TRPV2 Calcium Channel Gene Expression and Outcomes in Gastric Cancer Patients: A Clinically Relevant Association
Abstract
:1. Introduction
2. Experimental Section
2.1. Selection of Ca2+ Regulator Genes
2.2. Collection of Gene Expression Datasets
2.3. Evaluation of Overall and Progression-Free Survival
2.4. Differential Gene Expression Analysis among Tumor Stages
2.5. Differential Gene Expression Analysis
2.6. Evaluation of Protein Expression in Tissue Sections
2.7. Statistical Analysis
3. Results
3.1. Role of Ca2+ Regulator Genes on Gastric Cancer Prognosis
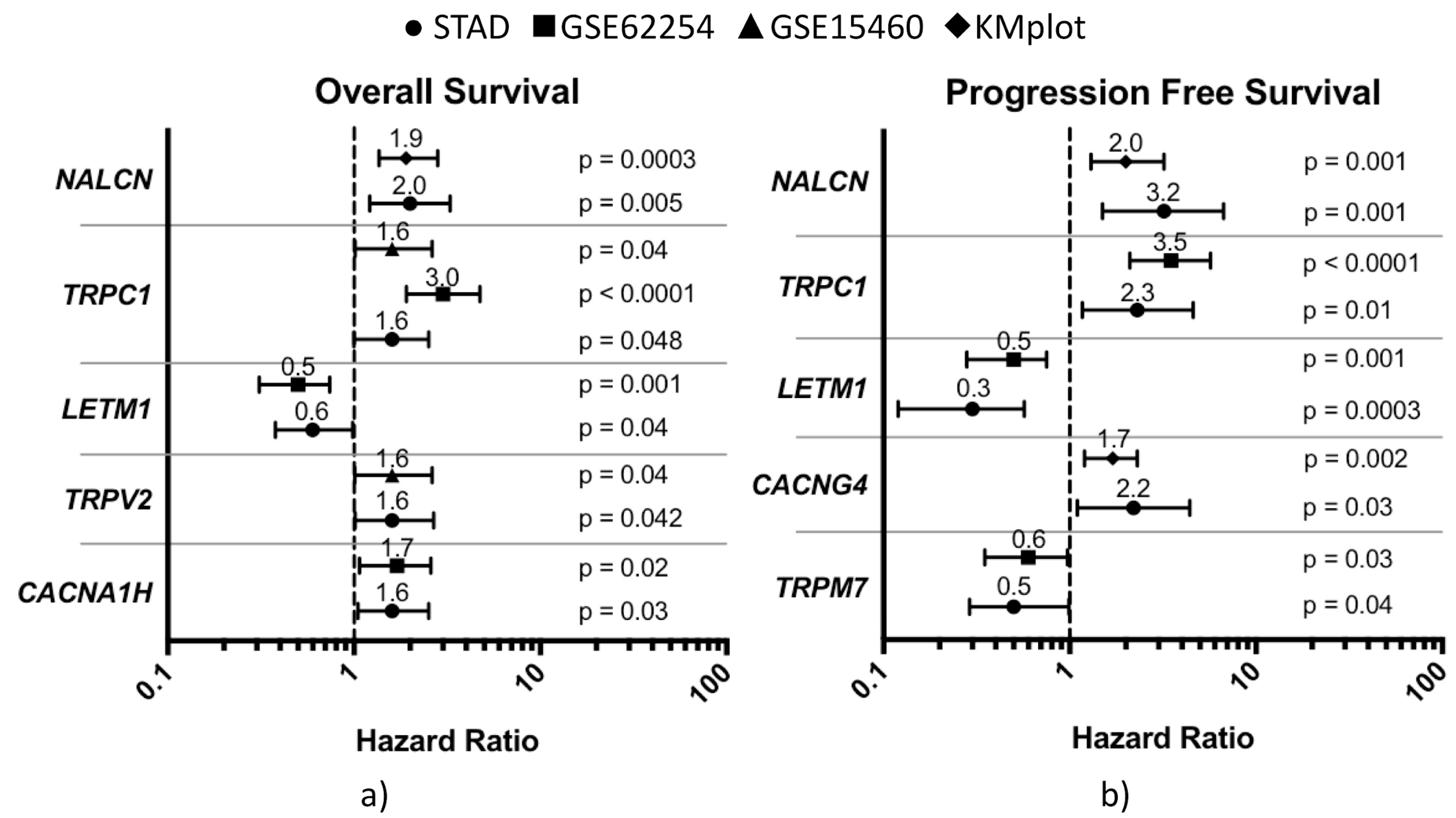
3.2. Cross-Match Validation of Prognostic CaRGs
3.3. Impact of CaRGs on OS and PFS of Different Clinicopathological Subgroups of Patients
3.4. Differential Prognostic CaRG Expression among Tumor Stages
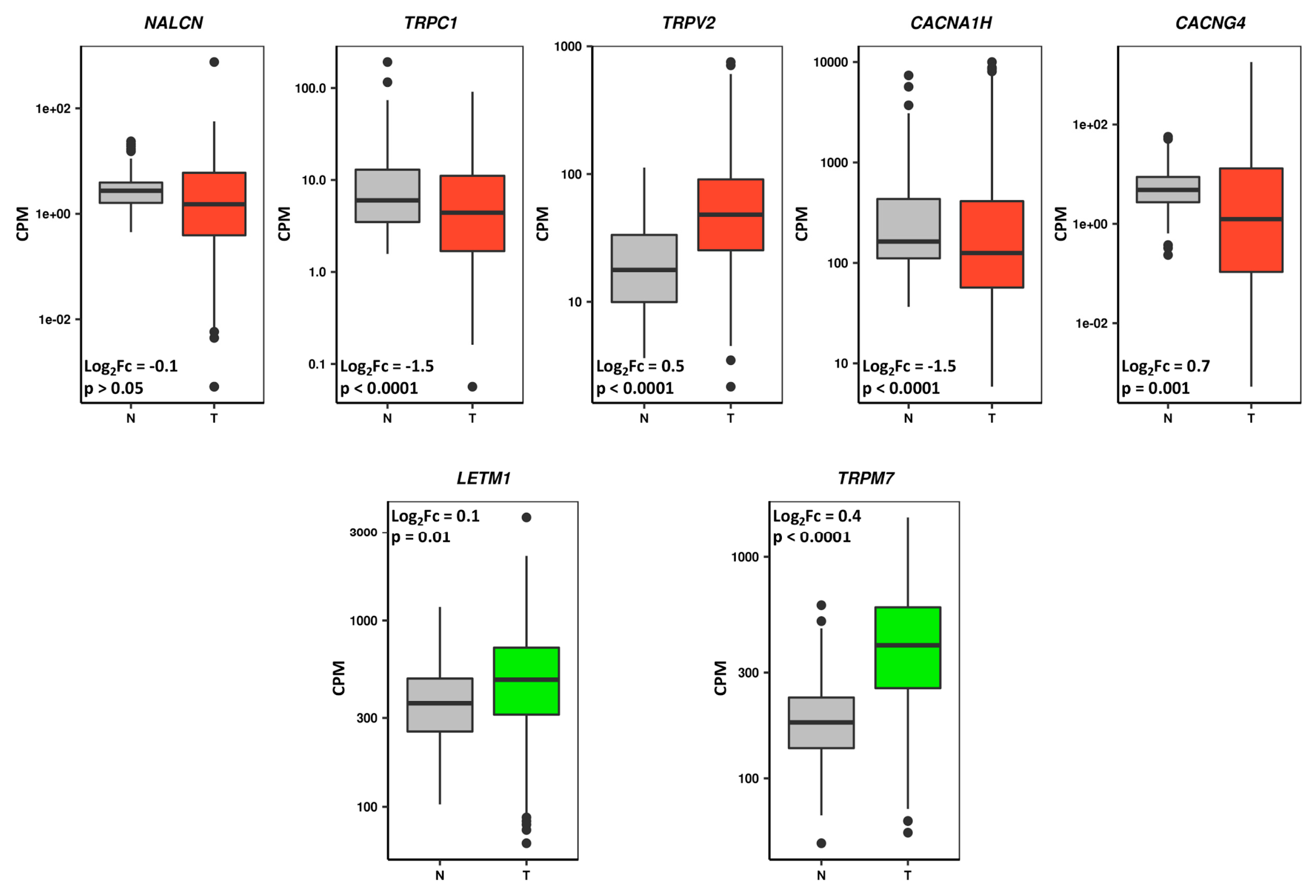
3.5. Prognostic CaRGs in Tumor and Normal Stomach Mucosa Tissue Samples
3.6. Prognostic CaRG Protein Expression in Normal Stomach and Gastric Cancer IHC-Stained Tissue Sections
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- GASTRIC (Global Advanced/Adjuvant Stomach Tumor Research International Collaboration) Group; Oba, K.; Paoletti, X.; Bang, Y.-J.; Bleiberg, H.; Burzykowski, T.; Fuse, N.; Michiels, S.; Morita, S.; Ohashi, Y.; et al. Role of chemotherapy for advanced/recurrent gastric cancer: An individual-patient-data meta-analysis. Eur. J. Cancer 2013, 49, 1565–1577. [Google Scholar] [PubMed]
- Charalampakis, N.; Economopoulou, P.; Kotsantis, I.; Tolia, M.; Schizas, D.; Liakakos, T.; Elimova, E.; Ajani, J.A.; Psyrri, A. Medical management of gastric cancer: A 2017 update. Cancer Med. 2018, 7, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Moiseyenko, V.M.; Tjulandin, S.; Majlis, A.; Constenla, M.; Boni, C.; Rodrigues, A.; Fodor, M.; Chao, Y.; Voznyi, E.; et al. Phase III Study of Docetaxel and Cisplatin Plus Fluorouracil Compared with Cisplatin and Fluorouracil as First-Line Therapy for Advanced Gastric Cancer: A Report of the V325 Study Group. J. Clin. Oncol. 2006, 24, 4991–4997. [Google Scholar] [CrossRef]
- Cunningham, D.; Starling, N.; Rao, S.; Iveson, T.; Nicolson, M.; Coxon, F.; Middleton, G.; Daniel, F.; Oates, J.; Norman, A.R. Capecitabine and Oxaliplatin for Advanced Esophagogastric Cancer. N. Engl. J. Med. 2008, 358, 36–46. [Google Scholar] [CrossRef]
