Lipid Modifications in Cilia Biology
Abstract
:1. Background
2. Palmitoylation
3. N-Myristoylation
4. Double Acylation: Myristoylation and Palmitoylation
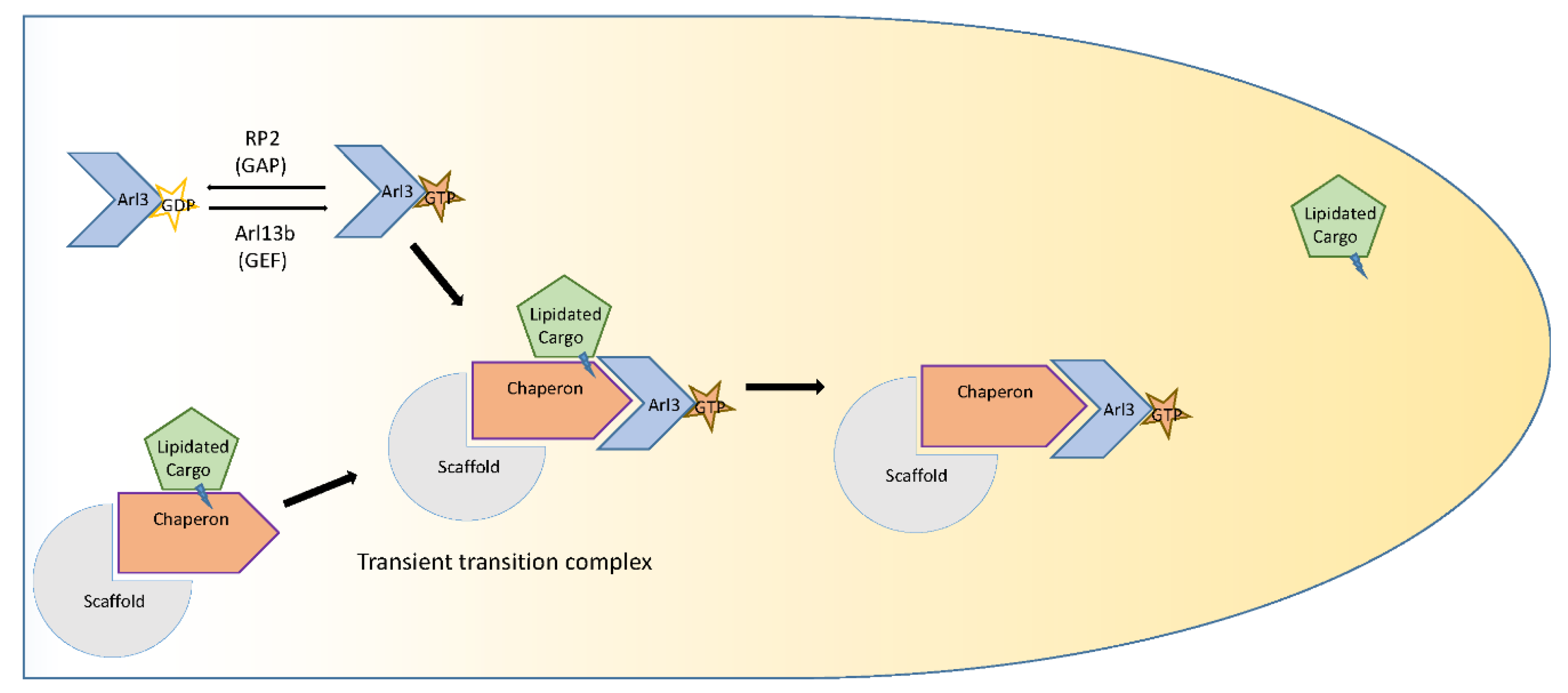
5. Prenylation: Farnesylation and Geranylgeranylation
6. Conclusion and Future Questions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ADCY3 | Adenylate cyclase 3 |
| ADPKD | Autosomal dominant polycystic kidney disease |
| AIPL1 | Aryl-hydrocarbon-interacting protein-like 1 |
| AMPK | AMP-regulated kinase |
| APT | Acyl protein thioesterase |
| Arl2 | ADP ribosylation factor-like protein 2 |
| Arl3 | ADP ribosylation factor-like protein 3 |
| Arl13b | ADP ribosylation factor-like protein 13B |
| Arf4 | ADP ribosylation factor 4 |
| ASAP1 | ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| BBS | Bardet–Biedl syndrome |
| CC | Coiled-coil |
| CePPEF | Caenorhabditis elegans protein phosphatase with EF-hands |
| CIL-7 | A ciliary protein |
| CTS | Cilia targeting sequence |
| eNOS | Endothelial nitric oxide synthase |
| EV | Extracellular vesicles |
| FCaBP | Flagellar calcium-binding protein |
| GAP | GTPase activating protein |
| GEF | Guanine nucleotide exchange factor |
| GNGT1 | Guanine nucleotide-binding protein G(T) subunit gamma-T1 |
| GPCR | G-protein-coupled receptors |
| GRK1 | G-protein-coupled receptor kinase 1 |
| GTP | Guanosine triphosphate |
| GTPase | Guanosine triphosphatase |
| Hh | Hedgehog |
| hTERT RPE-1 | hTERT-immortalized retinal pigment epithelial cell line, hTERT RPE-1, was derived by transfecting the RPE-340 cell line with the pGRN145 hTERT-expressing plasmid |
| IFT | Intraflagellar transport |
| IMCD3 | Inner medullary collecting duct3 |
| INPP5E | Inositol polyphosphate-5-phosphatase E |
| INPP5B | Inositol polyphosphate-5-phosphatase B |
| JS | Joubert syndrome |
| KIF17 | Kinesin family member 17 |
| mTORC1 | Mammalian target of rapamycin complex 1 |
| MKS | Meckel syndrome |
| nlCTS | N-terminal dual lipidation-coupled ciliary targeting signal |
| NPHP | Nephronophthisis |
| PATs | Palmitoyl acyl transferases |
| PC1 | Polycystin 1 |
| PC2 | Polycystin 2 |
| PDE6δ | Phosphodiesterase 6δ |
| PLD | Phospholipase D |
| PKD | Polycystic kidney disease |
| PKHD1 | Human autosomal recessive polycystic kidney disease gene |
| PPEF | Protein phosphatases with EF-hand domains |
| Ptc-1 | Patched 1 |
| PTM | Post-translational modifications |
| REP-1 | Rab escort protein-1 |
| RLD | RCC1-like domain |
| RFX3 | Regulatory factor X3 |
| RP2 | Retinitis pigmentosa protein |
| RPGR | Retinitis pigmentosa GTPase regulator |
| TPRs | Tetratricopeptide repeats |
| TZ | Transition zone |
| Smo | Smoothened |
| STPK | Serine/threonine protein kinase |
References
- Bürkle, A. Posttranslational Modification. In Encyclopedia of Genetics; Brenner, S., Miller, J.H., Eds.; Academic Press: New York, NY, USA, 2001; p. 1533. [Google Scholar]
- Satir, P.; Pedersen, L.B.; Christensen, S.T. The primary cilium at a glance. J. Cell Sci. 2010, 123, 499–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsiao, Y.-C.; Tuz, K.; Ferland, R.J. Trafficking in and to the primary cilium. Cilia 2012, 1, 4. [Google Scholar] [CrossRef] [PubMed]
- Nachury, M.V.; Seeley, E.S.; Jin, H. Trafficking to the ciliary membrane: How to get across the periciliary diffusion barrier? Annu. Rev. Cell Dev. Biol. 2010, 26, 59–87. [Google Scholar] [CrossRef] [PubMed]
