New Era of Endoscopic Ultrasound-Guided Tissue Acquisition: Next-Generation Sequencing by Endoscopic Ultrasound-Guided Sampling for Pancreatic Cancer
Abstract
:1. Introduction
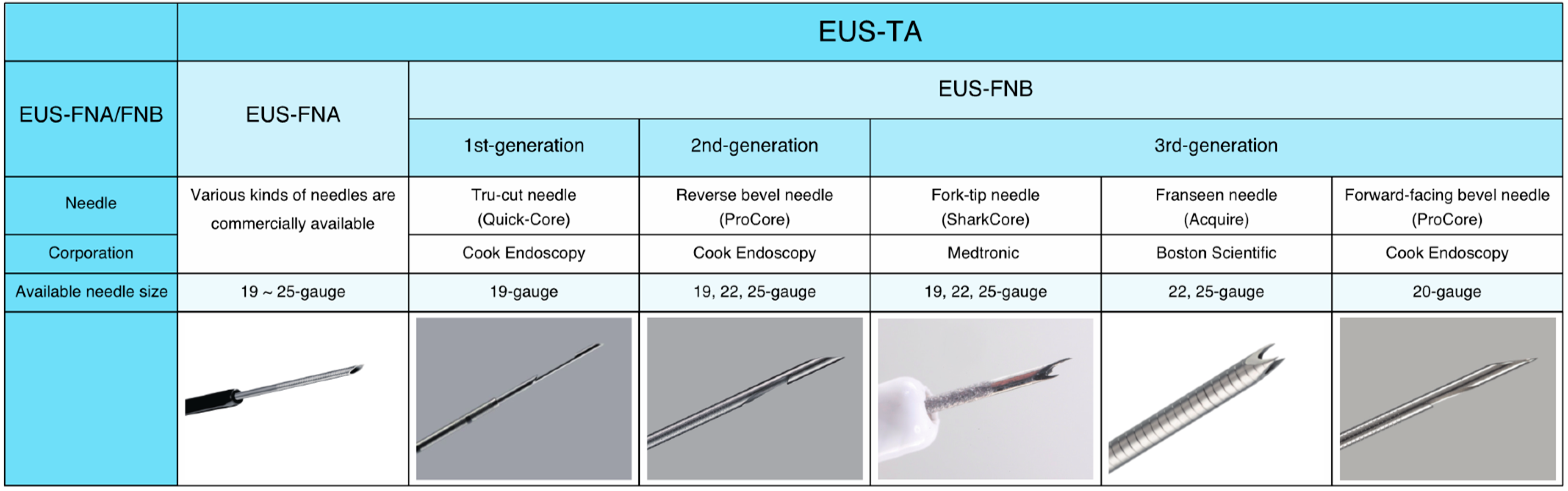
2. Endoscopic Ultrasound-Guided Tissue Acquisition (EUS-TA)
3. Next-Generation Sequencing (NGS)
4. Genetic Markers in Pancreatic Cancer
5. Clinical Utility of EUS-TA for NGS
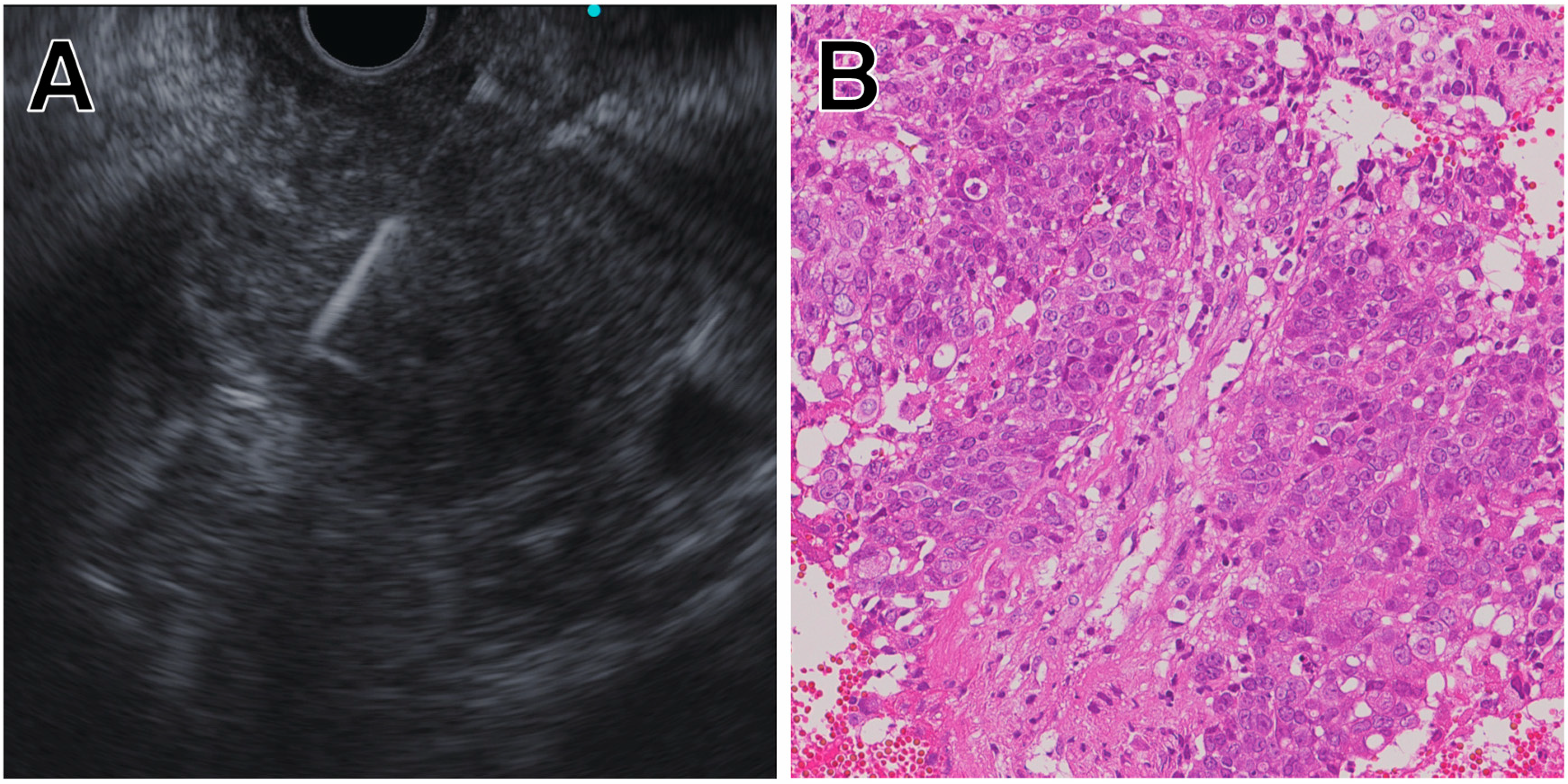
5.1. Solid Pancreatic Masses
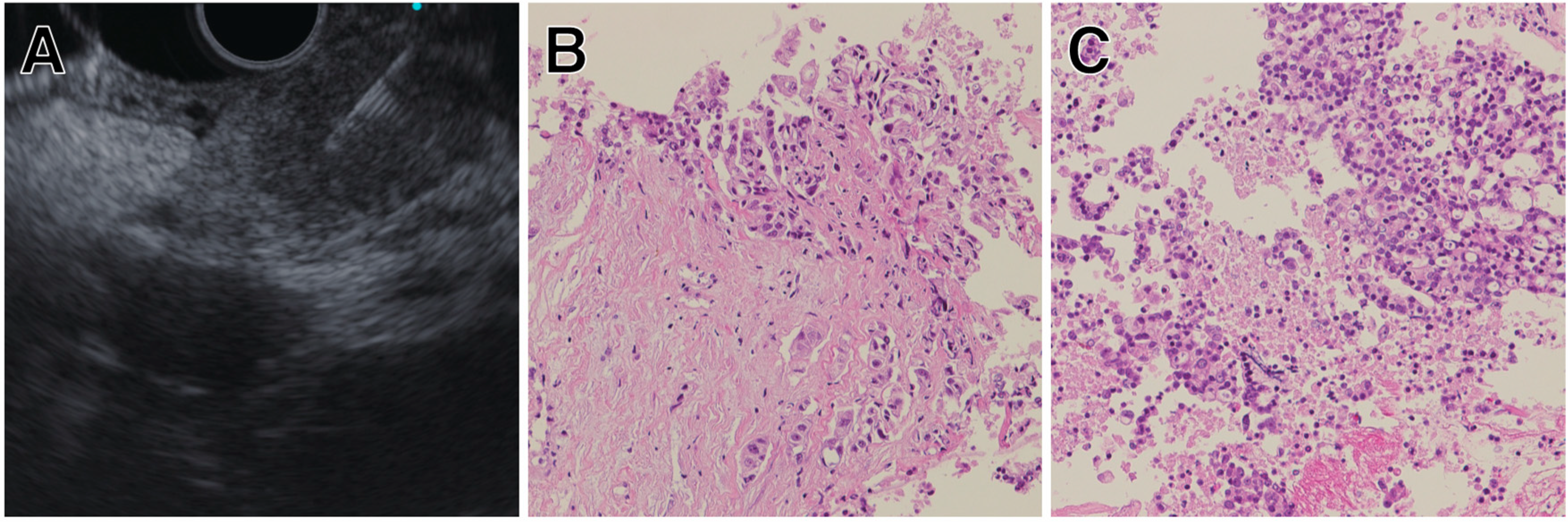
5.2. Pancreatic Cystic Lesions
6. MicroRNAs (miRNA)
7. How to Obtain Adequate Samples for NGS via EUS-TA?
7.1. Target Site
7.2. Rapid On-Site Evaluation (ROSE)
7.3. Which Should We Choose, EUS-FNA or EUS-FNB?
7.4. Needle Size: Small versus Large
7.5. Technique: Suction vs, Non-Suction (Slow-Pull Technique)
8. Conclusions
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [PubMed]
- Burris, H.A., 3rd; Moore, M.J.; Andersen, J.; Green, M.R.; Rothenberg, M.L.; Modiano, M.R.; Cripps, M.C.; Portenoy, R.K.; Storniolo, A.M.; Tarassoff, P.; et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: A randomized trial. J. Clin. Oncol. 1997, 15, 2403–2413. [Google Scholar] [PubMed]
- Moore, M.J.; Goldstein, D.; Hamm, J.; Figer, A.; Hecht, J.R.; Gallinger, S.; Au, H.J.; Murawa, P.; Walde, D.; Wolff, R.A.; et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: A phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J. Clin. Oncol. 2007, 25, 1960–1966. [Google Scholar] [PubMed]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouche, O.; Guimbaud, R.; Becouarn, Y.; Adenis, A.; Raoul, J.L.; Gourgou-Bourgade, S.; de la Fouchardiere, C.; et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [PubMed]
- Von Hoff, D.D.; Ervin, T.; Arena, F.P.; Chiorean, E.G.; Infante, J.; Moore, M.; Seay, T.; Tjulandin, S.A.; Ma, W.W.; Saleh, M.N.; et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med. 2013, 369, 1691–1703. [Google Scholar] [PubMed]
- Rawla, P.; Sunkara, T.; Gaduputi, V. Epidemiology of pancreatic cancer: Global trends, etiology and risk factors. World J. Oncol. 2019, 10, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Rosell, R.; Moran, T.; Queralt, C.; Porta, R.; Cardenal, F.; Camps, C.; Majem, M.; Lopez-Vivanco, G.; Isla, D.; Provencio , M.; et al. Screening for epidermal growth factor receptor mutations in lung cancer. N. Engl. J. Med. 2009, 361, 958–967. [Google Scholar] [PubMed]
- Moore, K.; Colombo, N.; Scambia, G.; Kim, B.G.; Oaknin, A.; Friedlander, M.; Lisyanskaya, A.; Floquet, A.; Leary, A.; Sonke, G.S.; et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med. 2018, 379, 2495–2505. [Google Scholar] [PubMed]