- Bang, Y.-J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef]
- Koizumi, W.; Narahara, H.; Hara, T.; Takagane, A.; Akiya, T.; Takagi, M.; Miyashita, K.; Nishizaki, T.; Kobayashi, O.; Takiyama, W.; et al. S-1 plus cisplatin versus S-1 alone for first-line treatment of advanced gastric cancer (SPIRITS trial): A phase III trial. Lancet Oncol. 2008, 9, 215–221. [Google Scholar] [CrossRef]
- Fuchs, C.S.; Tomasek, J.; Yong, C.J.; Dumitru, F.; Passalacqua, R.; Goswami, C.; Safran, H.; dos Santos, L.V.; Aprile, G.; Ferry, D.R.; et al. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): An international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet 2014, 383, 31–39. [Google Scholar] [CrossRef]
- Li, J.; Qin, S.; Xu, J.; Xiong, J.; Wu, C.; Bai, Y.; Liu, W.; Tong, J.; Liu, Y.; Xu, R.; et al. Randomized, double-blind, placebo-controlled phase iii trial of apatinib in patients with chemotherapy-refractory advanced or metastatic adenocarcinoma of the stomach or gastroesophageal junction. J. Clin. Oncol. 2016, 34, 1448–1454. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Biological and Therapeutic Impact of Intratumor Heterogeneity in Cancer Evolution. Cancer Cell 2015, 27, 15–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raschellà, G.; Melino, G.; Gambacurta, A. Cell death in cancer in the era of precision medicine. Genes Immun. 2018. [Google Scholar] [CrossRef]
- Bass, A.J.; Thorsson, V.; Shmulevich, I.; Reynolds, S.M.; Miller, M.; Bernard, B.; Hinoue, T.; Laird, P.W.; Curtis, C.; Shen, H.; et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [Green Version]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.-M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Berridge, M.J.; Lipp, P.; Bootman, M.D. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000, 1, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Seas, A.; Kiyani, M.; Ji, K.S.Y.; Bell, H.N. A temporal examination of calcium signaling in cancer—From tumorigenesis, to immune evasion, and metastasis. Cell Biosci. 2018, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Monteith, G.R.; Davis, F.M.; Roberts-Thomson, S.J. Calcium Channels and Pumps in Cancer: Changes and Consequences. J. Biol. Chem. 2012, 287, 31666–31673. [Google Scholar] [CrossRef]