- Reiter, J.F.; Leroux, M.R. Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Boil. 2017, 18, 533–547. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gonzalo, F.R.; Corbit, K.C.; Sirerol-Piquer, M.S.; Ramaswami, G.; Otto, E.A.; Noriega, T.R.; Seol, A.D.; Robinson, J.F.; Bennett, C.L.; Josifova, D.J.; et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet. 2011, 43, 776–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberson, E.C.; Dowdle, W.E.; Ozanturk, A.; Garcia-Gonzalo, F.R.; Li, C.; Halbritter, J.; Elkhartoufi, N.; Porath, J.D.; Cope, H.; Ashley-Koch, A.; et al. TMEM231, mutated in orofaciodigital and Meckel syndromes, organizes the ciliary transition zone. J. Cell Boil. 2015, 209, 129–142. [Google Scholar] [CrossRef]
- Kee, H.L.; Dishinger, J.F.; Blasius, T.L.; Liu, C.-J.; Margolis, B.; Verhey, K.J. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nature 2012, 14, 431–437. [Google Scholar] [CrossRef] [Green Version]
- Dishinger, J.F.; Kee, H.L.; Jenkins, P.M.; Fan, S.; Hurd, T.W.; Hammond, J.W.; Truong, Y.N.; Margolis, B.; Martens, J.R.; Verhey, K.J. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat. Cell Biol. 2010, 12, 703–710. [Google Scholar] [CrossRef]
- Hurd, T.W.; Fan, S.; Margolis, B.L. Localization of retinitis pigmentosa 2 to cilia is regulated by Importin beta2. J. Cell Sci. 2011, 124, 718–726. [Google Scholar] [CrossRef]
- Fan, S.; Margolis, B. The Ran importin system in cilia trafficking. Organogenesis 2011, 7, 147–153. [Google Scholar] [CrossRef] [Green Version]
- Madugula, V.; Lu, L. A ternary complex comprising transportin1, Rab8 and the ciliary targeting signal directs proteins to ciliary membranes. J. Cell Sci. 2016, 129, 3922–3934. [Google Scholar] [CrossRef]
- Nachury, M.V.; Loktev, A.V.; Zhang, Q.; Westlake, C.J.; Peränen, J.; Merdes, A.; Slusarski, D.C.; Scheller, R.H.; Bazan, J.F.; Sheffield, V.C.; et al. A Core Complex of BBS Proteins Cooperates with the GTPase Rab8 to Promote Ciliary Membrane Biogenesis. Cell 2007, 129, 1201–1213. [Google Scholar] [CrossRef] [Green Version]
- Jin, H.; White, S.R.; Shida, T.; Schulz, S.; Aguiar, M.; Gygi, S.P.; Bazan, J.F.; Nachury, M.V. The Conserved Bardet-Biedl Syndrome Proteins Assemble a Coat that Traffics Membrane Proteins to Cilia. Cell 2010, 141, 1208–1219. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Lechtreck, K.F. The Bardet–Biedl syndrome protein complex is an adapter expanding the cargo range of intraflagellar transport trains for ciliary export. Proc. Natl. Acad. Sci. USA 2018, 115, E934–E943. [Google Scholar] [CrossRef]
- Schou, K.B.; Pedersen, L.B.; Christensen, S.T. Ins and outs of GPCR signaling in primary cilia. EMBO Rep. 2015, 16, 1099–1113. [Google Scholar] [CrossRef] [Green Version]
- Berbari, N.F.; Johnson, A.D.; Lewis, J.S.; Askwith, C.C.; Mykytyn, K. Identification of Ciliary Localization Sequences within the Third Intracellular Loop of G Protein-coupled Receptors. Mol. Boil. Cell 2008, 19, 1540–1547. [Google Scholar] [CrossRef] [Green Version]
- Geneva, I.I.; Tan, H.Y.; Calvert, P.D. Untangling ciliary access and enrichment of two rhodopsin-like receptors using quantitative fluorescence microscopy reveals cell-specific sorting pathways. Mol. Biol. Cell 2016, 28, 554–566. [Google Scholar] [CrossRef]
- Roy, K.; Jerman, S.; Jozsef, L.; McNamara, T.; Onyekaba, G.; Sun, Z.; Marin, E.P. Palmitoylation of the ciliary GTPase ARL13b is necessary for its stability and its role in cilia formation. J. Boil. Chem. 2017, 292, 17703–17717. [Google Scholar] [CrossRef] [Green Version]
- Souther, C.; Emmer, B.T.; Toriello, K.M.; Olson, C.L.; Epting, C.L.; Engman, D.M. Identification of a palmitoyl acyltransferase required for protein sorting to the flagellar membrane. J. Cell Sci. 2009, 122, 867–874. [Google Scholar]
- Kumeta, M.; Panina, Y.; Yamazaki, H.; Takeyasu, K.; Yoshimura, S.H. N-terminal dual lipidation-coupled molecular targeting into the primary cilium. Genes Cells Devoted Mol. Cell. Mech. 2018, 23, 715–723. [Google Scholar] [CrossRef]
- Marshall, C. Protein prenylation: A mediator of protein-protein interactions. Science 1993, 259, 1865–1866. [Google Scholar] [CrossRef]