- Cecchini, M.; Rubin, E.H.; Blumenthal, G.M.; Ayalew, K.; Burris, H.A.; Russell-Einhorn, M.; Dillon, H.; Lyerly, H.K.; Reaman, G.H.; Boerner, S.; et al. Challenges with novel clinical trial designs: Master protocols. Clin. Cancer Res. 2019, 25, 2049–2057. [Google Scholar]
- Woodcock, J.; LaVange, L.M. Master Protocols to study multiple therapies, multiple diseases, or both. N. Engl. J. Med. 2017, 377, 62–70. [Google Scholar] [CrossRef]
- Heestand, G.M.; Kurzrock, R. Molecular landscape of pancreatic cancer: Implications for current clinical trials. Oncotarget 2015, 6, 4553–4561. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.; Marabelle, A.; Kim, T.W.; Geva, R.; Van Cutsem, E.; André, T.; Ascierto, P.A.; Maio, M.; Delord, J.-P.; Gottfried, M.; et al. Efficacy of pembrolizumab in phase 2 KEYNOTE-164 and KEYNOTE-158 studies of microsatellite instability high cancers. Ann. Oncol. 2017, 28. [Google Scholar] [CrossRef]
- DiMagno, E.P.; Buxton, J.L.; Regan, P.T.; Hattery, R.R.; Wilson, D.A.; Suarez, J.R.; Green, P.S. Ultrasonic endoscope. Lancet 1980, 1, 629–631. [Google Scholar] [CrossRef]
- Gress, F.G. The early history of interventional endoscopic ultrasound. Gastrointest. Endosc. Clin. N. Am. 2017, 27, 547–550. [Google Scholar]
- Vilmann, P.; Jacobsen, G.K.; Henriksen, F.W.; Hancke, S. Endoscopic ultrasonography with guided fine needle aspiration biopsy in pancreatic disease. Gastrointest. Endosc. 1992, 38, 172–173. [Google Scholar] [PubMed]
- Levy, M.J.; Wiersema, M.J. EUS-guided Trucut biopsy. Gastrointest. Endosc. 2005, 62, 417–426. [Google Scholar]
- Hebert-Magee, S.; Bae, S.; Varadarajulu, S.; Ramesh, J.; Frost, A.R.; Eloubeidi, M.A.; Eltoum, I.A. The presence of a cytopathologist increases the diagnostic accuracy of endoscopic ultrasound-guided fine needle aspiration cytology for pancreatic adenocarcinoma: A meta-analysis. Cytopathology 2013, 24, 159–171. [Google Scholar] [PubMed]
- Hewitt, M.J.; McPhail, M.J.; Possamai, L.; Dhar, A.; Vlavianos, P.; Monahan, K.J. EUS-guided FNA for diagnosis of solid pancreatic neoplasms: A meta-analysis. Gastrointest. Endosc. 2012, 75, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Puli, S.R.; Bechtold, M.L.; Buxbaum, J.L.; Eloubeidi, M.A. How good is endoscopic ultrasound-guided fine-needle aspiration in diagnosing the correct etiology for a solid pancreatic mass? A meta-analysis and systematic review. Pancreas 2013, 42, 20–26. [Google Scholar] [CrossRef]