- Tajbakhsh, A.; Pasdar, A.; Rezaee, M.; Fazeli, M.; Soleimanpour, S.; Hassanian, S.M.; FarshchiyanYazdi, Z.; Rad, T.Y.; Ferns, G.A.; Avan, A. The current status and perspectives regarding the clinical implication of intracellular calcium in breast cancer. J. Cell. Physiol. 2018, 233, 5623–5641. [Google Scholar] [CrossRef]
- Bong, A.H.L.; Monteith, G.R. Calcium signaling and the therapeutic targeting of cancer cells. Biochim. Biophys. Acta—Mol. Cell Res. 2018, 1865, 1786–1794. [Google Scholar] [CrossRef]
- Foyouzi-Youssefi, R.; Arnaudeau, S.; Borner, C.; Kelley, W.L.; Tschopp, J.; Lew, D.P.; Demaurex, N.; Krause, K.H. Bcl-2 decreases the free Ca2+ concentration within the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2000, 97, 5723–5728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- VanHouten, J.; Sullivan, C.; Bazinet, C.; Ryoo, T.; Camp, R.; Rimm, D.L.; Chung, G.; Wysolmerski, J. PMCA2 regulates apoptosis during mammary gland involution and predicts outcome in breast cancer. Proc. Natl. Acad. Sci. USA 2010, 107, 11405–11410. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Gou, W.; Yang, X.; Wang, G.; Takahashi, H.; Yu, M.; Mao, X.; Takano, Y.; Zheng, H. Aberrant SERCA3 expression is closely linked to pathogenesis, invasion, metastasis, and prognosis of gastric carcinomas. Tumor Biol. 2012, 33, 1845–1854. [Google Scholar] [CrossRef]
- Fornaro, L.; Vivaldi, C.; Lin, D.; Xue, H.; Falcone, A.; Wang, Y.; Crea, F.; Bootman, M.D. Prognostic relevance of a T-type calcium channels gene signature in solid tumours: A correlation ready for clinical validation. PLoS ONE 2017, 12, e0182818. [Google Scholar] [CrossRef] [PubMed]
- Almasi, S.; Kennedy, B.E.; El-Aghil, M.; Sterea, A.M.; Gujar, S.; Partida-Sánchez, S.; El Hiani, Y. TRPM2 channel–mediated regulation of autophagy maintains mitochondrial function and promotes gastric cancer cell survival via the JNK-signaling pathway. J. Biol. Chem. 2018, 293, 3637–3650. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; Wang, H.; Huang, H.; Sun, L.; Dong, S.; Huang, N.; Shi, M.; Bin, J.; Liao, Y.; Liao, W. Elevated Orai1 and STIM1 expressions upregulate MACC1 expression to promote tumor cell proliferation, metabolism, migration, and invasion in human gastric cancer. Cancer Lett. 2016, 381, 31–40. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Szász, A.M.; Lánczky, A.; Nagy, Á.; Förster, S.; Hark, K.; Green, J.E.; Boussioutas, A.; Busuttil, R.; Szabó, A.; Győrffy, B.; et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1065 patients. Oncotarget 2016, 7, 49322–49333. [Google Scholar] [CrossRef]
