Interleukin-18 Is a Prognostic Biomarker Correlated with CD8+ T Cell and Natural Killer Cell Infiltration in Skin Cutaneous Melanoma
Abstract
:1. Introduction
2. Experimental Section
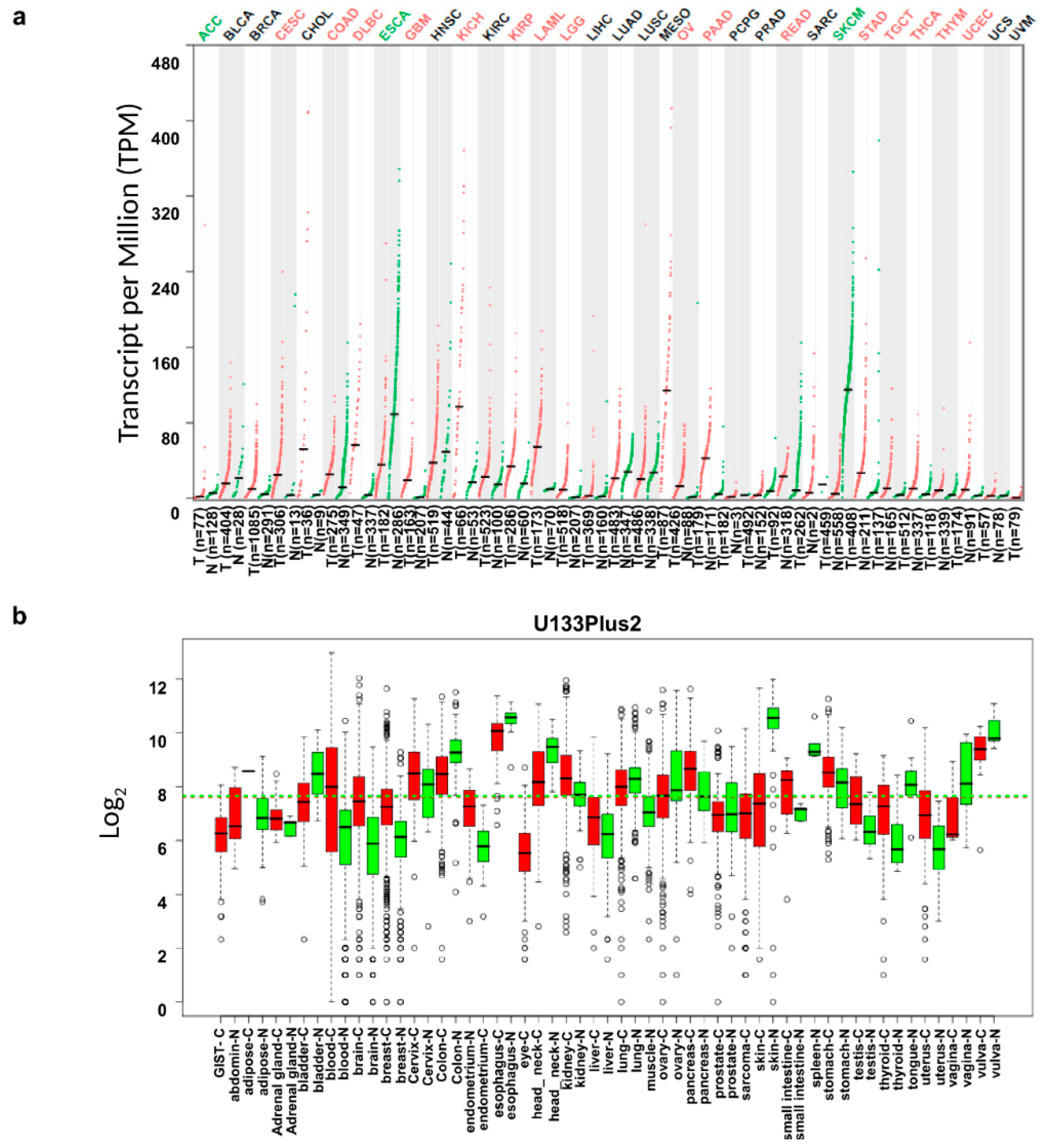
2.1. Comparison of Interleukin-18 (IL18) mRNA Expression in Various Types of Tumors and Their Normal Tissue Counterparts
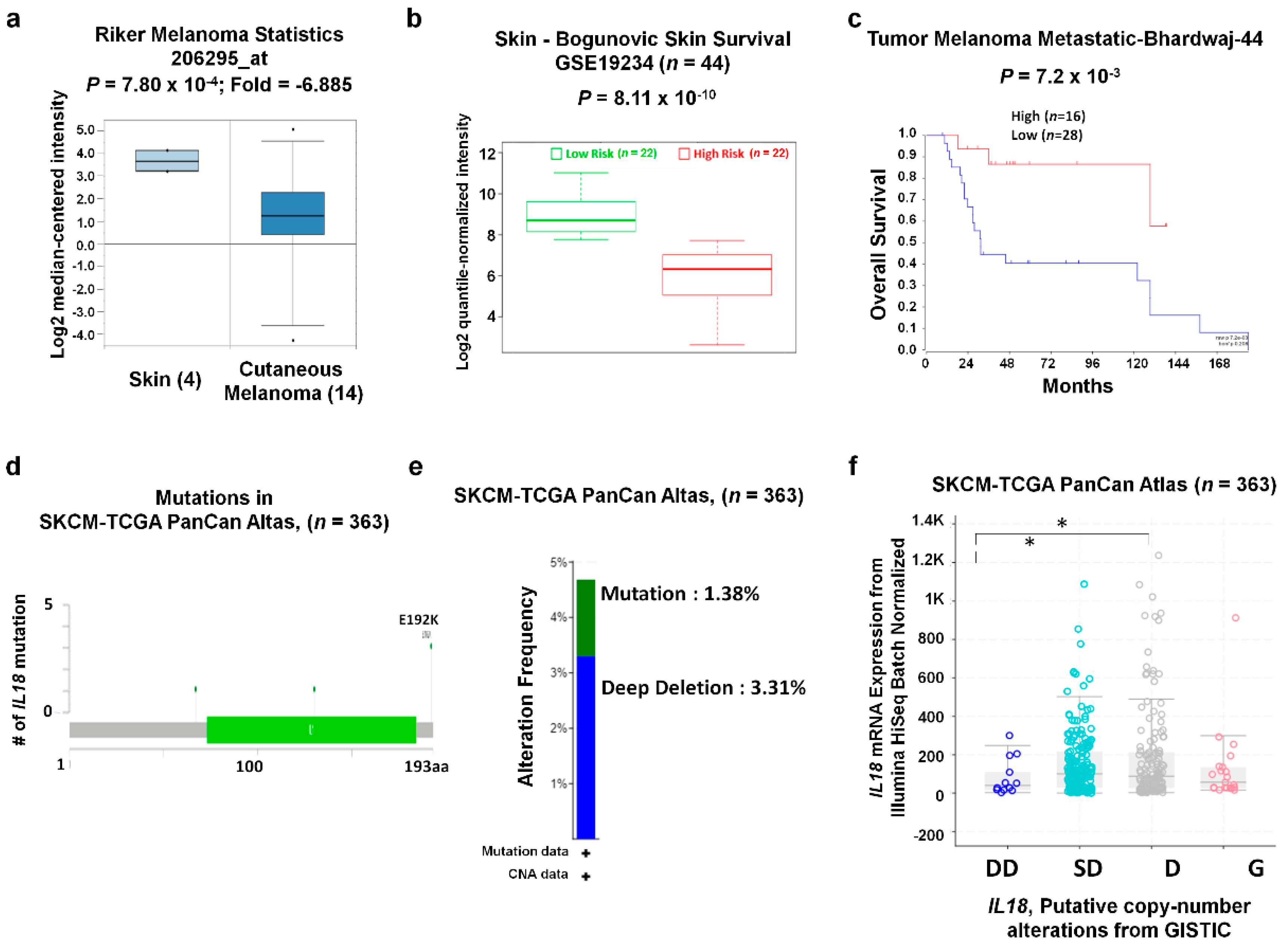
2.2. Analysis of Correlation between IL18 mRNA Expression and Patient Survival in Various Tumors
2.3. Analysis of IL18 Gene Mutations and Copy Number Alterations (CNA) in Skin Cutaneous Melanoma (SKCM)
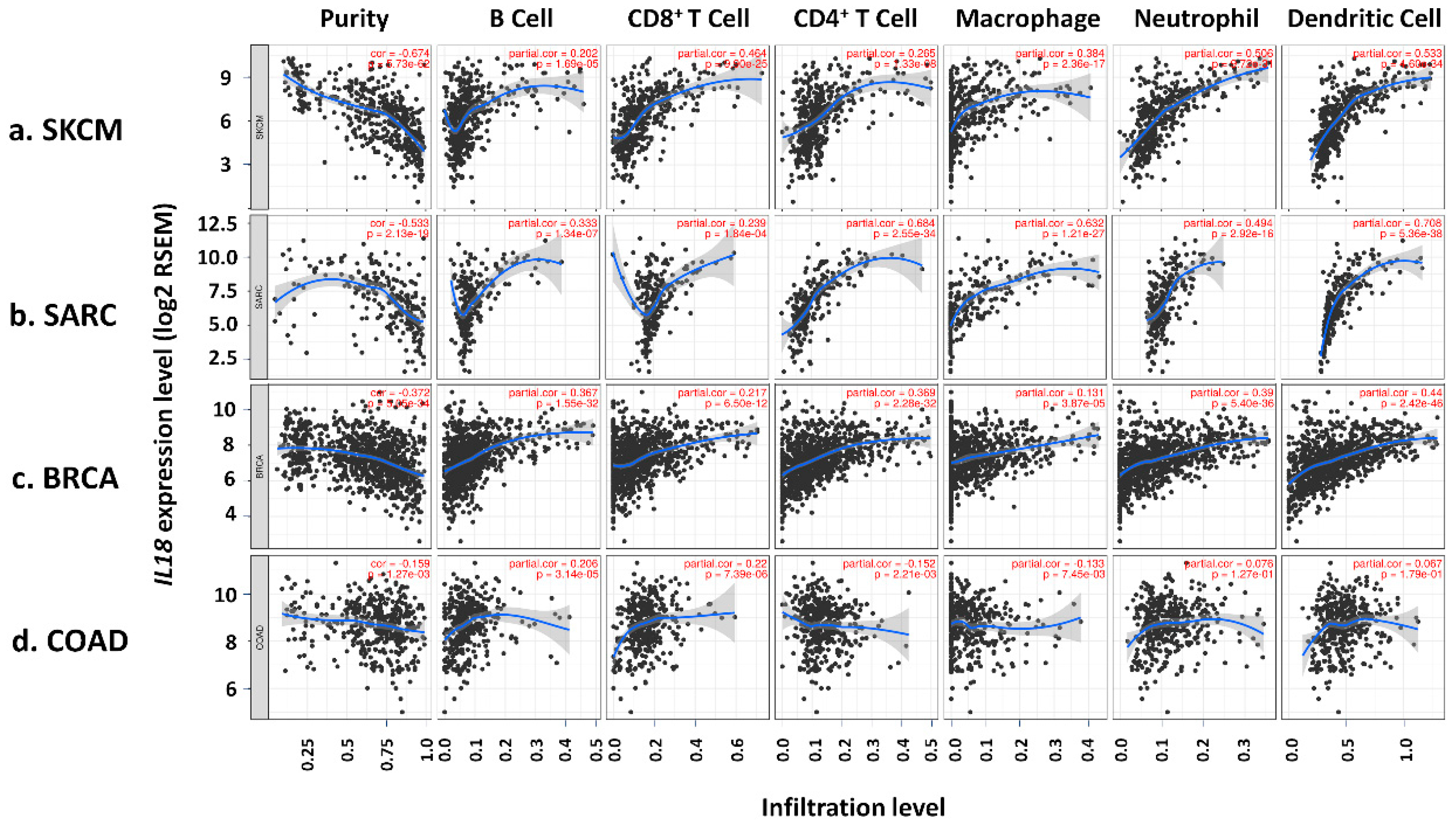
2.4. Analysis of the Correlation between IL18 Expression and the Immune Cell Infiltration
3. Results
3.1. IL18 mRNA Expression Levels in Various Types of Cancer
3.2. Correlation between IL18 Expression and Patient Survival Rates in Various Types of Cancers
3.3. IL18 Expression Pattern and Patient Survival in SKCM
3.4. Correlation of IL18 Expression with Immune Infiltrates in SKCM
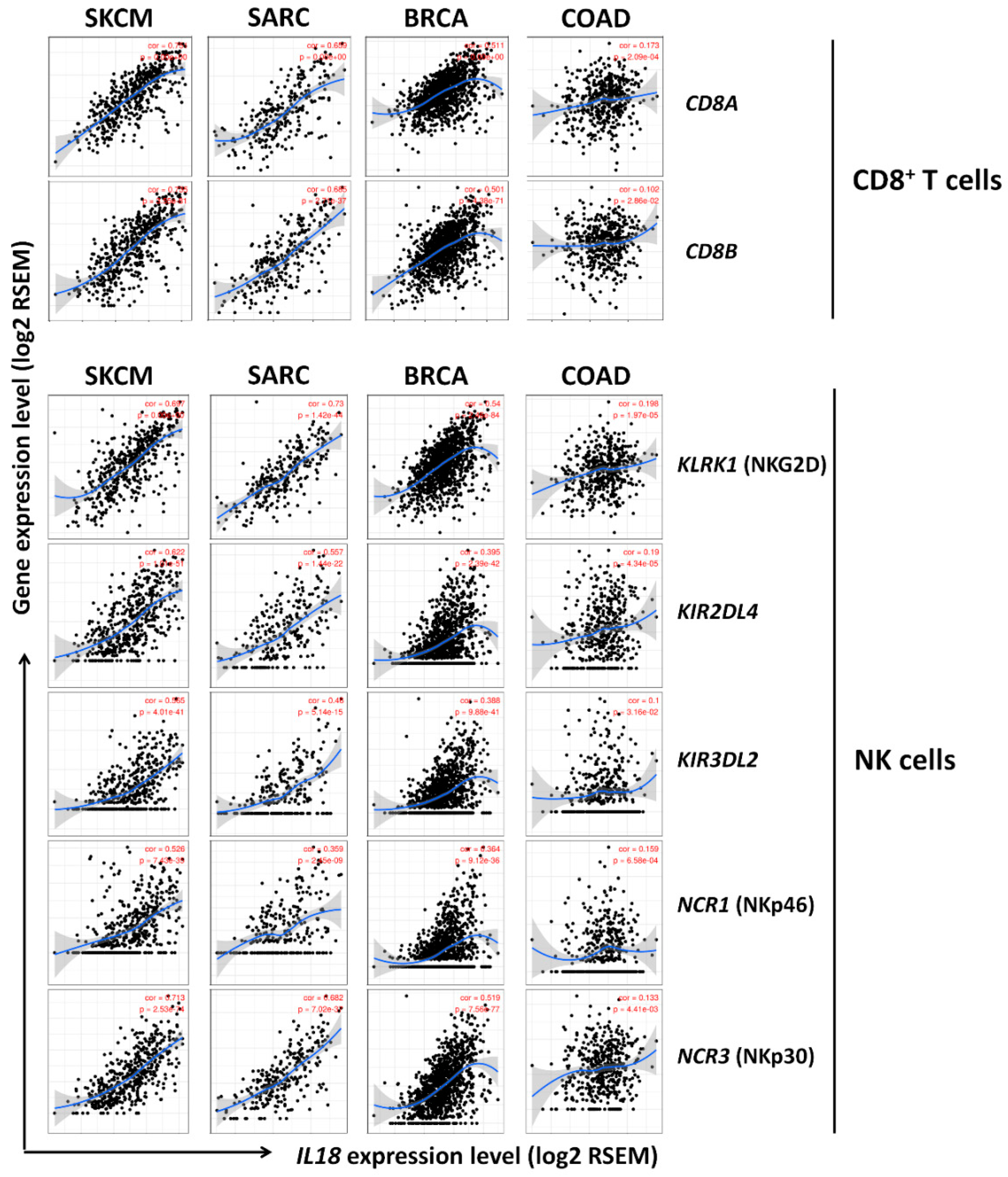
3.5. Correlation between IL18 Expression and Various Subsets of Immune Cells in Melanoma
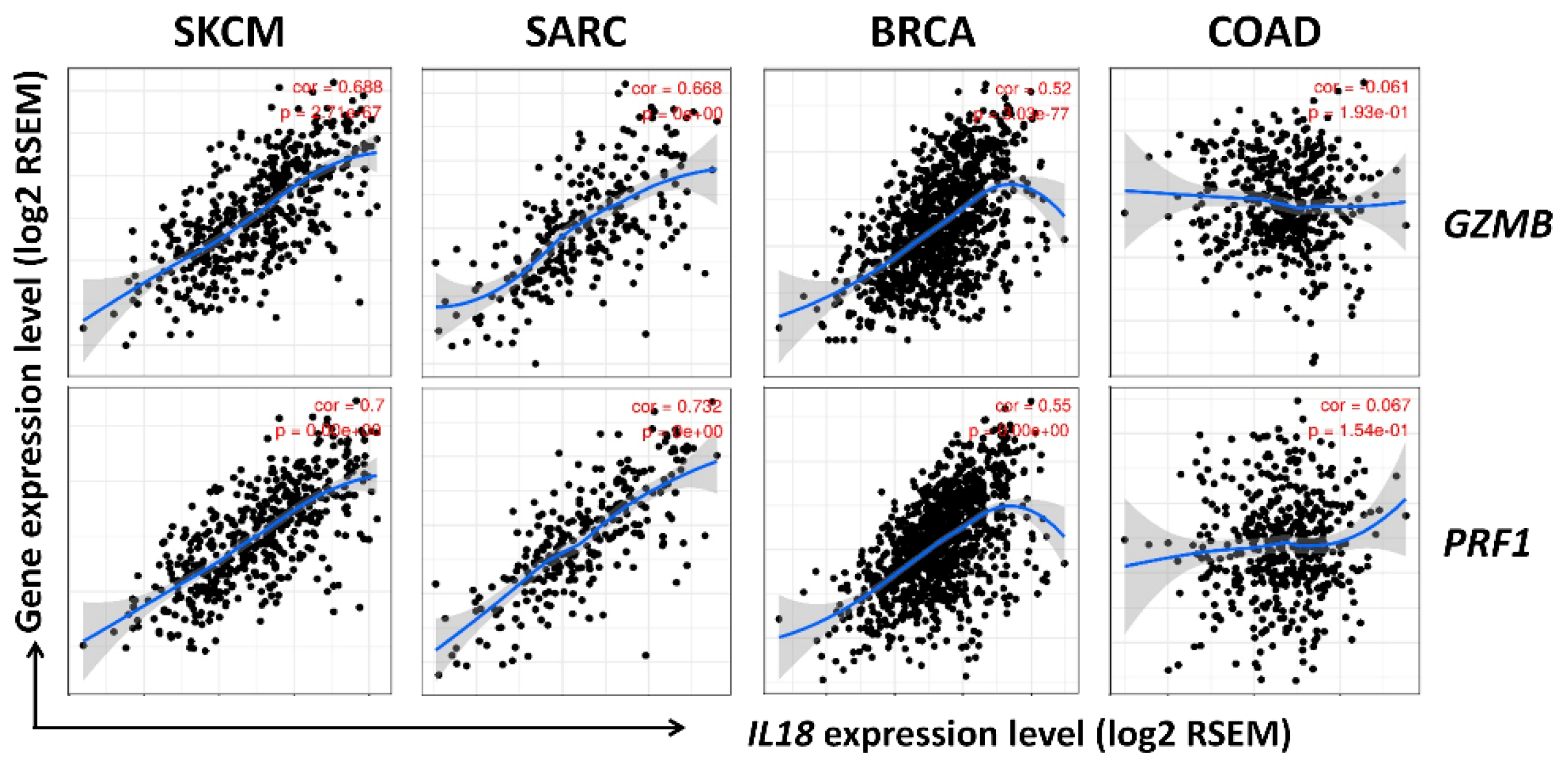
3.6. Correlation between IL18 Expression and Gene Expression of Cytolytic Molecules, Granzymeb and Perforin
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Nakamura, K.; Okamura, H.; Wada, M.; Nagata, K.; Tamura, T. Endotoxin-induced serum factor that stimulates gamma interferon production. Infect. Immun. 1989, 57, 590–595. [Google Scholar]
