Blockade of STAT3 Causes Severe In Vitro and In Vivo Maturation Defects in Intestinal Organoids Derived from Human Embryonic Stem Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals And Reagents
2.2. Differentiation And Culture of hIOs From hESCs
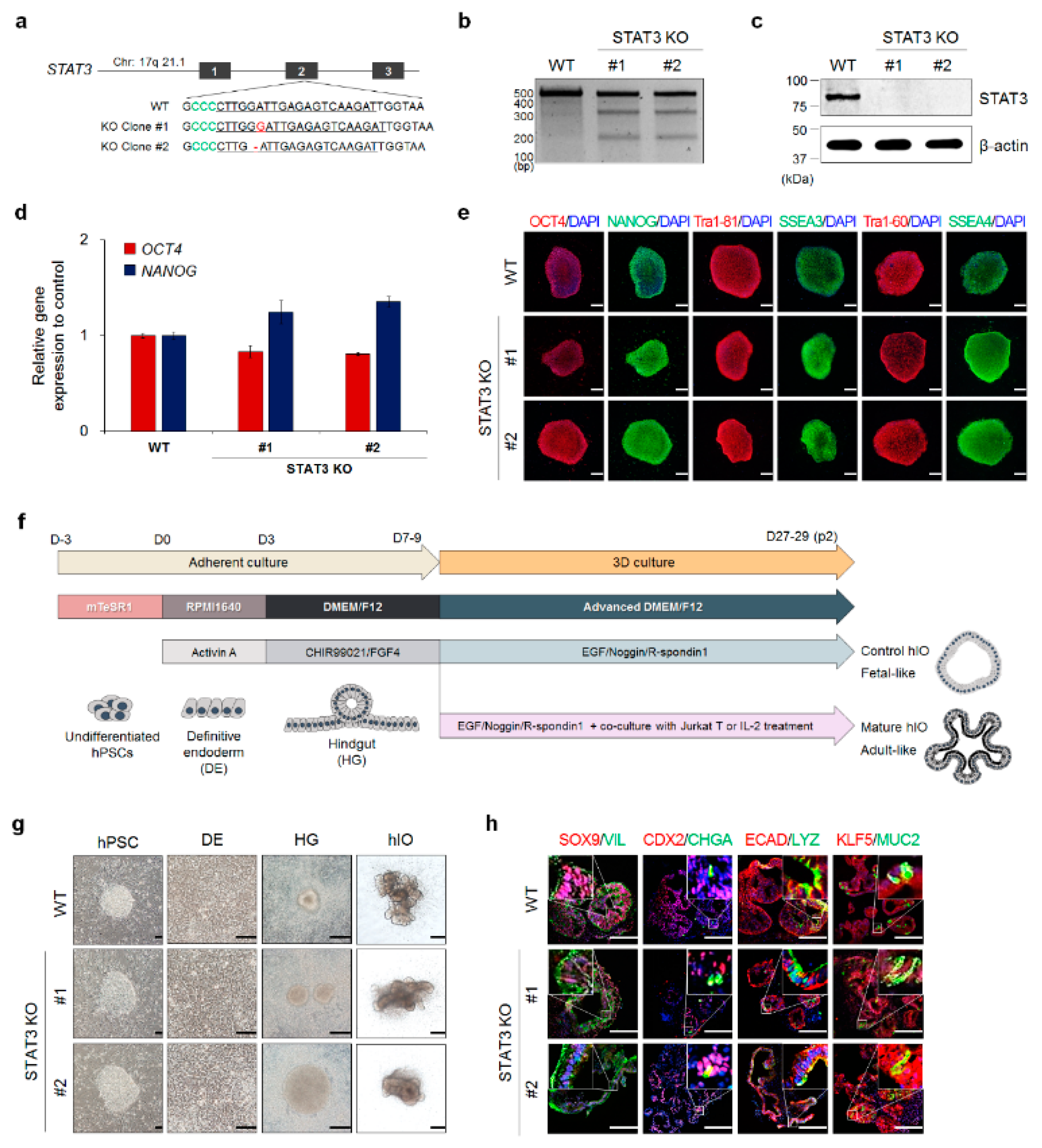
2.3. Generation And Validation of STAT3 KO hESC Lines
2.4. Quantitative Real-Time PCR (qPCR)
2.5. Immunofluorescence And Hematoxylin-Eosin (H&E) Staining
2.6. Transmission Electron Microscopy (TEM)
2.7. Transcriptome Analysis By Microarray
2.8. Transplantation
2.9. Western Blotting
2.10. Fluo-4 AM Assay
3. Results
3.1. Treatment With STAT3 Inhibitors Abrogated The In Vitro Maturation Of hESC-Derived hIOs
3.2. Generation And Validation of STAT3 KO hESC Lines
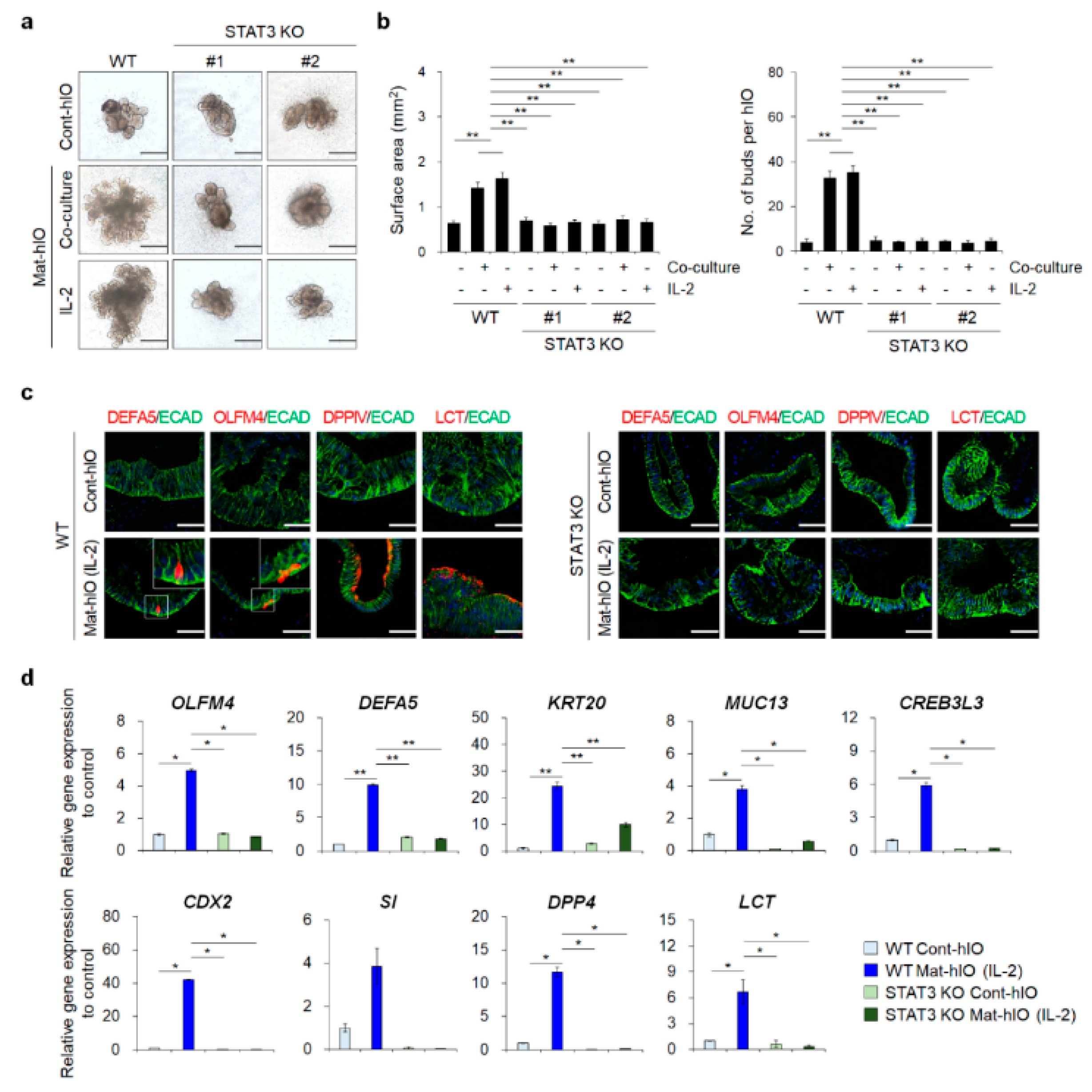
3.3. STAT3 KO hESC Lines Showed A Severe hIO Maturation Disadvantage In Vitro
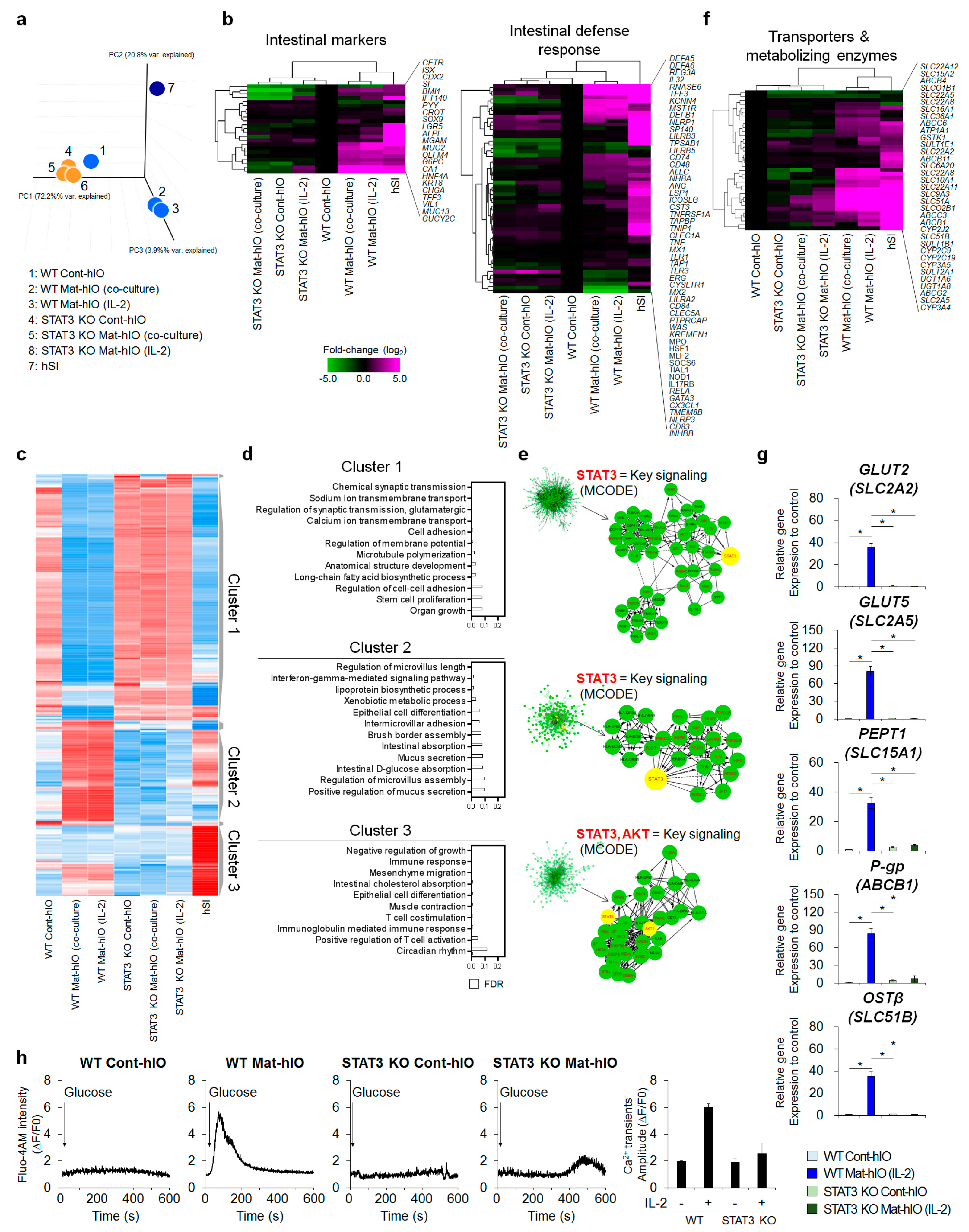
3.4. Transcriptome Comparisons Revealed That STAT3 Is Necessary for In Vitro hIO Maturation
3.5. STAT3 KO hIOs Did Not Grow And Maintain upon In Vivo Transplantation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Date, S.; Sato, T. Mini-gut organoids: Reconstitution of the stem cell niche. Annu. Rev. Cell Dev. Biol. 2015, 31, 269–289. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Clevers, H. Growing self-organizing mini-guts from a single intestinal stem cell: Mechanism and applications. Science 2013, 340, 1190–1194. [Google Scholar] [CrossRef]
- Fair, K.L.; Colquhoun, J.; Hannan, N.R.F. Intestinal organoids for modelling intestinal development and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20170217. [Google Scholar] [CrossRef] [PubMed]