- Martin, D.D.; Beauchamp, E.; Berthiaume, L.G. Post-translational myristoylation: Fat matters in cellular life and death. Biochimie 2011, 93, 18–31. [Google Scholar] [CrossRef]
- Resh, M.D. Fatty acylation of proteins: New insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta Bioenerg. 1999, 1451, 1–16. [Google Scholar] [CrossRef]
- Liu, J.; García-Cardeña, G.; Sessa, W.C. Palmitoylation of Endothelial Nitric Oxide Synthase Is Necessary for Optimal Stimulated Release of Nitric Oxide: Implications for Caveolae Localization†. Biochemistry 1996, 35, 13277–13281. [Google Scholar] [CrossRef]
- Farazi, T.A.; Waksman, G.; Gordon, J.I. The Biology and Enzymology of ProteinN-Myristoylation. J. Boil. Chem. 2001, 276, 39501–39504. [Google Scholar] [CrossRef] [Green Version]
- Tao, B.; Bu, S.; Yang, Z.; Siroky, B.; Kappes, J.C.; Kispert, A.; Guay-Woodford, L.M. Cystin Localizes to Primary Cilia via Membrane Microdomains and a Targeting Motif. J. Am. Soc. Nephrol. 2009, 20, 2570–2580. [Google Scholar] [CrossRef]
- A Chen, C.; Manning, D.R. Regulation of G proteins by covalent modification. Oncogene 2001, 20, 1643–1652. [Google Scholar] [CrossRef] [Green Version]
- Roosing, S.; Collin, R.W.J.; Hollander, A.I.D.; Cremers, F.P.M.; Siemiatkowska, A.M. Prenylation defects in inherited retinal diseases. J. Med. Genet. 2014, 51, 143–151. [Google Scholar] [CrossRef]
- Wang, M.; Casey, P.J. Protein prenylation: Unique fats make their mark on biology. Nat. Rev. Mol. Cell Boil. 2016, 17, 110–122. [Google Scholar] [CrossRef]
- Follit, J.A.; Li, L.; Vucica, Y.; Pazour, G.J. The cytoplasmic tail of fibrocystin contains a ciliary targeting sequence. J. Cell Boil. 2010, 188, 21–28. [Google Scholar] [CrossRef] [Green Version]
- Ward, C.J. Cellular and subcellular localization of the ARPKD protein; fibrocystin is expressed on primary cilia. Hum. Mol. Genet. 2003, 12, 2703–2710. [Google Scholar] [CrossRef]
- Duldulao, N.A.; Lee, S.; Sun, Z. Cilia localization is essential for in vivo functions of the Joubert syndrome protein Arl13b/Scorpion. Development 2009, 136, 4033–4042. [Google Scholar] [CrossRef] [Green Version]
- Deretic, D.; Wang, J. Molecular assemblies that control rhodopsin transport to the cilia. Vis. Res. 2012, 75, 5–10. [Google Scholar] [CrossRef] [Green Version]
- Tam, B.M. Identification of an Outer Segment Targeting Signal in the COOH Terminus of Rhodopsin Using Transgenic Xenopus laevis. J. Cell Boil. 2000, 151, 1369–1380. [Google Scholar] [CrossRef]
- I Papac, D.; Thornburg, K.R.; E Büllesbach, E.; Crouch, R.K.; Knapp, D.R. Palmitylation of a G-protein coupled receptor. Direct analysis by tandem mass spectrometry. J. Boil. Chem. 1992, 267, 16889–16894. [Google Scholar]
- Nemet, I.; Ropelewski, P.; Imanishi, Y. Chapter Three—Rhodopsin Trafficking and Mistrafficking: Signals, Molecular Components, and Mechanisms. In Progress in Molecular Biology and Translational Science; Wu, G., Ed.; Academic Press: New York, NY, USA, 2015; pp. 39–71. [Google Scholar]
- Su, X.; Wu, M.; Yao, G.; El-Jouni, W.; Luo, C.; Tabari, A.; Zhou, J. Regulation of polycystin-1 ciliary trafficking by motifs at its C-terminus and polycystin-2 but not by cleavage at the GPS site. J. Cell Sci. 2015, 128, 4063–4073. [Google Scholar] [CrossRef]
- Chen, B.; Niu, J.; Kreuzer, J.; Zheng, B.; Jarugumilli, G.K.; Haas, W.; Wu, X. Auto-fatty acylation of transcription factor RFX3 regulates ciliogenesis. Proc. Natl. Acad. Sci. USA 2018, 115, E8403–E8412. [Google Scholar] [CrossRef]
- El Zein, L.; Ait-Lounis, A.; Morle, L.; Thomas, J.; Chhin, B.; Spassky, N.; Reith, W.; Durand, B.; Carlton, J.G.; Bujny, M.V.; et al. RFX3 governs growth and beating efficiency of motile cilia in mouse and controls the expression of genes involved in human ciliopathies. J. Cell Sci. 2009, 122, 3180–3189. [Google Scholar] [CrossRef] [Green Version]