- Okamura, H.; Tsutsi, H.; Komatsu, T.; Yutsudo, M.; Hakura, A.; Tanimoto, T.; Torigoe, K.; Okura, T.; Nukada, Y.; Hattori, K.; et al. Cloning of a new cytokine that induces IFN-gamma production by T cells. Nature 1995, 378, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Puren, A.J.; Fantuzzi, G.; Dinarello, C.A. Gene expression, synthesis, and secretion of interleukin 18 and interleukin 1β are differentially regulated in human blood mononuclear cells and mouse spleen cells. Proc. Natl. Acad. Sci. USA 1999, 96, 2256–2261. [Google Scholar] [CrossRef] [PubMed]
- Lebel-Binay, S.; Berger, A.; Zinzindohoue, F.; Cugnenc, P.; Thiounn, N.; Fridman, W.H.; Pages, F. Interleukin-18: Biological properties and clinical implications. Eur. Cytokine Netw. 2000, 11, 15–26. [Google Scholar]
- Park, S.; Cheon, S.; Cho, D. The dual effects of interleukin-18 in tumor progression. Cell. Mol. Immunol. 2007, 4, 329–335. [Google Scholar] [PubMed]
- Vidal-Vanaclocha, F.; Fantuzzi, G.; Mendoza, L.; Fuentes, A.M.; Anasagasti, M.J.; Martin, J.; Carrascal, T.; Walsh, P.; Reznikov, L.L.; Kim, S.H.; et al. IL-18 regulates IL-1beta-dependent hepatic melanoma metastasis via vascular cell adhesion molecule-1. Proc. Natl. Acad. Sci. USA 2000, 97, 734–739. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Cheon, S.; Jung, M.K.; Song, S.B.; Kim, D.; Kim, H.J.; Park, H.; Bang, S.I.; Cho, D. Interleukin-18 enhances breast cancer cell migration via down-regulation of claudin-12 and induction of the p38 MAPK pathway. Biochem. Biophys. Res. Commun. 2015, 459, 379–386. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute’s Surveillance, Epidemiology, and End Results (SEER) Program Website. Available online: https://seer.cancer.gov/statfacts/html/melan.html (accessed on 1 October 2019).