- Spence, J.R.; Mayhew, C.N.; Rankin, S.A.; Kuhar, M.F.; Vallance, J.E.; Tolle, K.; Hoskins, E.E.; Kalinichenko, V.V.; Wells, S.I.; Zorn, A.M.; et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011, 470, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.M.; Spence, J.R. How to make an intestine. Development 2014, 141, 752–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finkbeiner, S.R.; Hill, D.R.; Altheim, C.H.; Dedhia, P.H.; Taylor, M.J.; Tsai, Y.H.; Chin, A.M.; Mahe, M.M.; Watson, C.L.; Freeman, J.J.; et al. Transcriptome-wide analysis reveals hallmarks of human intestine development and maturation in vitro and in vivo. Stem Cell Rep. 2015, 4, 1140–1155. [Google Scholar] [CrossRef] [PubMed]
- Fordham, R.P.; Yui, S.; Hannan, N.R.; Soendergaard, C.; Madgwick, A.; Schweiger, P.J.; Nielsen, O.H.; Vallier, L.; Pedersen, R.A.; Nakamura, T.; et al. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell 2013, 13, 734–744. [Google Scholar] [CrossRef]
- Watson, C.L.; Mahe, M.M.; Munera, J.; Howell, J.C.; Sundaram, N.; Poling, H.M.; Schweitzer, J.I.; Vallance, J.E.; Mayhew, C.N.; Sun, Y.; et al. An in vivo model of human small intestine using pluripotent stem cells. Nat. Med. 2014, 20, 1310–1314. [Google Scholar] [CrossRef] [Green Version]
- Jung, K.B.; Lee, H.; Son, Y.S.; Lee, M.O.; Kim, Y.D.; Oh, S.J.; Kwon, O.; Cho, S.; Cho, H.S.; Kim, D.S.; et al. Interleukin-2 induces the in vitro maturation of human pluripotent stem cell-derived intestinal organoids. Nat. Commun. 2018, 9, 3039. [Google Scholar] [CrossRef]
- Neufert, C.; Pickert, G.; Zheng, Y.; Wittkopf, N.; Warntjen, M.; Nikolaev, A.; Ouyang, W.; Neurath, M.F.; Becker, C. Activation of epithelial stat3 regulates intestinal homeostasis. Cell Cycle 2010, 9, 652–655. [Google Scholar] [CrossRef]
- Pickert, G.; Neufert, C.; Leppkes, M.; Zheng, Y.; Wittkopf, N.; Warntjen, M.; Lehr, H.A.; Hirth, S.; Weigmann, B.; Wirtz, S.; et al. Stat3 links il-22 signaling in intestinal epithelial cells to mucosal wound healing. J. Exp. Med. 2009, 206, 1465–1472. [Google Scholar] [CrossRef] [PubMed]