- Ooi, C.H.; Ivanova, T.; Wu, J.; Lee, M.; Tan, I.B.; Tao, J.; Ward, L.; Koo, J.H.; Gopalakrishnan, V.; Zhu, Y.; et al. Oncogenic Pathway Combinations Predict Clinical Prognosis in Gastric Cancer. PLoS Genet. 2009, 5, e1000676. [Google Scholar] [CrossRef] [Green Version]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef] [Green Version]
- Uhlén, M.; Björling, E.; Agaton, C.; Szigyarto, C.A.-K.; Amini, B.; Andersen, E.; Andersson, A.-C.; Angelidou, P.; Asplund, A.; Asplund, C.; et al. A Human Protein Atlas for Normal and Cancer Tissues Based on Antibody Proteomics. Mol. Cell. Proteom. 2005, 4, 1920–1932. [Google Scholar] [CrossRef] [Green Version]
- Irizarry, R.A.; Hobbs, B.; Collin, F.; Beazer-Barclay, Y.D.; Antonellis, K.J.; Scherf, U.; Speed, T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003, 4, 249–264. [Google Scholar] [CrossRef] [Green Version]
- Risso, D.; Schwartz, K.; Sherlock, G.; Dudoit, S. GC-content normalization for RNA-Seq data. BMC Bioinform. 2011, 12, 480. [Google Scholar] [CrossRef] [PubMed]
- Risso, D.; Ngai, J.; Speed, T.P.; Dudoit, S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014, 32, 896–902. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [Green Version]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008. [Google Scholar]
- Nabissi, M.; Morelli, M.B.; Amantini, C.; Farfariello, V.; Ricci-Vitiani, L.; Caprodossi, S.; Arcella, A.; Santoni, M.; Giangaspero, F.; De Maria, R.; et al. TRPV2 channel negatively controls glioma cell proliferation and resistance to Fas-induced apoptosis in ERK-dependent manner. Carcinogenesis 2010, 31, 794–803. [Google Scholar] [CrossRef] [Green Version]
- Caterina, M.J.; Rosen, T.A.; Tominaga, M.; Brake, A.J.; Julius, D. A capsaicin-receptor homologue with a high threshold for noxious heat. Nature 1999, 398, 436–441. [Google Scholar] [CrossRef]
- Liberati, S.; Morelli, M.B.; Amantini, C.; Santoni, M.; Nabissi, M.; Cardinali, C.; Santoni, G. Advances in transient receptor potential vanilloid-2 channel expression and function in tumor growth and progression. Curr. Protein Pept. Sci. 2014, 15, 732–737. [Google Scholar] [CrossRef]