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Soengas, M.S.; Lowe, S.W. Apoptosis and melanoma chemoresistance. Oncogene 2003, 22, 3138–3151. [Google Scholar] [CrossRef]
- Fu, Q.; Chen, N.; Ge, C.; Li, R.; Li, Z.; Zeng, B.; Li, C.; Wang, Y.; Xue, Y.; Song, X.; et al. Prognostic value of tumor-infiltrating lymphocytes in melanoma: A systematic review and meta-analysis. Oncoimmunology 2019, 8, 1593806. [Google Scholar] [CrossRef]
- Jung, M.K.; Song, H.K.; Kim, K.E.; Hur, D.Y.; Kim, T.; Bang, S.; Park, H.; Cho, D.H. IL-18 enhances the migration ability of murine melanoma cells through the generation of ROI and the MAPK pathway. Immunol. Lett. 2006, 107, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Kim, E.Y.; Yun, J.S.; Park, Y.L.; Do, S.I.; Chae, S.W.; Park, C.H. The prognostic and predictive value of tumor-infiltrating lymphocytes and hematologic parameters in patients with breast cancer. BMC Cancer 2018, 18, 938. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Zakka, L.R.; Mihm, M.C., Jr.; Schatton, T. Tumour-infiltrating lymphocytes in melanoma prognosis and cancer immunotherapy. Pathology 2016, 48, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, S.M.; Paish, E.C.; Powe, D.G.; Macmillan, R.D.; Grainge, M.J.; Lee, A.H.; Ellis, I.O.; Green, A.R. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J. Clin. Oncol. 2011, 29, 1949–1955. [Google Scholar] [CrossRef]
- Schatton, T.; Scolyer, R.A.; Thompson, J.F.; Mihm, M.C., Jr. Tumor-infiltrating lymphocytes and their significance in melanoma prognosis. Methods Mol. Biol. 2014, 1102, 287–324. [Google Scholar] [CrossRef]