- Lindemans, C.A.; Calafiore, M.; Mertelsmann, A.M.; O’Connor, M.H.; Dudakov, J.A.; Jenq, R.R.; Velardi, E.; Young, L.F.; Smith, O.M.; Lawrence, G.; et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature 2015, 528, 560–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthews, J.R.; Sansom, O.J.; Clarke, A.R. Absolute requirement for stat3 function in small-intestine crypt stem cell survival. Cell Death Differ. 2011, 18, 1934–1943. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, K. Role of stat3 in inflammatory bowel disease. World J. Gastroenterol. 2008, 14, 5110–5114. [Google Scholar] [CrossRef] [PubMed]
- Atreya, R.; Mudter, J.; Finotto, S.; Mullberg, J.; Jostock, T.; Wirtz, S.; Schutz, M.; Bartsch, B.; Holtmann, M.; Becker, C.; et al. Blockade of interleukin 6 trans signaling suppresses t-cell resistance against apoptosis in chronic intestinal inflammation: Evidence in crohn disease and experimental colitis in vivo. Nat. Med. 2000, 6, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, W.T.; Warren, C.R.; Cowan, C.A. Genome editing in human pluripotent stem cells: Approaches, pitfalls, and solutions. Cell Stem Cell 2016, 18, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Son, M.Y.; Choi, H.; Han, Y.M.; Cho, Y.S. Unveiling the critical role of rex1 in the regulation of human stem cell pluripotency. Stem Cells 2013, 31, 2374–2387. [Google Scholar] [CrossRef] [PubMed]
- McCracken, K.W.; Howell, J.C.; Wells, J.M.; Spence, J.R. Generating human intestinal tissue from pluripotent stem cells in vitro. Nat. Protoc. 2011, 6, 1920–1928. [Google Scholar] [CrossRef] [Green Version]
- Son, M.Y.; Sim, H.; Son, Y.S.; Jung, K.B.; Lee, M.O.; Oh, J.H.; Chung, S.K.; Jung, C.R.; Kim, J. Distinctive genomic signature of neural and intestinal organoids from familial parkinson’s disease patient-derived induced pluripotent stem cells. Neuropathol. Appl. Neurobiol. 2017, 43, 584–603. [Google Scholar] [CrossRef]
- Ahn, J.; Ahn, J.H.; Yoon, S.; Nam, Y.S.; Son, M.Y.; Oh, J.H. Human three-dimensional in vitro model of hepatic zonation to predict zonal hepatotoxicity. J. Biol. Eng. 2019, 13, 22. [Google Scholar] [CrossRef] [Green Version]