- Yoshinaga, S.; Itoi, T.; Yamao, K.; Yasuda, I.; Irisawa, A.; Imaoka, H.; Tsuchiya, T.; Doi, S.; Yamabe, A.; Murakami, Y.; et al. Safety and efficacy of endoscopic ultrasound-guided fine needle aspiration for pancreatic masses: A prospective multicenter study. Dig. Endosc. 2019. E-pub ahead of print. [Google Scholar] [CrossRef]
- Van Riet, P.A.; Larghi, A.; Attili, F.; Rindi, G.; Nguyen, N.Q.; Ruszkiewicz, A.; Kitano, M.; Chikugo, T.; Aslanian, H.; Farrell, J.; et al. A multicenter randomized trial comparing a 25-gauge EUS fine-needle aspiration device with a 20-gauge EUS fine-needle biopsy device. Gastrointest. Endosc. 2019, 89, 329–339. [Google Scholar] [PubMed]
- El Chafic, A.H.; Loren, D.; Siddiqui, A.; Mounzer, R.; Cosgrove, N.; Kowalski, T. Comparison of FNA and fine-needle biopsy for EUS-guided sampling of suspected GI stromal tumors. Gastrointest. Endosc. 2017, 86, 510–515. [Google Scholar] [PubMed]
- Standards of Practice Committee; Faulx, A.L.; Kothari, S.; Acosta, R.D.; Agrawal, D.; Bruining, D.H.; Chandrasekhara, V.; Eloubeidi, M.A.; Fanelli, R.D.; Gurudu, S.R.; et al. The role of endoscopy in subepithelial lesions of the GI tract. Gastrointest. Endosc. 2017, 85, 1117–1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Ying, K.; Shi, L.; Zhang, L.; Zhou, L. Combined endobronchial and endoscopic ultrasound-guided fine needle aspiration for mediastinal lymph node staging of lung cancer: A meta-analysis. Eur. J. Cancer 2013, 49, 1860–1867. [Google Scholar] [CrossRef] [PubMed]
- Puli, S.R.; Batapati Krishna Reddy, J.; Bechtold, M.L.; Ibdah, J.A.; Antillon, D.; Singh, S.; Olyaee, M.; Antillon, M.R. Endoscopic ultrasound: It’s accuracy in evaluating mediastinal lymphadenopathy? A meta-analysis and systematic review. World J. Gastroenterol. 2008, 14, 3028–3037. [Google Scholar] [PubMed]
- Sadeghi, A.; Mohamadnejad, M.; Islami, F.; Keshtkar, A.; Biglari, M.; Malekzadeh, R.; Eloubeidi, M.A. Diagnostic yield of EUS-guided FNA for malignant biliary stricture: A systematic review and meta-analysis. Gastrointest. Endosc. 2016, 83, 290–298.e1. [Google Scholar] [PubMed]
- Yamao, K.; Sawaki, A.; Takahashi, K.; Imaoka, H.; Ashida, R.; Mizuno, N. EUS-guided choledochoduodenostomy for palliative biliary drainage in case of papillary obstruction: Report of 2 cases. Gastrointest. Endosc. 2006, 64, 663–667. [Google Scholar] [CrossRef] [PubMed]
- Artifon, E.L.; Aparicio, D.; Paione, J.B.; Lo, S.K.; Bordini, A.; Rabello, C.; Otoch, J.P.; Gupta, K. Biliary drainage in patients with unresectable, malignant obstruction where ERCP fails: Endoscopic ultrasonography-guided choledochoduodenostomy versus percutaneous drainage. J. Clin. Gastroenterol. 2012, 46, 768–774. [Google Scholar]