- Pepinsky, R.B.; Zeng, C.; Wen, D.; Rayhorn, P.; Baker, D.P.; Williams, K.P.; Bixler, S.A.; Ambrose, C.M.; Garber, E.A.; Miatkowski, K.; et al. Identification of a Palmitic Acid-modified Form of Human Sonic hedgehog. J. Boil. Chem. 1998, 273, 14037–14045. [Google Scholar] [CrossRef] [Green Version]
- Willert, K.; Brown, J.D.; Danenberg, E.; Duncan, A.W.; Weissman, I.L.; Reya, T.; Yates, J.R., 3rd; Nusse, R. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 2003, 423, 448–452. [Google Scholar] [CrossRef]
- Nakata, K.; Shiba, D.; Kobayashi, D.; Yokoyama, T. Targeting of Nphp3 to the primary cilia is controlled by an N-terminal myristoylation site and coiled-coil domains. Cytoskeleton 2012, 69, 221–234. [Google Scholar] [CrossRef]
- Maguire, J.E.; Silva, M.; Nguyen, K.C.Q.; Hellen, E.; Kern, A.D.; Hall, D.H.; Barr, M.M. Myristoylated CIL-7 regulates ciliary extracellular vesicle biogenesis. Mol. Boil. Cell 2015, 26, 2823–2832. [Google Scholar] [CrossRef]
- Ramulu, P. Cellular and Subcellular Localization, N-terminal Acylation, and Calcium Binding of Caenorhabditis elegans Protein Phosphatase with EF-hands. J. Boil. Chem. 2001, 276, 25127–25135. [Google Scholar] [CrossRef] [Green Version]
- Hardcastle, A.J.; Grayson, C.; Spackman, L.; Willison, K.R.; Chapple, J.P.; Cheetham, M.E. Mutations in the N-terminus of the X-linked retinitis pigmentosa protein RP2 interfere with the normal targeting of the protein to the plasma membrane. Hum. Mol. Genet. 2000, 9, 1919–1926. [Google Scholar]
- Luo, N.; Kumar, A.; Conwell, M.; Weinreb, R.N.; Anderson, R.; Sun, Y. Compensatory Role of Inositol 5-Phosphatase INPP5B to OCRL in Primary Cilia Formation in Oculocerebrorenal Syndrome of Lowe. PLoS ONE 2013, 8, e66727. [Google Scholar] [CrossRef]
- Williams, C.; Choudhury, R.; McKenzie, E.; Lowe, M. Targeting of the type II inositol polyphosphate 5-phosphatase INPP5B to the early secretory pathway. J. Cell Sci. 2007, 120, 3941–3951. [Google Scholar] [CrossRef] [Green Version]
- Dutta, N.; Seo, S. RPGR, a prenylated retinal ciliopathy protein, is targeted to cilia in a prenylation- and PDE6D-dependent manner. Boil. Open 2016, 5, 1283–1289. [Google Scholar] [CrossRef] [Green Version]
- Humbert, M.C.; Weihbrecht, K.; Searby, C.C.; Li, Y.; Pope, R.M.; Sheffield, V.C.; Seo, S. ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc. Natl. Acad. Sci. USA 2012, 109, 19691–19696. [Google Scholar] [CrossRef] [Green Version]
- Nozaki, S.; Katoh, Y.; Terada, M.; Michisaka, S.; Funabashi, T.; Takahashi, S.; Kontani, K.; Nakayama, K. Regulation of ciliary retrograde protein trafficking by the Joubert syndrome proteins ARL13B and INPP5E. J. Cell Sci. 2017, 130, 563. [Google Scholar] [CrossRef]
- Fansa, E.K.; Kosling, S.K.; Zent, E.; Wittinghofer, A.; Ismail, S. PDE6delta-mediated sorting of INPP5E into the cilium is determined by cargo-carrier affinity. Nat. Commun. 2016, 7, 11366. [Google Scholar] [CrossRef]
- Boom, A. Post-translational Modification of Human Brain Type I Inositol-1,4,5-trisphosphate 5-Phosphatase by Farnesylation. J. Boil. Chem. 1996, 271, 10419–10424. [Google Scholar] [Green Version]
- Guan, X.; Fierke, C.A. Understanding Protein Palmitoylation: Biological Significance and Enzymology, Science China. Chemistry 2011, 54, 1888–1897. [Google Scholar]
- Jia, L.; Chisari, M.; Maktabi, M.H.; Sobieski, C.; Zhou, H.; Konopko, A.M.; Martin, B.R.; Mennerick, S.J.; Blumer, K.J. A Mechanism Regulating G Protein-coupled Receptor Signaling That Requires Cycles of Protein Palmitoylation and Depalmitoylation. J. Boil. Chem. 2014, 289, 6249–6257. [Google Scholar] [CrossRef] [Green Version]