- Zhou, K.; Zhang, S.-S.; Yan, Y.; Zhao, S. Overexpression of transient receptor potential vanilloid 2 is associated with poor prognosis in patients with esophageal squamous cell carcinoma. Med. Oncol. 2014, 31, 17. [Google Scholar] [CrossRef]
- Liu, G.; Xie, C.; Sun, F.; Xu, X.; Yang, Y.; Zhang, T.; Deng, Y.; Wang, D.; Huang, Z.; Yang, L.; et al. Clinical significance of transient receptor potential vanilloid 2 expression in human hepatocellular carcinoma. Cancer Genet. Cytogenet. 2010, 197, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Caprodossi, S.; Lucciarini, R.; Amantini, C.; Nabissi, M.; Canesin, G.; Ballarini, P.; Di Spilimbergo, A.; Cardarelli, M.A.; Servi, L.; Mammana, G.; et al. Transient Receptor Potential Vanilloid Type 2 (TRPV2) Expression in Normal Urothelium and in Urothelial Carcinoma of Human Bladder: Correlation with the Pathologic Stage. Eur. Urol. 2008, 54, 612–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monet, M.; Lehen’kyi, V.; Gackiere, F.; Firlej, V.; Vandenberghe, M.; Roudbaraki, M.; Gkika, D.; Pourtier, A.; Bidaux, G.; Slomianny, C.; et al. Role of Cationic Channel TRPV2 in Promoting Prostate Cancer Migration and Progression to Androgen Resistance. Cancer Res. 2010, 70, 1225–1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiozaki, A.; Kudou, M.; Ichikawa, D.; Fujiwara, H.; Shimizu, H.; Ishimoto, T.; Arita, T.; Kosuga, T.; Konishi, H.; Komatsu, S.; et al. Esophageal cancer stem cells are suppressed by tranilast, a TRPV2 channel inhibitor. J. Gastroenterol. 2018, 53, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Lao, Y.; Chang, D.C. Calcium and Apoptosis. In Handbook of Neurochemistry and Molecular Neurobiology; Springer US: Boston, MA, USA, 2009; pp. 597–622. [Google Scholar]




| GENE | Study | Cases | Δmedian | HR | 95% CI | Log-Rank P |
|---|---|---|---|---|---|---|
| Intestinal Type | ||||||
| TRPV2 | GSE15460 | 70 | −80.1 | 1.9 | 1.0–3.7 | 0.04 |
| STAD | 86 | −12.3 * | 2.1 | 1.1–5.4 | 0.04 | |
| ATP2B3 | GSE62254 | 74 | 21.8 | 0.4 | 0.2–0.9 | 0.01 |
| STAD | 86 | −35.2 | 2.4 | 1.0–4.2 | 0.03 | |
| Diffuse Type | ||||||
| CACNB3 | KMplot | 52 | 23.3 | 0.5 | 0.2–1.0 | 0.04 |
| STAD | 38 | −36.2 * | 5.2 | 1.5–18.5 | 0.04 | |
| GRIN2B | GSE62254 | 72 | 24.5 * | 0.5 | 0.2–0.9 | 0.02 |
| STAD | 38 | −31.5 * | 4.1 | 1.4–11.3 | 0.004 | |
| CACNA2D4 | GSE15460 | 44 | 44.2 | 0.4 | 0.2–0.9 | 0.03 |
| STAD | 38 | −13.3 | 2.9 | 1.0–8.8 | 0.046 | |
| M Stage = 0 | ||||||