- Hara, K.; Yamao, K.; Hijioka, S.; Mizuno, N.; Imaoka, H.; Tajika, M.; Kondo, S.; Tanaka, T.; Haba, S.; Takeshi, O.; et al. Prospective clinical study of endoscopic ultrasound-guided choledochoduodenostomy with direct metallic stent placement using a forward-viewing echoendoscope. Endoscopy 2013, 45, 392–396. [Google Scholar]
- Artifon, E.L.; Marson, F.P.; Gaidhane, M.; Kahaleh, M.; Otoch, J.P. Hepaticogastrostomy or choledochoduodenostomy for distal malignant biliary obstruction after failed ERCP: Is there any difference? Gastrointest. Endosc. 2015, 81, 950–959. [Google Scholar]
- Lee, T.H.; Choi, J.H.; Park do, H.; Song, T.J.; Kim, D.U.; Paik, W.H.; Hwangbo, Y.; Lee, S.S.; Seo, D.W.; Lee, S.K.; et al. Similar efficacies of endoscopic ultrasound-guided transmural and percutaneous drainage for malignant distal biliary obstruction. Clin. Gastroenterol. Hepatol. 2016, 14, 1011–1019.e3. [Google Scholar]
- Fabbri, C.; Luigiano, C.; Lisotti, A.; Cennamo, V.; Virgilio, C.; Caletti, G.; Fusaroli, P. Endoscopic ultrasound-guided treatments: Are we getting evidence based--A systematic review. World J. Gastroenterol. 2014, 20, 8424–8448. [Google Scholar] [CrossRef]
- Varadarajulu, S.; Bang, J.Y.; Sutton, B.S.; Trevino, J.M.; Christein, J.D.; Wilcox, C.M. Equal efficacy of endoscopic and surgical cystogastrostomy for pancreatic pseudocyst drainage in a randomized trial. Gastroenterology 2013, 145, 583–590.e1. [Google Scholar]
- Park, D.H.; Lee, S.S.; Moon, S.H.; Choi, S.Y.; Jung, S.W.; Seo, D.W.; Lee, S.K.; Kim, M.H. Endoscopic ultrasound-guided versus conventional transmural drainage for pancreatic pseudocysts: A prospective randomized trial. Endoscopy 2009, 41, 842–848. [Google Scholar] [CrossRef]
- Varadarajulu, S.; Christein, J.D.; Tamhane, A.; Drelichman, E.R.; Wilcox, C.M. Prospective randomized trial comparing EUS and EGD for transmural drainage of pancreatic pseudocysts (with videos). Gastrointest. Endosc. 2008, 68, 1102–1111. [Google Scholar]
- Puli, S.R.; Reddy, J.B.; Bechtold, M.L.; Antillon, M.R.; Brugge, W.R. EUS-guided celiac plexus neurolysis for pain due to chronic pancreatitis or pancreatic cancer pain: A meta-analysis and systematic review. Dig. Dis. Sci. 2009, 54, 2330–2337. [Google Scholar]