- Jung, K.B.; Son, Y.S.; Lee, H.; Jung, C.R.; Kim, J.; Son, M.Y. Transcriptome dynamics of human pluripotent stem cell-derived contracting cardiomyocytes using an embryoid body model with fetal bovine serum. Mol. Biosyst. 2017, 13, 1565–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, K.B.; Lee, H.; Son, Y.S.; Lee, J.H.; Cho, H.S.; Lee, M.O.; Oh, J.H.; Lee, J.; Kim, S.; Jung, C.R.; et al. In vitro and in vivo imaging and tracking of intestinal organoids from human induced pluripotent stem cells. FASEB J. 2018, 32, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.O.; Jung, K.B.; Jo, S.J.; Hyun, S.A.; Moon, K.S.; Seo, J.W.; Kim, S.H.; Son, M.Y. Modelling cardiac fibrosis using three-dimensional cardiac microtissues derived from human embryonic stem cells. J. Biol. Eng. 2019, 13, 15. [Google Scholar] [CrossRef] [PubMed]
- Torre, D.; Lachmann, A.; Ma’ayan, A. Biojupies: Automated Generation of Interactive Notebooks for RNA-Seq Data Analysis in the Cloud. Cell Syst. 2018, 7, 556–561.e553. [Google Scholar] [CrossRef] [PubMed]
- Helander, H.F.; Fandriks, L. Surface area of the digestive tract—revisited. Scand. J. Gastroenterol. 2014, 49, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Poling, H.M.; Wu, D.; Brown, N.; Baker, M.; Hausfeld, T.A.; Huynh, N.; Chaffron, S.; Dunn, J.C.Y.; Hogan, S.P.; Wells, J.M.; et al. Mechanically induced development and maturation of human intestinal organoids in vivo. Nat. Biomed. Eng. 2018, 2, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M.; Mizutani, T.; Mochizuki, W.; Matsumoto, T.; Nozaki, K.; Sakamaki, Y.; Ichinose, S.; Okada, Y.; Tanaka, T.; Watanabe, M.; et al. Small intestinal stem cell identity is maintained with functional paneth cells in heterotopically grafted epithelium onto the colon. Genes Dev. 2014, 28, 1752–1757. [Google Scholar] [CrossRef]