| CACNA1I | GSE62254 | 136 | 20.3 | 0.5 | 0.3–0.8 | 0.003 |
| STAD | 172 | −44.1 | 1.7 | 1.0–2.9 | 0.048 | |
| ATP2B3 | GSE62254 | 136 | 14.1 | 0.6 | 0.4–1.0 | 0.049 |
| STAD | 172 | −46.8 | 2.0 | 1.2–3.3 | 0.005 | |
| CACNA1H | GSE62254 | 136 | −12.5 | 1.6 | 1.0–2.6 | 0.047 |
| STAD | 152 | −43.5 | 1.8 | 1.1–2.9 | 0.01 | |
| SLC24A3 | GSE62254 | 136 | −18.3 | 2.0 | 1.2–3.2 | 0.006 |
| STAD | 172 | −41.0 | 1.7 | 1.0–2.7 | 0.04 | |
| NALCN | KMplot | 93 | −29 | 2.3 | 1.4–4.0 | 0.001 |
| STAD | 172 | −43.3 | 2.1 | 1.2–3.6 | 0.006 | |
| N Stage = 1+2+3 | ||||||
| ATP2A1 | GSE15460 | 69 | 67.5 | 0.4 | 0.2–0.8 | 0.008 |
| STAD | 130 | 20.0 | 0.6 | 0.3–0.9 | 0.02 | |
| ATP13A2 | GSE62254 | 130 | 38.7 | 0.5 | 0.3–0.8 | 0.007 |
| GSE15460 | 68 | 69.4 | 0.5 | 0.2–0.9 | 0.03 | |
| TRPC1 | GSE62254 | 130 | −30.9 * | 2.7 | 1.7–4.3 | <0.0001 |
| GSE15460 | 68 | −13.3 | 1.9 | 1.0–3.6 | 0.04 | |
| NALCN | KMplot | 89 | −17.3 | 2.1 | 1.3–3.5 | 0.003 |
| STAD | 130 | −49.8 | 1.8 | 1.0–3.1 | 0.03 | |
| Stage IV | ||||||
| CACNA2D1 | GSE62254 | 38 | −18.0 | 2.5 | 1.2–5.0 | 0.01 |
| STAD | 20 | −65.6 | 5.0 | 1.5–16.8 | 0.004 | |
| SLC3A2 | GSE15460 | 36 | −20.1 | 2.8 | 1.2–6.6 | 0.01 |
| STAD | 20 | 19.9 | 0.2 | 0.1–0.7 | 0.005 | |
| Treated with Adjuvant Chemotherapy | ||||||
| TRPV2 | KMplot | 76 | −2.8 | 1.7 | 1–2.9 | 0.03 |
| STAD | 14 | −9.9 * | ∞ | 1–∞ | 0.04 | |
| CACNB1 | KMplot | 78 | −10.7 | 1.8 | 1.1–3 | 0.02 |
| STAD | 14 | −13.9 * | 4.9 | 0.9–26.1 | 0.04 | |
| TRPC1 | KMplot | 76 | 5.3 | 0.5 | 0.3–0.8 | 0.008 |
| STAD | 14 | −11.8 * | 7.4 | 0.8–67.6 | 0.04 | |
| ITPR1 | KMplot | 76 | 10.6 | 0.4 | 0.2–0.7 | <0.0001 |
| STAD | 14 | −12.6 * | ∞ | 1.8–∞ | 0.01 | |
| Only Surgery | ||||||
| ATP2A1 | GSE62254 | 110 | 54.2 | 0.6 | 0.3–1.0 | 0.04 |
| STAD | 20 | 11.2 | 0.2 | 0.04–1.0 | 0.04 | |
| LETM1 | GSE62254 | 110 | 63.8 | 0.4 | 0.3–0.7 | <0.0001 |
| STAD | 20 | 8.2 | 0.2 | 0.04–1.0 | 0.03 | |
| NALCN | KMplot | 89 | −15 | 2 | 1.1–3.4 | 0.008 |
| STAD | 20 | −13.9 | 4.9 | 1.0–24.0 | 0.03 | |
| GENE | Study | Cases | Δmedian | HR | 95% CI | Log-Rank P |
|---|---|---|---|---|---|---|
| Intestinal Type | ||||||
| ATP13A3 | KMplot | 63 | −74.9 | 2.7 | 1.3–5.5 | 0.005 |
| STAD | 66 | 17.3 * | 0.2 | 0.04–1.0 | 0.03 | |
| SLC24A4 | KMplot | 66 | 71.1 | 0.5 | 0.2–0.9 | 0.02 |
| STAD | 66 | −39.3 * | 10.4 | 1.3–82.7 | 0.006 | |
| Diffuse Type | ||||||