- Wyse, J.M.; Carone, M.; Paquin, S.C.; Usatii, M.; Sahai, A.V. Randomized, double-blind, controlled trial of early endoscopic ultrasound-guided celiac plexus neurolysis to prevent pain progression in patients with newly diagnosed, painful, inoperable pancreatic cancer. J. Clin. Oncol. 2011, 29, 3541–3546. [Google Scholar] [CrossRef]
- LeBlanc, J.K.; Al-Haddad, M.; McHenry, L.; Sherman, S.; Juan, M.; McGreevy, K.; Johnson, C.; Howard, T.J.; Lillemoe, K.D.; DeWitt, J. A prospective, randomized study of EUS-guided celiac plexus neurolysis for pancreatic cancer: One injection or two? Gastrointest. Endosc. 2011, 74, 1300–1307. [Google Scholar]
- Doi, S.; Yasuda, I.; Kawakami, H.; Hayashi, T.; Hisai, H.; Irisawa, A.; Mukai, T.; Katanuma, A.; Kubota, K.; Ohnishi, T.; et al. Endoscopic ultrasound-guided celiac ganglia neurolysis vs. celiac plexus neurolysis: A randomized multicenter trial. Endoscopy 2013, 45, 362–369. [Google Scholar] [PubMed]
- Santosh, D.; Lakhtakia, S.; Gupta, R.; Reddy, D.N.; Rao, G.V.; Tandan, M.; Ramchandani, M.; Guda, N.M. Clinical trial: A randomized trial comparing fluoroscopy guided percutaneous technique vs. endoscopic ultrasound guided technique of coeliac plexus block for treatment of pain in chronic pancreatitis. Aliment. Pharmacol. Ther. 2009, 29, 979–984. [Google Scholar]
- Gress, F.; Schmitt, C.; Sherman, S.; Ikenberry, S.; Lehman, G. A prospective randomized comparison of endoscopic ultrasound- and computed tomography-guided celiac plexus block for managing chronic pancreatitis pain. Am. J. Gastroenterol. 1999, 94, 900–905. [Google Scholar] [CrossRef]
- Rios Castellanos, E.; Seron, P.; Gisbert, J.P.; Bonfill Cosp, X. Endoscopic injection of cyanoacrylate glue versus other endoscopic procedures for acute bleeding gastric varices in people with portal hypertension. Cochrane Database Syst. Rev. 2015, 5, CD010180. [Google Scholar] [CrossRef]
- De Paulo, G.A.; Ardengh, J.C.; Nakao, F.S.; Ferrari, A.P. Treatment of esophageal varices: A randomized controlled trial comparing endoscopic sclerotherapy and EUS-guided sclerotherapy of esophageal collateral veins. Gastrointest. Endosc. 2006, 63, 396–402. [Google Scholar] [CrossRef]
- Romero-Castro, R.; Ellrichmann, M.; Ortiz-Moyano, C.; Subtil-Inigo, J.C.; Junquera-Florez, F.; Gornals, J.B.; Repiso-Ortega, A.; Vila-Costas, J.; Marcos-Sanchez, F.; Munoz-Navas, M.; et al. EUS-guided coil versus cyanoacrylate therapy for the treatment of gastric varices: A multicenter study (with videos). Gastrointest. Endosc. 2013, 78, 711–721. [Google Scholar] [CrossRef]
- Binmoeller, K.F.; Weilert, F.; Shah, J.N.; Kim, J. EUS-guided transesophageal treatment of gastric fundal varices with combined coiling and cyanoacrylate glue injection (with videos). Gastrointest. Endosc. 2011, 74, 1019–1025. [Google Scholar] [CrossRef]