- Letca, A.F.; Ungureanu, L.; Senila, S.C.; Grigore, L.E.; Pop, S.; Fechete, O.; Vesa, S.C.; Cosgarea, R. Regression and Sentinel Lymph Node Status in Melanoma Progression. Med. Sci. Monit. 2018, 24, 1359–1365. [Google Scholar] [CrossRef]
- De Moll, E.H.; Fu, Y.; Qian, Y.; Perkins, S.H.; Wieder, S.; Gnjatic, S.; Remark, R.; Bernardo, S.G.; Moskalenko, M.; Yao, J.; et al. Immune biomarkers are more accurate in prediction of survival in ulcerated than in non-ulcerated primary melanomas. Cancer Immunol. Immunother. 2015, 64, 1193–1203. [Google Scholar] [CrossRef]
- Reissfelder, C.; Stamova, S.; Gossmann, C.; Braun, M.; Bonertz, A.; Walliczek, U.; Grimm, M.; Rahbari, N.N.; Koch, M.; Saadati, M.; et al. Tumor-specific cytotoxic T lymphocyte activity determines colorectal cancer patient prognosis. J. Clin. Investig. 2015, 125, 739–751. [Google Scholar] [CrossRef]
- Seo, A.N.; Lee, H.J.; Kim, E.J.; Kim, H.J.; Jang, M.H.; Lee, H.E.; Kim, Y.J.; Kim, J.H.; Park, S.Y. Tumour-infiltrating CD8+ lymphocytes as an independent predictive factor for pathological complete response to primary systemic therapy in breast cancer. Br. J. Cancer 2013, 109, 2705–2713. [Google Scholar] [CrossRef]
- Konjevic, G.M.; Vuletic, A.M.; Mirjacic Martinovic, K.M.; Larsen, A.K.; Jurisic, V.B. The role of cytokines in the regulation of NK cells in the tumor environment. Cytokine 2019, 117, 30–40. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- GEPIA (Gene Expression Profiling Interactive Analysis). Available online: http://gepia.cancer-pku.cn/ (accessed on 1 October 2019).
