journal of Clinical Medicine, 8(11), 1906. https://doi.org/10.3390/jcm8111906