- Roychowdhury, S.; Chinnaiyan, A.M. Translating cancer genomes and transcriptomes for precision oncology. CA Cancer J. Clin. 2016, 66, 75–88. [Google Scholar] [CrossRef]
- Janiaud, P.; Serghiou, S.; Ioannidis, J.P.A. New clinical trial designs in the era of precision medicine: An overview of definitions, strengths, weaknesses, and current use in oncology. Cancer Treat. Rev. 2019, 73, 20–30. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [Green Version]
- Voelkerding, K.V.; Dames, S.A.; Durtschi, J.D. Next-generation sequencing: From basic research to diagnostics. Clin. Chem. 2009, 55, 641–658. [Google Scholar] [CrossRef]
- Mardis, E.R.; Wilson, R.K. Cancer genome sequencing: A review. Hum. Mol. Genet. 2009, 18, R163–R168. [Google Scholar] [CrossRef]
- Aziz, N.; Zhao, Q.; Bry, L.; Driscoll, D.K.; Funke, B.; Gibson, J.S.; Grody, W.W.; Hegde, M.R.; Hoeltge, G.A.; Leonard, D.G.; et al. College of American Pathologists’ laboratory standards for next-generation sequencing clinical tests. Arch. Pathol. Lab. Med. 2015, 139, 481–493. [Google Scholar] [CrossRef]
- Luthra, R.; Chen, H.; Roy-Chowdhuri, S.; Singh, R.R. Next-generation sequencing in clinical molecular diagnostics of cancer: Advantages and challenges. Cancers 2015, 7, 2023–2036. [Google Scholar] [CrossRef]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 2004, 431, 931–945. [Google Scholar] [CrossRef]
- Kamisawa, T.; Wood, L.D.; Itoi, T.; Takaori, K. Pancreatic cancer. The Lancet 2016, 388, 73–85. [Google Scholar] [CrossRef]
- Wang, X.; Gao, J.; Ren, Y.; Gu, J.; Du, Y.; Chen, J.; Jin, Z.; Zhan, X.; Li, Z.; Huang, H.; et al. Detection of KRAS gene mutations in endoscopic ultrasound-guided fine-needle aspiration biopsy for improving pancreatic cancer diagnosis. Am J. Gastroenterol. 2011, 106, 2104–2111. [Google Scholar] [CrossRef]
- Bournet, B.; Souque, A.; Senesse, P.; Assenat, E.; Barthet, M.; Lesavre, N.; Aubert, A.; O’Toole, D.; Hammel, P.; Levy, P.; et al. Endoscopic ultrasound-guided fine-needle aspiration biopsy coupled with KRAS mutation assay to distinguish pancreatic cancer from pseudotumoral chronic pancreatitis. Endoscopy 2009, 41, 552–557. [Google Scholar] [CrossRef]
- Ogura, T.; Yamao, K.; Sawaki, A.; Mizuno, N.; Hara, K.; Hijioka, S.; Niwa, Y.; Tajika, M.; Kondo, S.; Shimizu, Y.; et al. Clinical impact of K-ras mutation analysis in EUS-guided FNA specimens from pancreatic masses. Gastrointest. Endosc. 2012, 75, 769–774. [Google Scholar] [CrossRef]