- Shin, G.; Kang, T.W.; Yang, S.; Baek, S.J.; Jeong, Y.S.; Kim, S.Y. GENT: Gene expression database of normal and tumor tissues. Cancer Inform. 2011, 10, 149–157. [Google Scholar] [CrossRef] [PubMed]
- GENT (Gene Expression database across Normal and Tumor). Available online: http://medical-genome.kribb.re.kr/GENT/ (accessed on 1 October 2019).
- Vivian, J.; Rao, A.A.; Nothaft, F.A.; Ketchum, C.; Armstrong, J.; Novak, A.; Pfeil, J.; Narkizian, J.; Deran, A.D.; Musselman-Brown, A.; et al. Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 2017, 35, 314–316. [Google Scholar] [CrossRef]
- Bolstad, B.M.; Irizarry, R.A.; Astrand, M.; Speed, T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003, 19, 185–193. [Google Scholar] [CrossRef]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.R.; Kalyana-Sundaram, S.; Mahavisno, V.; Varambally, R.; Yu, J.; Briggs, B.B.; Barrette, T.R.; Anstet, M.J.; Kincead-Beal, C.; Kulkarni, P.; et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007, 9, 166–180. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.R.; Yu, J.; Shanker, K.; Deshpande, N.; Varambally, R.; Ghosh, D.; Barrette, T.; Pandey, A.; Chinnaiyan, A.M. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia 2004, 6, 1–6. [Google Scholar] [CrossRef]
- OncomineTM Research Edition. Available online: https://www.oncomine.org/resource/login.html (accessed on 1 October 2019).
- Anaya, J. OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput. Sci. 2016, 2, e67. [Google Scholar] [CrossRef]
- Anaya, J. OncoLnc. Available online: http://www.oncolnc.org/ (accessed on 1 October 2019).
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416. [Google Scholar] [CrossRef]
- Aguirre-Gamboa, R.; Gomez-Rueda, H.; Martinez-Ledesma, E.; Martinez-Torteya, A.; Chacolla-Huaringa, R.; Rodriguez-Barrientos, A.; Tamez-Pena, J.G.; Trevino, V. SurvExpress: An online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS ONE 2013, 8, e74250. [Google Scholar] [CrossRef]
- SurvExpress. Available online: http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp (accessed on 1 October 2019).
- R2: Genomics Analysis and Visualization Platform. Available online: http://r2.amc.nl or https://hgserver1.amc.nl/cgi-bin/r2/main.cgi (accessed on 1 October 2019).
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- cBioPortal for Cancer Genomics. Available online: http://www.cbioportal.org/ (accessed on 1 October 2019).
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017, 77, e108–e110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- TIMER: Tumor IMmune Estimation Resource. Available online: https://cistrome.shinyapps.io/timer/ (accessed on 1 October 2019).
- Pan, J.H.; Zhou, H.; Cooper, L.; Huang, J.L.; Zhu, S.B.; Zhao, X.X.; Ding, H.; Pan, Y.L.; Rong, L. LAYN Is a Prognostic Biomarker and Correlated With Immune Infiltrates in Gastric and Colon Cancers. Front. Immunol. 2019, 10, 6. [Google Scholar] [CrossRef] [Green Version]
- Gentles, A.J.; Newman, A.M.; Liu, C.L.; Bratman, S.V.; Feng, W.; Kim, D.; Nair, V.S.; Xu, Y.; Khuong, A.; Hoang, C.D.; et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015, 21, 938–945. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Tian, Z.; Wei, H. Developmental and Functional Control of Natural Killer Cells by Cytokines. Front. Immunol. 2017, 8, 930. [Google Scholar] [CrossRef]
- Baginska, J.; Viry, E.; Paggetti, J.; Medves, S.; Berchem, G.; Moussay, E.; Janji, B. The critical role of the tumor microenvironment in shaping natural killer cell-mediated anti-tumor immunity. Front. Immunol. 2013, 4, 490. [Google Scholar] [CrossRef] [Green Version]
- Boivin, W.A.; Cooper, D.M.; Hiebert, P.R.; Granville, D.J. Intracellular versus extracellular granzyme B in immunity and disease: Challenging the dogma. Lab. Investig. 2009, 89, 1195–1220. [Google Scholar] [CrossRef] [Green Version]
- Andersen, M.H.; Schrama, D.; Thor Straten, P.; Becker, J.C. Cytotoxic T cells. J. Investig. Dermatol. 2006, 126, 32–41. [Google Scholar] [CrossRef] [Green Version]
- Inoue, N.; Li, W.; Fujimoto, Y.; Matsushita, Y.; Katagiri, T.; Okamura, H.; Miyoshi, Y. High Serum Levels of Interleukin-18 Are Associated With Worse Outcomes in Patients With Breast Cancer. Anticancer Res. 2019, 39, 5009–5018. [Google Scholar] [CrossRef]
- Tas, F.; Tilgen Yasasever, C.; Karabulut, S.; Tastekin, D.; Duranyildiz, D. Clinical significance of serum interleukin-18 (IL-18) levels in patients with gastric cancer. Biomed. Pharmacother. 2015, 70, 19–23. [Google Scholar] [CrossRef]