| P2RX1 | GSE62254 | 68 | 28.2 * | 0.3 | 0.2–0.7 | 0.001 |
| STAD | 32 | 31.3 | 0.3 | 0.1–1.0 | 0.04 | |
| LETM1 | GSE62254 | 68 | 23.8 * | 0.5 | 0.2–0.9 | 0.03 |
| STAD | 32 | 11.4 * | 0.1 | 0.02–1.1 | 0.03 | |
| M Stage = 0 | ||||||
| LETM1 | GSE62254 | 128 | 27.6 * | 0.4 | 0.2–0.7 | 0.0005 |
| STAD | 146 | 13.3 * | 0.3 | 0.1–0.7 | 0.002 | |
| TRPC1 | GSE62254 | 128 | −37.2 * | 4.0 | 2.3–7.0 | <0.0001 |
| STAD | 146 | −9.1 | 2.4 | 1.2–5.0 | 0.01 | |
| NALCN | KMplot | 93 | −23.4 | 2.3 | 1.4–3.8 | 0.001 |
| STAD | 146 | −8.8 * | 3.3 | 1.5–7.3 | 0.001 | |
| N Stage = 1+2+3 | ||||||
| CACNA1F | GSE62254 | 122 | 20.2 * | 0.5 | 0.3–0.9 | 0.01 |
| STAD | 104 | 25.5 | 0.4 | 0.2–1.0 | 0.046 | |
| LETM1 | GSE62254 | 122 | 27.2 * | 0.4 | 0.3–0.7 | 0.002 |
| STAD | 104 | 12.2 * | 0.3 | 0.1–0.8 | 0.009 | |
| CHRNA10 | GSE62254 | 122 | 16.6 * | 0.6 | 0.4–1.0 | 0.04 |
| STAD | 104 | 31.0 * | 0.5 | 0.2–1.0 | 0.04 | |
| TRPC1 | GSE62254 | 122 | −32.9 * | 3.0 | 1.8–5.0 | <0.0001 |
| STAD | 104 | −40.4 | 2.4 | 1.1–5.4 | 0.02 | |
| P2RX1 | GSE62254 | 122 | 16.2 * | 0.6 | 0.3–0.9 | 0.03 |
| KMplot | 88 | 10.9 | 0.5 | 0.3–0.9 | 0.02 | |
| TRPV3 | KMplot | 89 | 10.2 | 0.6 | 0.3–0.9 | 0.02 |
| STAD | 104 | −27.8 * | 2.8 | 1.2–6.5 | 0.01 | |
| Stage II | ||||||
| LETM1 | GSE62254 | 44 | 24.1 * | 0.3 | 0.1–1.0 | 0.04 |
| STAD | 56 | 19.3 * | 0.1 | 0.03–0.7 | 0.004 | |
| Stage III | ||||||
| MCOLN2 | GSE62254 | 46 | 35.4 * | 0.3 | 0.1–0.7 | 0.005 |
| STAD | 66 | 32.2 * | 0.3 | 0.1–1.0 | 0.04 | |
| LETM1 | GSE62254 | 46 | 26.1 * | 0.3 | 0.1–0.8 | 0.008 |
| STAD | 66 | 6.4 | 0.3 | 0.1–0.9 | 0.02 | |
| TRPC1 | GSE62254 | 46 | −38.3 * | 4.0 | 1.7–9.6 | 0.0007 |
| STAD | 66 | −21.3 | 5.1 | 1.4–18.3 | 0.006 | |
| ATP2B3 | GSE62254 | 46 | 24.2 * | 0.4 | 0.2–1.0 | 0.04 |
| STAD | 66 | −10.8 * | 3.1 | 1.1–8.9 | 0.03 | |
| Treated with Adjuvant Chemotherapy | ||||||
| MCOLN2 | GSE62254 | 36 | 26.5 * | 0.1 | 0.01–0.9 | 0.01 |
| KMplot | 17 | n/a | 0.2 | 0.03–0.9 | 0.02 | |
| ORAI3 | GSE62254 | 36 | −32.2 * | 12.3 | 1.6–96.1 | 0.002 |
| KMplot | 76 | 3.8 | 0.6 | 0.3–0.9 | 0.03 | |
| TRPA1 | KMplot | 78 | −4.2 | 1.8 | 1.1–3.0 | 0.02 |
| STAD | 14 | 15 | 0.2 | 0.05–0.9 | 0.02 | |
| TRPC1 | KMplot | 76 | 4.2 | 0.5 | 0.3–0.8 | 0.008 |
| STAD | 14 | −13.5 | 3.8 | 1.0–15.4 | 0.04 | |
| Only Surgery | ||||||
| RYR3 | GSE62254 | 100 | −18.7 * | 1.9 | 1.0–3.3 | 0.02 |