- Reicher, S.; Boyar, F.Z.; Albitar, M.; Sulcova, V.; Agersborg, S.; Nga, V.; Zhou, Y.; Li, G.; Venegas, R.; French, S.W.; et al. Fluorescence in situ hybridization and K-ras analyses improve diagnostic yield of endoscopic ultrasound-guided fine-needle aspiration of solid pancreatic masses. Pancreas 2011, 40, 1057–1062. [Google Scholar] [CrossRef]
- Tada, M.; Komatsu, Y.; Kawabe, T.; Sasahira, N.; Isayama, H.; Toda, N.; Shiratori, Y.; Omata, M. Quantitative analysis of K-ras gene mutation in pancreatic tissue obtained by endoscopic ultrasonography-guided fine needle aspiration: Clinical utility for diagnosis of pancreatic tumor. Am J. Gastroenterol. 2002, 97, 2263–2270. [Google Scholar] [CrossRef]
- Maluf-Filho, F.; Kumar, A.; Gerhardt, R.; Kubrusly, M.; Sakai, P.; Hondo, F.; Matuguma, S.E.; Artifon, E.; Monteiro da Cunha, J.E.; Cesar Machado, M.C.; et al. Kras mutation analysis of fine needle aspirate under EUS guidance facilitates risk stratification of patients with pancreatic mass. J. Clin. Gastroenterol. 2007, 41, 906–910. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamao, K.; Okubo, K.; Sawaki, A.; Mizuno, N.; Ashida, R.; Koshikawa, T.; Ueyama, Y.; Kasugai, K.; Hase, S.; et al. Differential diagnosis of pancreatic cancer and focal pancreatitis by using EUS-guided FNA. Gastrointest. Endosc. 2005, 61, 76–79. [Google Scholar] [CrossRef]
- Pellise, M.; Castells, A.; Gines, A.; Sole, M.; Mora, J.; Castellvi-Bel, S.; Rodriguez-Moranta, F.; Fernandez-Esparrach, G.; Llach, J.; Bordas, J.M.; et al. Clinical usefulness of KRAS mutational analysis in the diagnosis of pancreatic adenocarcinoma by means of endosonography-guided fine-needle aspiration biopsy. Aliment. Pharmacol. Ther. 2003, 17, 1299–1307. [Google Scholar] [CrossRef]
- Ogura, T.; Yamao, K.; Hara, K.; Mizuno, N.; Hijioka, S.; Imaoka, H.; Sawaki, A.; Niwa, Y.; Tajika, M.; Kondo, S.; et al. Prognostic value of K-ras mutation status and subtypes in endoscopic ultrasound-guided fine-needle aspiration specimens from patients with unresectable pancreatic cancer. J. Gastroenterol. 2013, 48, 640–646. [Google Scholar] [CrossRef]
- Kyriakis, J.M.; App, H.; Zhang, X.F.; Banerjee, P.; Brautigan, D.L.; Rapp, U.R.; Avruch, J. Raf-1 activates MAP kinase-kinase. Nature 1992, 358, 417–421. [Google Scholar] [CrossRef]
- Bonni, A.; Brunet, A.; West, A.E.; Datta, S.R.; Takasu, M.A.; Greenberg, M.E. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science 1999, 286, 1358–1362. [Google Scholar] [CrossRef]