- Aicart-Ramos, C.; Valero, R.A.; Rodriguez-Crespo, I. Protein palmitoylation and subcellular trafficking. Biochim. Biophys. Acta 2011, 1808, 2981–2994. [Google Scholar] [CrossRef] [Green Version]
- Salaun, C.; Gould, G.W.; Chamberlain, L.H. The SNARE proteins SNAP-25 and SNAP-23 display different affinities for lipid rafts in PC12 cells. Regulation by distinct cysteine-rich domains. J. Biol. Chem. 2005, 280, 1236–1240. [Google Scholar] [CrossRef]
- Badgandi, H.B.; Hwang, S.-H.; Shimada, I.S.; Loriot, E.; Mukhopadhyay, S. Tubby family proteins are adapters for ciliary trafficking of integral membrane proteins. J. Cell Boil. 2017, 216, 743–760. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Wen, X.-H.; Ablonczy, Z.; Crouch, R.K.; Makino, C.L.; Lem, J. Enhanced Shutoff of Phototransduction in Transgenic Mice Expressing Palmitoylation-deficient Rhodopsin. J. Boil. Chem. 2005, 280, 24293–24300. [Google Scholar] [CrossRef]
- Qanbar, R.; Bouvier, M. Role of palmitoylation/depalmitoylation reactions in G-protein-coupled receptor function. Pharmacol. Ther. 2003, 97, 1–33. [Google Scholar] [CrossRef]
- Park, P.S.-H.; Sapra, K.T.; Jastrzebska, B.; Maeda, T.; Maeda, A.; Pulawski, W.; Kono, M.; Lem, J.; Crouch, R.K.; Filipek, S.; et al. Modulation of Molecular Interactions and Function by Rhodopsin Palmitylation. Biochem. 2009, 48, 4294–4304. [Google Scholar] [CrossRef] [Green Version]
- Hargrave, P.A.; Hamm, H.E.; Hofmann, K.P. Interaction of rhodopsin with the G-protein, transducin, BioEssays: News and reviews in molecular. Cell. Dev. Biol. 1993, 15, 43–50. [Google Scholar]
- Tanimoto, Y.; Okada, K.; Hayashi, F.; Morigaki, K. Evaluating the Raftophilicity of Rhodopsin Photoreceptor in a Patterned Model Membrane. Biophys. J. 2015, 109, 2307–2316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seno, K.; Hayashi, F. Palmitoylation is a prerequisite for dimerization-dependent raftophilicity of rhodopsin. J. Boil. Chem. 2017, 292, 15321–15328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeda, A.; Okano, K.; Park, P.S.-H.; Lem, J.; Crouch, R.K.; Maeda, T.; Palczewski, K. Palmitoylation stabilizes unliganded rod opsin. Proc. Natl. Acad. Sci. USA 2010, 107, 8428–8433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, K.; Marin, E.P. Polycystin-1, the product of the polycystic kidney disease gene PKD1, is post-translationally modified by palmitoylation. Mol. Biol. Rep. 2018, 45, 1515–1521. [Google Scholar] [CrossRef] [PubMed]
- Bangs, F.; Anderson, K.V. Primary Cilia and Mammalian Hedgehog Signaling. Cold Spring Harb. Perspect. Biol. 2017, 9, a028175. [Google Scholar] [CrossRef] [PubMed]
- Mann, R.K.; Beachy, P.A. Novel Lipid Modifications of Secreted Protein Signals. Annu. Rev. Biochem. 2004, 73, 891–923. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.P.; Rayhorn, P.; Chi-Rosso, G.; A Garber, E.; Strauch, K.L.; Horan, G.S.; O Reilly, J.; Baker, D.P.; Taylor, F.R.; Koteliansky, V.; et al. Functional antagonists of sonic hedgehog reveal the importance of the N terminus for activity. J. Cell Sci. 1999, 112, 4405–4414. [Google Scholar] [PubMed]
- Gao, X.; Arenas-Ramirez, N.; Scales, S.J.; Hannoush, R.N. Membrane targeting of palmitoylated Wnt and Hedgehog revealed by chemical probes. FEBS Lett. 2011, 585, 2501–2506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takada, R.; Satomi, Y.; Kurata, T.; Ueno, N.; Norioka, S.; Kondoh, H.; Takao, T.; Takada, S. Monounsaturated Fatty Acid Modification of Wnt Protein: Its Role in Wnt Secretion. Dev. Cell 2006, 11, 791–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergmann, C.; Fliegauf, M.; Brüchle, N.O.; Frank, V.; Olbrich, H.; Kirschner, J.; Schermer, B.; Schmedding, I.; Kispert, A.; Kränzlin, B.; et al. Loss of Nephrocystin-3 Function Can Cause Embryonic Lethality, Meckel-Gruber-like Syndrome, Situs Inversus, and Renal-Hepatic-Pancreatic Dysplasia. Am. J. Hum. Genet. 