- Cho, Y.Y.; Jeong, H.U.; Kim, J.H.; Lee, H.S. Effect of honokiol on the induction of drug-metabolizing enzymes in human hepatocytes. Drug Des. Dev. Ther. 2014, 8, 2137–2145. [Google Scholar] [Green Version]
- Takebe, T.; Enomura, M.; Yoshizawa, E.; Kimura, M.; Koike, H.; Ueno, Y.; Matsuzaki, T.; Yamazaki, T.; Toyohara, T.; Osafune, K.; et al. Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation. Cell Stem Cell 2015, 16, 556–565. [Google Scholar] [CrossRef]
- Takebe, T.; Sekine, K.; Enomura, M.; Koike, H.; Kimura, M.; Ogaeri, T.; Zhang, R.R.; Ueno, Y.; Zheng, Y.W.; Koike, N.; et al. Vascularized and functional human liver from an ipsc-derived organ bud transplant. Nature 2013, 499, 481–484. [Google Scholar] [CrossRef]
- Varzideh, F.; Pahlavan, S.; Ansari, H.; Halvaei, M.; Kostin, S.; Feiz, M.S.; Latifi, H.; Aghdami, N.; Braun, T.; Baharvand, H. Human cardiomyocytes undergo enhanced maturation in embryonic stem cell-derived organoid transplants. Biomaterials 2019, 192, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Asai, A.; Aihara, E.; Watson, C.; Mourya, R.; Mizuochi, T.; Shivakumar, P.; Phelan, K.; Mayhew, C.; Helmrath, M.; Takebe, T.; et al. Paracrine signals regulate human liver organoid maturation from induced pluripotent stem cells. Development 2017, 144, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Acuna, R.; Garcia, A.J. Engineered materials to model human intestinal development and cancer using organoids. Exp. Cell Res. 2019, 377, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Workman, M.J.; Mahe, M.M.; Trisno, S.; Poling, H.M.; Watson, C.L.; Sundaram, N.; Chang, C.F.; Schiesser, J.; Aubert, P.; Stanley, E.G.; et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat. Med. 2017, 23, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Forster, R.; Chiba, K.; Schaeffer, L.; Regalado, S.G.; Lai, C.S.; Gao, Q.; Kiani, S.; Farin, H.F.; Clevers, H.; Cost, G.J.; et al. Human intestinal tissue with adult stem cell properties derived from pluripotent stem cells. Stem Cell Rep. 2014, 2, 838–852. [Google Scholar] [CrossRef] [PubMed]