- Carbone, A.; Vizio, B.; Novarino, A.; Mauri, F.A.; Geuna, M.; Robino, C.; Brondino, G.; Prati, A.; Giacobino, A.; Campra, D.; et al. IL-18 paradox in pancreatic carcinoma: Elevated serum levels of free IL-18 are correlated with poor survival. J. Immunother. 2009, 32, 920–931. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Byun, D.; Kim, T.S.; Kim, Y.I.; Kang, J.S.; Hahm, E.S.; Kim, S.H.; Lee, W.J.; Song, H.K.; Yoon, D.Y.; et al. Enhanced IL-18 expression in common skin tumors. Immunol. Lett. 2001, 79, 215–219. [Google Scholar] [CrossRef]
- Valcarcel, M.; Carrascal, T.; Crende, O.; Vidal-Vanaclocha, F. IL-18 regulates melanoma VLA-4 integrin activation through a Hierarchized sequence of inflammatory factors. J. Investig. Dermatol. 2014, 134, 470–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, S.; Salim, N.; Robertson, M.J. Interleukin-18: Biology and role in the immunotherapy of cancer. Curr. Med. Chem. 2010, 17, 3353–3357. [Google Scholar] [CrossRef]
- Mailliard, R.B.; Alber, S.M.; Shen, H.; Watkins, S.C.; Kirkwood, J.M.; Herberman, R.B.; Kalinski, P. IL-18-induced CD83+CCR7+ NK helper cells. J. Exp. Med. 2005, 202, 941–953. [Google Scholar] [CrossRef] [Green Version]
- Song, H.; Kim, K.E.; Hur, D.; Lim, J.S.; Yang, Y.; Cho, B.J.; Kim, C.H.; Kim, T.; Bang, S.; Lee, W.J.; et al. IL-18 enhances ULBP2 expression through the MAPK pathway in leukemia cells. Immunol. Lett. 2008, 120, 103–107. [Google Scholar] [CrossRef]
- Ni, J.; Miller, M.; Stojanovic, A.; Garbi, N.; Cerwenka, A. Sustained effector function of IL-12/15/18-preactivated NK cells against established tumors. J. Exp. Med. 2012, 209, 2351–2365. [Google Scholar] [CrossRef]
- Choi, I.K.; Lee, J.S.; Zhang, S.N.; Park, J.; Sonn, C.H.; Lee, K.M.; Yun, C.O. Oncolytic adenovirus co-expressing IL-12 and IL-18 improves tumor-specific immunity via differentiation of T cells expressing IL-12Rbeta2 or IL-18Ralpha. Gene. Ther. 2011, 18, 898–909. [Google Scholar] [CrossRef] [Green Version]
- Martinovic, K.M.; Babovic, N.; Dzodic, R.; Jurisic, V.; Matkovic, S.; Konjevic, G. Favorable in vitro effects of combined IL-12 and IL-18 treatment on NK cell cytotoxicity and CD25 receptor expression in metastatic melanoma patients. J. Transl. Med. 2015, 13, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, J.; Feige, K.; Wunderlin, P.; Hodl, A.; Meli, M.L.; Seltenhammer, M.; Grest, P.; Nicolson, L.; Schelling, C.; Heinzerling, L.M. Double-blind placebo-controlled study with interleukin-18 and interleukin-12-encoding plasmid DNA shows antitumor effect in metastatic melanoma in gray horses. J. Immunother. 2011, 34, 58–64. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Cao, H.; Liu, N.; Xu, K.; Ding, M.; Mao, L.J. Oncolytic adenovirus expressing interleukin-18 improves antitumor activity of dacarbazine for malignant melanoma. Drug Des. Dev. Ther. 2016, 10, 3755–3761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinarello, C.A. Interleukin-18. Methods 1999, 1, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Son, Y.I.; Dallal, R.M.; Mailliard, R.B.; Egawa, S.; Jonak, Z.L.; Lotze, M.T. Interleukin-18 (IL-18) synergizes with IL-2 to enhance cytotoxicity, interferon-gamma production, and expansion of natural killer cells. Cancer Res. 2001, 61, 884–888. [Google Scholar] [PubMed]
- Coughlin, C.M.; Salhany, K.E.; Wysocka, M.; Aruga, E.; Kurzawa, H.; Chang, A.E.; Hunter, C.A.; Fox, J.C.; Trinchieri, G.; Lee, W.M. Interleukin-12 and interleukin-18 synergistically induce murine tumor regression which involves inhibition of angiogenesis. J. Clin. Invest. 1998, 101, 1441–1452. [Google Scholar] [CrossRef] [PubMed]
- Dranoff, G. Cytokines in cancer pathogenesis and cancer therapy. Nat. Rev. Cancer 2004, 4, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Fergusson, J.R.; Smith, K.E.; Fleming, V.M.; Rajoriya, N.; Newell, E.W.; Simmons, R.; Marchi, E.; Bjorkander, S.; Kang, Y.H.; Swadling, L.; et al. CD161 defines a transcriptional and functional phenotype across distinct human T cell lineages. Cell Rep. 2014, 9, 1075–1088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiossone, L.; Dumas, P.Y.; Vienne, M.; Vivier, E. Natural killer cells and other innate lymphoid cells in cancer. Nat. Rev. Immunol. 2018, 18, 671–688. [Google Scholar] [CrossRef] [PubMed]
- Church, S.E.; Galon, J. Regulation of CTL Infiltration Within the Tumor Microenvironment. Adv. Exp. Med. Biol. 2017, 1036, 33–49. [Google Scholar] [CrossRef]
- Martinez-Lostao, L.; Anel, A.; Pardo, J. How Do Cytotoxic Lymphocytes Kill Cancer Cells? Clin. Cancer. Res. 2015, 21, 5047–5056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.C.; Lanier, L.L. NK cell development, homeostasis and function: Parallels with CD8(+) T cells. Nat. Rev. Immunol. 2011, 11, 645–657. [Google Scholar] [CrossRef]