2008, 82, 959–970. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Dai, J.; Attanasio, M.; Hildebrandt, F. Nephrocystin-3 is required for ciliary function in zebrafish embryos. Am. J. Physiol. Physiol. 2010, 299, F55–F62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Constantine, R.; Vorobiev, S.; Chen, Y.; Seetharaman, J.; Huang, Y.J.; Xiao, R.; Montelione, G.T.; Gerstner, C.D.; Davis, M.W.; et al. UNC119 is required for G protein trafficking in sensory neurons. Nat. Neurosci. 2011, 14, 874–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, K.J.; Baye, L.M.; Olivier-Mason, A.; Mukhopadhyay, S.; Sang, L.; Kwong, M.; Wang, W.; Pretorius, P.R.; Sheffield, V.C.; Sengupta, P.; et al. An ARL3–UNC119–RP2 GTPase cycle targets myristoylated NPHP3 to the primary cilium. Genes Dev. 2011, 25, 2347–2360. [Google Scholar] [CrossRef] [PubMed]
- A Ismail, S.; Chen, Y.-X.; Miertzschke, M.; Vetter, I.R.; Koerner, C.; Wittinghofer, A. Structural basis for Arl3-specific release of myristoylated ciliary cargo from UNC119. EMBO J. 2012, 31, 4085–4094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Constantine, R.; Zhang, H.; Gerstner, C.D.; Frederick, J.M.; Baehr, W. Uncoordinated (UNC)119: Coordinating the trafficking of myristoylated proteins. Vis. Res. 2012, 75, 26–32. [Google Scholar] [CrossRef] [Green Version]
- Jaiswal, M.; Fansa, E.K.; Kösling, S.K.; Mejuch, T.; Waldmann, H.; Wittinghofer, A. Novel Biochemical and Structural Insights into the Interaction of Myristoylated Cargo with Unc119 Protein and Their Release by Arl2/3. J. Boil. Chem. 2016, 291, 20766–20778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, B.; Wu, N.; Yang, J.-M.; Gould, S.J. Protein Targeting to Exosomes/Microvesicles by Plasma Membrane Anchors. J. Boil. Chem. 2011, 286, 14383–14395. [Google Scholar] [CrossRef] [Green Version]
- Tull, D.; Vince, J.E.; Callaghan, J.M.; Naderer, T.; Spurck, T.; McFadden, G.I.; Currie, G.; Ferguson, K.; Bacic, A.; McConville, M.J.; et al. SMP-1, a Member of a New Family of Small Myristoylated Proteins in Kinetoplastid Parasites, Is Targeted to the Flagellum Membrane in Leishmania. Mol. Boil. Cell 2004, 15, 4775–4786. [Google Scholar] [CrossRef]
- Zang, J.; Neuhauss, S.C.F. The Binding Properties and Physiological Functions of Recoverin. Front. Mol. Neurosci. 2018, 11, 473. [Google Scholar] [CrossRef]
- Dizhoor, A.; Ray, S.; Kumar, S.; Niemi, G.; Spencer, M.; Brolley, D.; Walsh, K.; Philipov, P.; Hurley, J.; Stryer, L. Recoverin: A calcium sensitive activator of retinal rod guanylate cyclase. Science 1991, 251, 915–918. [Google Scholar] [CrossRef]
- Godsel, L.M.; Engman, D.M. Flagellar protein localization mediated by a calcium-myristoyl/palmitoyl switch mechanism. EMBO J. 1999, 18, 2057–2065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zozulya, S.; Stryer, L. Calcium-myristoyl protein switch. Proc. Natl. Acad. Sci. USA 1992, 89, 11569–11573. [Google Scholar] [CrossRef] [PubMed]
- Porumb, T.; Ames, J.B.; Tanaka, T.; Ikura, M.; Stryer, L. Amino-terminal Myristoylation Induces Cooperative Calcium Binding to Recoverin. J. Boil. Chem. 1995, 270, 4526–4533. [Google Scholar] [Green Version]
- Lechtreck, K.-F.; Johnson, E.C.; Sakai, T.; Cochran, D.; Ballif, B.A.; Rush, J.; Pazour, G.J.; Ikebe, M.; Witman, G.B. The Chlamydomonas reinhardtii; BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J. Cell Biol. 2009, 187, 1117. [Google Scholar] [CrossRef] [PubMed]