| STAD | 22 | 7.2 | 0.4 | 0.1–1.0 | 0.048 | |
| GRIN3A | GSE62254 | 100 | 23.4 * | 0.5 | 0.3–0.8 | 0.009 |
| STAD | 22 | 28.5 | 0.3 | 0.1–1.0 | 0.03 | |
| LETM1 | GSE62254 | 100 | 27.4 * | 0.5 | 0.3–0.8 | 0.004 |
| STAD | 22 | 12.4 | 0.2 | 0.08–0.8 | 0.009 | |
| GENE | Prognostic Value | Log2Fc | Mean CPM in Normal Samples | Mean CPM in Tumor Samples | p Value | FDR |
|---|---|---|---|---|---|---|
| SLC24A3 | Negative | 0.6 | 17.1 | 32.8 | <0.0001 | <0.0001 |
| CACNB1 | Negative | −0.3 | 15.1 | 15.9 | 0.0001 | 0.0002 |
| ATP2A1 | Positive | −0.7 | 2.0 | 1.0 | <0.0001 | <0.0001 |
| ATP13A2 | Positive | 1.0 | 259.6 | 771.6 | <0.0001 | <0.0001 |
| P2RX1 | Positive | −1.6 | 22.5 | 3.6 | <0.0001 | <0.0001 |
| CACNA1F | Positive | −1.0 | 1.5 | 0.4 | <0.0001 | <0.0001 |
| CHRNA10 | Positive | −1.1 | 1.2 | 0.5 | <0.0001 | <0.0001 |
| GRIN3A | Positive | 1.0 | 0.3 | 1.0 | <0.0001 | <0.0001 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zoppoli, P.; Calice, G.; Laurino, S.; Ruggieri, V.; La Rocca, F.; La Torre, G.; Ciuffi, M.; Amendola, E.; De Vita, F.; Petrillo, A.; et al. TRPV2 Calcium Channel Gene Expression and Outcomes in Gastric Cancer Patients: A Clinically Relevant Association. J. Clin. Med. 2019, 8, 662. https://doi.org/10.3390/jcm8050662
Zoppoli P, Calice G, Laurino S, Ruggieri V, La Rocca F, La Torre G, Ciuffi M, Amendola E, De Vita F, Petrillo A, et al. TRPV2 Calcium Channel Gene Expression and Outcomes in Gastric Cancer Patients: A Clinically Relevant Association. Journal of Clinical Medicine. 2019; 8(5):662. https://doi.org/10.3390/jcm8050662
Chicago/Turabian StyleZoppoli, Pietro, Giovanni Calice, Simona Laurino, Vitalba Ruggieri, Francesco La Rocca, Giuseppe La Torre, Mario Ciuffi, Elena Amendola, Ferdinando De Vita, Angelica Petrillo, and et al. 2019. "TRPV2 Calcium Channel Gene Expression and Outcomes in Gastric Cancer Patients: A Clinically Relevant Association" Journal of Clinical Medicine 8, no. 5: 662. https://doi.org/10.3390/jcm8050662
APA StyleZoppoli, P., Calice, G., Laurino, S., Ruggieri, V., La Rocca, F., La Torre, G., Ciuffi, M., Amendola, E., De Vita, F., Petrillo, A., Napolitano, G., Falco, G., & Russi, S. (2019). TRPV2 Calcium Channel Gene Expression and Outcomes in Gastric Cancer Patients: A Clinically Relevant Association. Journal of Clinical Medicine, 8(5), 662. https://doi.org/10.3390/jcm8050662