- Hutchins, A.P.; Diez, D.; Takahashi, Y.; Ahmad, S.; Jauch, R.; Tremblay, M.L.; Miranda-Saavedra, D. Distinct transcriptional regulatory modules underlie stat3’s cell type-independent and cell type-specific functions. Nucleic Acids Res. 2013, 41, 2155–2170. [Google Scholar] [CrossRef] [PubMed]
- Mudter, J.; Weigmann, B.; Bartsch, B.; Kiesslich, R.; Strand, D.; Galle, P.R.; Lehr, H.A.; Schmidt, J.; Neurath, M.F. Activation pattern of signal transducers and activators of transcription (stat) factors in inflammatory bowel diseases. Am. J. Gastroenterol. 2005, 100, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Lovato, P.; Brender, C.; Agnholt, J.; Kelsen, J.; Kaltoft, K.; Svejgaard, A.; Eriksen, K.W.; Woetmann, A.; Odum, N. Constitutive stat3 activation in intestinal t cells from patients with crohn’s disease. J. Biol. Chem. 2003, 278, 16777–16781. [Google Scholar] [CrossRef]
- Takeda, K.; Clausen, B.E.; Kaisho, T.; Tsujimura, T.; Terada, N.; Forster, I.; Akira, S. Enhanced th1 activity and development of chronic enterocolitis in mice devoid of stat3 in macrophages and neutrophils. Immunity 1999, 10, 39–49. [Google Scholar] [CrossRef]
- Willson, T.A.; Jurickova, I.; Collins, M.; Denson, L.A. Deletion of intestinal epithelial cell stat3 promotes t-lymphocyte stat3 activation and chronic colitis following acute dextran sodium sulfate injury in mice. Inflamm. Bowel Dis. 2013, 19, 512–525. [Google Scholar] [CrossRef]
- Camporeale, A.; Poli, V. Il-6, il-17 and stat3: A holy trinity in auto-immunity? Front. Biosci. (Landmark Ed.) 2012, 17, 2306–2326. [Google Scholar] [CrossRef]
- Huang, S. Regulation of metastases by signal transducer and activator of transcription 3 signaling pathway: Clinical implications. Clin. Cancer Res. 2007, 13, 1362–1366. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Zhang, Z.G.; Tian, X.Q.; Sun, D.F.; Liang, Q.C.; Zhang, Y.J.; Lu, R.; Chen, Y.X.; Fang, J.Y. Inhibition of jak1, 2/stat3 signaling induces apoptosis, cell cycle arrest, and reduces tumor cell invasion in colorectal cancer cells. Neoplasia 2008, 10, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Snyder, M.; Huang, X.Y.; Zhang, J.J. Identification of novel direct stat3 target genes for control of growth and differentiation. J. Biol. Chem. 2008, 283, 3791–3798. [Google Scholar] [CrossRef] [PubMed]
- Bitar, K.N.; Raghavan, S.; Zakhem, E. Tissue engineering in the gut: Developments in neuromusculature. Gastroenterology 2014, 146, 1614–1624. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, K.B.; Kwon, O.; Lee, M.-O.; Lee, H.; Son, Y.S.; Habib, O.; Oh, J.-H.; Cho, H.-S.; Jung, C.-R.; Kim, J.; et al. Blockade of STAT3 Causes Severe In Vitro and In Vivo Maturation Defects in Intestinal Organoids Derived from Human Embryonic Stem Cells. J. Clin. Med. 2019, 8, 976. https://doi.org/10.3390/jcm8070976
Jung KB, Kwon O, Lee M-O, Lee H, Son YS, Habib O, Oh J-H, Cho H-S, Jung C-R, Kim J, et al. Blockade of STAT3 Causes Severe In Vitro and In Vivo Maturation Defects in Intestinal Organoids Derived from Human Embryonic Stem Cells. Journal of Clinical Medicine. 2019; 8(7):976. https://doi.org/10.3390/jcm8070976
Chicago/Turabian StyleJung, Kwang Bo, Ohman Kwon, Mi-Ok Lee, Hana Lee, Ye Seul Son, Omer Habib, Jung-Hwa Oh, Hyun-Soo Cho, Cho-Rok Jung, Janghwan Kim, and et al. 2019. "Blockade of STAT3 Causes Severe In Vitro and In Vivo Maturation Defects in Intestinal Organoids Derived from Human Embryonic Stem Cells" Journal of Clinical Medicine 8, no. 7: 976. https://doi.org/10.3390/jcm8070976
APA StyleJung, K. B., Kwon, O., Lee, M.-O., Lee, H., Son, Y. S., Habib, O., Oh, J.-H., Cho, H.-S., Jung, C.-R., Kim, J., & Son, M.-Y. (2019). Blockade of STAT3 Causes Severe In Vitro and In Vivo Maturation Defects in Intestinal Organoids Derived from Human Embryonic Stem Cells. Journal of Clinical Medicine, 8(7), 976. https://doi.org/10.3390/jcm8070976