- Vögler, O.; Barcelo, J.M.; Ribas, C.; Escribá, P.V. Membrane interactions of G proteins and other related proteins. Biochim. Biophys. Acta Biomembr. 2008, 1778, 1640–1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ismail, S.A.; Chen, Y.-X.; Rusinova, A.; Chandra, A.; Bierbaum, M.; Gremer, L.; Triola, G.; Waldmann, H.; Bastiaens, P.I.H.; Wittinghofer, A. Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat. Methods 2011, 7, 942–949. [Google Scholar] [CrossRef]
- Zhang, H.; Li, S.; Doan, T.; Rieke, F.; Detwiler, P.B.; Frederick, J.M.; Baehr, W. Deletion of PrBP/delta impedes transport of GRK1 and PDE6 catalytic subunits to photoreceptor outer segments. Proc. Natl. Acad. Sci. USA 2007, 104, 8857–8862. [Google Scholar] [CrossRef]
- Yan, D.; Swain, P.K.; Breuer, D.; Tucker, R.M.; Wu, W.; Fujita, R.; Rehemtulla, A.; Burke, D.; Swaroop, A. Biochemical Characterization and Subcellular Localization of the Mouse Retinitis Pigmentosa GTPase Regulator (mRpgr). J. Boil. Chem. 1998, 273, 19656–19663. [Google Scholar] [CrossRef] [Green Version]
- Watzlich, D.; Vetter, I.; Gotthardt, K.; Miertzschke, M.; Chen, Y.X.; Wittinghofer, A.; Ismail, S. The interplay between RPGR, PDEdelta and Arl2/3 regulate the ciliary targeting of farnesylated cargo. EMBO Rep. 2013, 14, 465–472. [Google Scholar] [CrossRef]
- Gotthardt, K.; Lokaj, M.; Koerner, C.; Falk, N.; Gießl, A.; Wittinghofer, A.; E Clapham, D. A G-protein activation cascade from Arl13B to Arl3 and implications for ciliary targeting of lipidated proteins. eLife 2015, 4, e11859. [Google Scholar] [CrossRef]
- Jacoby, M.; Cox, J.J.; Gayral, S.; Hampshire, D.J.; Ayub, M.; Blockmans, M.; Pernot, E.; Kisseleva, M.V.; Compère, P.; Schiffmann, S.N.; et al. INPP5E mutations cause primary cilium signaling defects, ciliary instability and ciliopathies in human and mouse. Nat. Genet. 2009, 41, 1027–1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alkanderi, S.; Molinari, E.; Shaheen, R.; Elmaghloob, Y.; Stephen, L.A.; Sammut, V.; Ramsbottom, S.A.; Srivastava, S.; Cairns, G.; Edwards, N.; et al. ARL3 Mutations Cause Joubert Syndrome by Disrupting Ciliary Protein Composition. Am. J. Hum. Genet. 2018, 103, 612–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Palmitoylation | N-Myristoylation | Prenylation | |
|---|---|---|---|
| Structure | 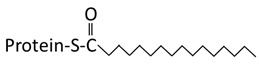 Saturated 16 carbon palmitate addition to cysteines on the targeted proteins via the formation of a thioester linkage |  Saturated 14 carbon myristoyl group addition by an amide bond to the α-amino group of an N-terminal glycine | 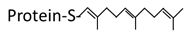 Transfer of either a farnesyl or a geranyl- geranyl moiety to the cysteine of the CaaX motif of the target protein |
| Reversibility | Reversible | Irreversible | Irreversible |
| Enzymes | A number of enzymes involved in the process: 24 Palmitoyltransferase (also known as PATs or DHHCs) and ~15 acyl protein thioesterase (APT) | N-myristoyltransferase | Farnesyltransferase, Geranylgeranyltransferase I, Geranylgeranyltransferase II |
| Motif | Can occur on cysteines located at different sites in a protein | N-myristoylation occurs at the N-terminus at MGxxxS motif | Occurs only at the CaaX boxes at the C-terminus |
| Example | eNOS, Arl13b [19,25] | Src, Cystin [26,27] | G-protein γ subunit, AIPL1 [28,29,30] |
| Protein | Site of Lipid Modification | Lipid Modification Identification Method | Construct Used to Identify Lipid Modification | Cilia Targeting Requirements (CTS and More) | Disease Association/Function | Reference |
|---|---|---|---|---|---|---|
| Fibrocystin | Palmitoylation on three conserved cysteine residues in CTS | Mutation and metabolic labeling | 193 residues of C-terminal | 18 intracellular aa flanking the transmembrane domain, including conserved cysteines | ARPKD | [31,32] |