- Zhao, Y.; Niu, C.; Cui, J. Gamma-delta (gammadelta) T cells: Friend or foe in cancer development? J. Transl. Med. 2018, 16, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, J.H.; Ley, T.J. Lymphocyte-mediated cytotoxicity. Annu. Rev. Immunol. 2002, 20, 323–370. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol. Biol. 2018, 1711, 243–259. [Google Scholar] [CrossRef]
- Cursons, J.; Souza-Fonseca-Guimaraes, F.; Foroutan, M.; Anderson, A.; Hollande, F.; Hediyeh-Zadeh, S.; Behren, A.; Huntington, N.D.; Davis, M.J. A Gene Signature Predicting Natural Killer Cell Infiltration and Improved Survival in Melanoma Patients. Cancer Immunol. Res. 2019, 7, 1162–1174. [Google Scholar] [CrossRef] [Green Version]
- Vantourout, P.; Hayday, A. Six-of-the-best: Unique contributions of gammadelta T cells to immunology. Nat. Rev. Immunol. 2013, 13, 88–100. [Google Scholar] [CrossRef] [Green Version]
- Maimela, N.R.; Liu, S.; Zhang, Y. Fates of CD8+ T cells in Tumor Microenvironment. Comput. Struct. Biotechnol. J. 2019, 17, 1–13. [Google Scholar] [CrossRef]
- Vivier, E.; Raulet, D.H.; Moretta, A.; Caligiuri, M.A.; Zitvogel, L.; Lanier, L.L.; Yokoyama, W.M.; Ugolini, S. Innate or adaptive immunity? The example of natural killer cells. Science 2011, 331, 44–49. [Google Scholar] [CrossRef] [Green Version]
- Terme, M.; Ullrich, E.; Aymeric, L.; Meinhardt, K.; Desbois, M.; Delahaye, N.; Viaud, S.; Ryffel, B.; Yagita, H.; Kaplanski, G.; et al. IL-18 induces PD-1-dependent immunosuppression in cancer. Cancer Res. 2011, 71, 5393–5399. [Google Scholar] [CrossRef] [Green Version]
- Park, I.H.; Yang, H.N.; Lee, K.J.; Kim, T.S.; Lee, E.S.; Jung, S.Y.; Kwon, Y.; Kong, S.Y. Tumor-derived IL-18 induces PD-1 expression on immunosuppressive NK cells in triple-negative breast cancer. Oncotarget 2017, 8, 32722–32730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herbst, R.S.; Soria, J.C.; Kowanetz, M.; Fine, G.D.; Hamid, O.; Gordon, M.S.; Sosman, J.A.; McDermott, D.F.; Powderly, J.D.; Gettinger, S.N.; et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014, 515, 563–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guirgis, H.M. The impact of PD-L1 on survival and value of the immune check point inhibitors in non-small-cell lung cancer; proposal, policies and perspective. J. Immunother. Cancer 2018, 6, 15. [Google Scholar] [CrossRef] [PubMed]

| Description | Gene Markers | SKMC | COAD | ||||||
|---|---|---|---|---|---|---|---|---|---|
| None | Purity | None | Purity | ||||||
| Cor | P | Cor | P | Cor | P | Cor | P | ||
| CD8+ T cell | CD8A | 0.748 | *** | 0.573 | *** | 0.166 | *** | 0.114 | * |
| CD8B | 0.732 | *** | 0.536 | *** | 0.070 | 0.047 | 0.041 | 0.249 | |
| T cell (general) | CD3D | 0.785 | *** | 0.596 | *** | 0.160 | *** | 0.104 | * |
| CD3E | 0.772 | *** | 0.572 | *** | 0.123 | ** | 0.056 | 0.114 | |
| CD2 | 0.805 | *** | 0.639 | *** | 0.218 | *** | 0.168 | *** | |
| B cell | CD19 | 0.547 | *** | 0.334 | *** | 0.108 | * | 0.048 | 0.175 |
| CD79A | 0.604 | *** | 0.374 | *** | 0.109 | * | 0.044 | 0.208 | |
| Monocyte | CD86 | 0.812 | *** | 0.677 | *** | 0.109 | * | 0.046 | 0.188 |
| CD115 (CSF1R) | 0.757 | *** | 0.615 | *** | −0.120 | * | −0.193 | *** | |
| TAM | CCL2 | 0.570 | *** | 0.330 | *** | −0.011 | 0.744 | −0.072 | 0.041 |
| CD68 | 0.471 | *** | 0.233 | *** | −0.048 | 0.174 | −0.100 | * | |
| IL10 | 0.678 | *** | 0.428 | *** | 0.100 | * | 0.057 | 0.106 | |
| M1 Macrophage | INOS (NOS2) | 0.030 | 0.524 | −0.086 | 0.065 | 0.235 | *** | 0.220 | *** |
| IRF5 | 0.623 | *** | 0.378 | *** | −0.149 | *** | −0.161 | *** | |
| COX2 (PTGS2) | 0.070 | 0.134 | −0.046 | 0.325 | 0.085 | 0.015 | 0.053 | 0.128 | |
| M2 Macrophage | CD163 | 0.618 | *** | 0.434 | *** | −0.018 | 0.605 | −0.089 | 0.011 |
| VSIG4 | 0.615 | *** | 0.459 | *** | −0.063 | 0.071 | −0.128 | 0.000 | |
| MS4A4A | 0.702 | *** | 0.540 | *** | 0.062 | 0.076 | 0.004 | 0.911 | |
| Neutrophils | CD66b (CEACAM8) | −0.070 | 0.136 | −0.052 | 0.263 | 0.087 | 0.013 | 0.105 | * |
| CD11b (ITGAM) | 0.661 | *** | 0.514 | *** | −0.114 | 0.001 | −0.194 | *** | |
| CCR7 | 0.688 | *** | 0.425 | *** | 0.068 | 0.053 | 0.003 | 0.928 | |
| Natural killer cell | KIR2DL1 | 0.359 | *** | 0.198 | *** | 0.071 | 0.043 | 0.042 | 0.228 |
| KIR2DL3 | 0.507 | *** | 0.284 | *** | −0.013 | 0.714 | −0.046 | 0.194 | |
| KIR2DL4 | 0.615 | *** | 0.421 | *** | 0.187 | *** | 0.146 | *** | |
| KIR3DL1 | 0.475 | *** | 0.264 | *** | 0.068 | 0.051 | 0.030 | 0.388 | |
| KIR3DL2 | 0.565 | *** | 0.340 | *** | 0.075 | 0.032 | 0.035 | 0.322 | |
| KIR3DL3 | 0.211 | *** | 0.149 | ** | 0.064 | 0.068 | 0.055 | 0.117 | |