| Arl13b | Palmitoylation in N-terminal region | Mutation and metabolic labeling | Full length protein | Multiple regions in the protein | JS | [19,33] |
| Rhodopsin | Palmitoylation in C-terminal cytoplasmic region | Enzymatic and chemical cleavage techniques, tandem mass spectrometry, mutations/deletions and knock-out mice | C-terminal fusion protein | VxPx and the FR (in IMCD3) | Inherited retinal degenerative diseases | [34,35,36] |
| PC1 | Palmitoylation in C-terminal region | Metabolic labeling and Biochemical assays | ~200-amino-acid C-terminal tail (CTT) | Multiple sites in the coiled-coil motif in the C-terminal tail including VxP motif and multiple cis-acting elements | Mutation cause ADPKD | [37,38] |
| RFX3 | Palmitoylation on a cysteine residue in the dimerization domain | Biochemical assays and mass spectrometry | Full length protein | - | Ciliopathies and metabolic disorders, like diabetes | [39,40] |
| Hedgehog | Palmitoylation in N-terminal cysteine | Mass spectrometry and metabolic labeling | Full length protein | - | Organ development and cancer | [41] |
| Wnt | Palmitoylation on a conserved cysteine in N-terminal region | Mass spectrometry, biochemical, enzymatic methods and mutations | Full length protein | - | Wnt signaling involved in animal development including proliferation of stem cells | [42] |
| Cystin | Myristoylation on glycine 2 | Mutations and metabolic labeling | Full length and various truncated mutants | AxEGG | PKD | [27] |
| NPHP3 | Myristoylation at the N-terminus | Metabolic labeling and mutations | Truncated N-terminal fusion proteins | N-terminal CC domain and the myristoylation site | Nephronophthisis | [43] |
| CIL-7 | Myristoylation at the N-terminus | Mutations | Full length protein | Myristoylation motif | PKD | [44] |
| CePPEF | N-terminal myristoylation and palmitoylation | Metabolic labeling and mutations | Full length protein and N-terminal recombinants | N-terminal region, palmitoylation is particularly important | Calcium regulation | [45] |
| Calflagin | Myristoylation at glycine 2 and palmitoylation at cysteine 3 in the N-terminal region | Biochemical assays, metabolic labeling and mutations | Full length protein | Palmitoylation | Calcium binding protein | [20] |
| RP2 | Myristoylation at glycine 2 and palmitoylation at cysteine 3 in the N-terminal region | Mutation | Truncated N-terminal fusion proteins | N-terminal dual lipidation-coupled ciliary targeting signal (nlCTS) | X-linked retinitis pigmentosa | [21,46] |
| INPP5B | Prenylated at its C-terminus CaaX | Mutation and knock downs | Full length protein | Prenylation | Important for retrograde trafficking | [47,48] |
| RPGR | Prenylated at its C-terminus CaaX | Mutation | Full length and deletion mutants | Two independent ciliary targeting signals: one within the RLD and the other near the C-terminus. | Inherited retinal degenerative diseases | [49] |
| INPP5E | prenylated at its C-terminus CaaX | Biochemical assays, enzymatic assays and mutations | Full length protein | FDRELYL (not sufficient, require other interactors) | JS and MORM syndrome | [50,51,52,53] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roy, K.; Marin, E.P. Lipid Modifications in Cilia Biology. J. Clin. Med. 2019, 8, 921. https://doi.org/10.3390/jcm8070921
Roy K, Marin EP. Lipid Modifications in Cilia Biology. Journal of Clinical Medicine. 2019; 8(7):921. https://doi.org/10.3390/jcm8070921
Chicago/Turabian StyleRoy, Kasturi, and Ethan P. Marin. 2019. "Lipid Modifications in Cilia Biology" Journal of Clinical Medicine 8, no. 7: 921. https://doi.org/10.3390/jcm8070921
APA StyleRoy, K., & Marin, E. P. (2019). Lipid Modifications in Cilia Biology. Journal of Clinical Medicine, 8(7), 921. https://doi.org/10.3390/jcm8070921