| KIR2DS4 | 0.442 | *** | 0.295 | *** | 0.061 | 0.081 | 0.043 | 0.217 | |
| KLRK1 (NKG2D) | 0.697 | *** | 0.516 | *** | 0.198 | ** | 0.138 | * | |
| NCR1 (NKp46) | 0.526 | *** | 0.362 | *** | 0.159 | * | 0.095 | 0.055 | |
| NCR2 (NKp44) | 0.255 | *** | 0.179 | ** | 0.105 | 0.024 | 0.092 | 0.065 | |
| NCR3 (NKp30) | 0.713 | *** | 0.503 | *** | 0.133 | 0.004 | 0.066 | 0.187 | |
| Dendritic cell | HLA-DPB1 | 0.790 | *** | 0.626 | *** | 0.025 | 0.468 | −0.051 | 0.150 |
| HLA-DQB1 | 0.725 | *** | 0.537 | *** | 0.068 | 0.051 | 0.020 | 0.573 | |
| HLA-DRA | 0.808 | *** | 0.658 | *** | 0.148 | *** | 0.093 | 0.008 | |
| HLA-DPA1 | 0.764 | *** | 0.610 | *** | 0.101 | * | 0.039 | 0.269 | |
| BDCA1 (CD1C) | 0.612 | *** | 0.396 | *** | 0.069 | 0.048 | 0.019 | 0.596 | |
| BDCA4 (NRP1) | 0.372 | *** | 0.217 | *** | −0.037 | 0.286 | −0.120 | * | |
| CD11c (ITGAX) | 0.608 | *** | 0.356 | *** | −0.049 | 0.164 | −0.132 | ** | |
| Th1 | T-bet (TBX21) | 0.737 | *** | 0.536 | *** | 0.087 | 0.013 | 0.026 | 0.464 |
| STAT4 | 0.754 | *** | 0.589 | *** | 0.173 | *** | 0.123 | 0.000 | |
| STAT1 | 0.549 | *** | 0.388 | *** | 0.164 | *** | 0.126 | 0.000 | |
| IFNγ (IFNG) | 0.724 | *** | 0.560 | *** | 0.167 | *** | 0.135 | 0.000 | |
| TNFα (TNF) | 0.711 | *** | 0.531 | *** | 0.064 | 0.066 | 0.027 | 0.445 | |
| Th2 | GATA3 | 0.833 | *** | 0.686 | *** | −0.102 | 0.003 | −0.172 | *** |
| STAT6 | 0.007 | 0.886 | −0.026 | 0.586 | −0.079 | 0.024 | −0.069 | 0.048 | |
| STAT5A | 0.181 | *** | 0.168 | ** | −0.141 | *** | −0.170 | *** | |
| IL13 | 0.227 | *** | 0.127 | * | 0.046 | 0.186 | 0.017 | 0.633 | |
| Tfh | BCL6 | 0.383 | *** | 0.268 | *** | −0.054 | 0.126 | −0.121 | 0.001 |
| IL21 | 0.487 | *** | 0.348 | *** | 0.008 | 0.809 | −0.013 | 0.711 | |
| Th17 | STAT3 | 0.300 | *** | 0.211 | *** | 0.065 | 0.065 | 0.026 | 0.459 |
| IL17A | −0.009 | 0.856 | −0.091 | 0.051 | 0.099 | * | 0.100 | 0.004 | |
| Treg | FOXP3 | 0.690 | *** | 0.462 | *** | −0.076 | 0.029 | −0.152 | *** |
| CCR8 | 0.682 | *** | 0.513 | *** | −0.035 | 0.314 | −0.096 | 0.006 | |
| STAT5B | 0.226 | *** | 0.249 | *** | −0.323 | *** | −0.329 | *** | |
| TGFβ (TGFB1) | 0.431 | *** | 0.207 | *** | −0.024 | 0.486 | −0.103 | 0.003 | |
| T cell exhaustion | PD1 (PDCD1) | 0.740 | *** | 0.555 | *** | 0.078 | 0.026 | 0.019 | 0.586 |
| CTLA4 | 0.454 | *** | 0.199 | *** | 0.062 | 0.079 | −0.002 | 0.950 | |
| LAG3 | 0.703 | *** | 0.512 | *** | 0.094 | 0.008 | 0.037 | 0.290 | |
| TIM3 (HAVCR2) | 0.777 | *** | 0.610 | *** | 0.084 | 0.016 | 0.021 | 0.543 | |
| Cell Type | Gene Markers | SKCM | COAD | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Tumor | Normal | Tumor | Normal | ||||||
| R | P | R | P | R | P | R | P | ||
| CD8+ T cells | CD8A | 0.63 | *** | −0.11 | * | −0.11 | 0.067 | 0.41 | *** |
| CD8B | 0.63 | *** | −0.13 | * | −0.008 | 0.089 | 0.38 | *** | |
| NK cells | KIR2DL1 | 0.22 | *** | −0.091 | 0.031 | 0.015 | 0.8 | 0.1 | 0.053 |
| KIR2DL3 | 0.3 | *** | −0.07 | 0.097 | 0.006 | 0.92 | 0.21 | *** | |
| KIR2DL4 | 0.44 | *** | −0.075 | 0.079 | 0.11 | 0.064 | 0.66 | *** | |
| KIR3DL1 | 0.25 | *** | −0.1 | 0.013 | 0.087 | 0.15 | 0.24 | *** | |
| KIR3DL2 | 0.42 | *** | −0.11 | * | 0.043 | 0.48 | 0.43 | *** | |
| KIR3DL3 | 0.19 | *** | 0.06 | 0.16 | 0.032 | 0.6 | 0.12 | 0.02 | |
| KIR2DS4 | 0.16 | ** | −0.16 | ** | 0.015 | 0.8 | 0.18 | ** | |
| KLRK1 (NKG2D) | 0.63 | *** | −0.11 | * | 0.094 | 0.12 | 0.45 | *** | |
| NCR1 (NKp46) | 0.29 | *** | −0.086 | 0.043 | 0.081 | 0.19 | 0.15 | * | |
| NCR2 (NKp44) | 0.075 | 0.11 | −0.018 | 0.67 | 0.21 | ** | 0.38 | *** | |
| NCR3 (NKp30) | 0.46 | *** | −0.11 | 0.011 | 0.083 | 0.17 | 0.26 | *** | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gil, M.; Kim, K.E. Interleukin-18 Is a Prognostic Biomarker Correlated with CD8+ T Cell and Natural Killer Cell Infiltration in Skin Cutaneous Melanoma. J. Clin. Med. 2019, 8, 1993. https://doi.org/10.3390/jcm8111993
Gil M, Kim KE. Interleukin-18 Is a Prognostic Biomarker Correlated with CD8+ T Cell and Natural Killer Cell Infiltration in Skin Cutaneous Melanoma. Journal of Clinical Medicine. 2019; 8(11):1993. https://doi.org/10.3390/jcm8111993
Chicago/Turabian StyleGil, Minchan, and Kyung Eun Kim. 2019. "Interleukin-18 Is a Prognostic Biomarker Correlated with CD8+ T Cell and Natural Killer Cell Infiltration in Skin Cutaneous Melanoma" Journal of Clinical Medicine 8, no. 11: 1993. https://doi.org/10.3390/jcm8111993
APA StyleGil, M., & Kim, K. E. (2019). Interleukin-18 Is a Prognostic Biomarker Correlated with CD8+ T Cell and Natural Killer Cell Infiltration in Skin Cutaneous Melanoma. Journal of Clinical Medicine, 8(11), 1993. https://doi.org/10.3390/jcm8111993