- Minden, A.; Lin, A.; McMahon, M.; Lange-Carter, C.; Derijard, B.; Davis, R.J.; Johnson, G.L.; Karin, M. Differential activation of ERK and JNK mitogen-activated protein kinases by Raf-1 and MEKK. Science 1994, 266, 1719–1723. [Google Scholar] [CrossRef]
- Biankin, A.V.; Waddell, N.; Kassahn, K.S.; Gingras, M.C.; Muthuswamy, L.B.; Johns, A.L.; Miller, D.K.; Wilson, P.J.; Patch, A.M.; Wu, J.; et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012, 491, 399–405. [Google Scholar] [CrossRef]
- Omori, Y.; Ono, Y.; Tanino, M.; Karasaki, H.; Yamaguchi, H.; Furukawa, T.; Enomoto, K.; Ueda, J.; Sumi, A.; Katayama, J.; et al. Pathways of progression from intraductal papillary mucinous neoplasm to pancreatic ductal adenocarcinoma based on molecular features. Gastroenterology 2019, 156, 647–661.e2. [Google Scholar] [CrossRef]
- Wood, L.D.; Hruban, R.H. Pathology and molecular genetics of pancreatic neoplasms. Cancer J. 2012, 18, 492–501. [Google Scholar] [CrossRef]
- Ruggeri, B.; Zhang, S.Y.; Caamano, J.; DiRado, M.; Flynn, S.D.; Klein-Szanto, A.J. Human pancreatic carcinomas and cell lines reveal frequent and multiple alterations in the p53 and Rb-1 tumor-suppressor genes. Oncogene 1992, 7, 1503–1511. [Google Scholar]
- Kanda, M.; Matthaei, H.; Wu, J.; Hong, S.M.; Yu, J.; Borges, M.; Hruban, R.H.; Maitra, A.; Kinzler, K.; Vogelstein, B.; et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 2012, 142, 730–733.e9. [Google Scholar] [CrossRef]
- Maitra, A.; Adsay, N.V.; Argani, P.; Iacobuzio-Donahue, C.; De Marzo, A.; Cameron, J.L.; Yeo, C.J.; Hruban, R.H. Multicomponent analysis of the pancreatic adenocarcinoma progression model using a pancreatic intraepithelial neoplasia tissue microarray. Mod. Pathol. 2003, 16, 902–912. [Google Scholar] [CrossRef]
- Pfeifer, J.D. Clinical next generation sequencing in cancer. Cancer Genet. 2013, 206, 409–412. [Google Scholar] [CrossRef]
- Kameta, E.; Sugimori, K.; Kaneko, T.; Ishii, T.; Miwa, H.; Sato, T.; Ishii, Y.; Sue, S.; Sasaki, T.; Yamashita, Y.; et al. Diagnosis of pancreatic lesions collected by endoscopic ultrasound-guided fine-needle aspiration using next-generation sequencing. Oncol. Lett. 2016, 12, 3875–3881. [Google Scholar] [CrossRef]
- De Biase, D.; Visani, M.; Baccarini, P.; Polifemo, A.M.; Maimone, A.; Fornelli, A.; Giuliani, A.; Zanini, N.; Fabbri, C.; Pession, A.; et al. Next generation sequencing improves the accuracy of KRAS mutation analysis in endoscopic ultrasound fine needle aspiration pancreatic lesions. PLoS ONE 2014, 9, e87651. [Google Scholar] [CrossRef]
- Valero, V., 3rd; Saunders, T.J.; He, J.; Weiss, M.J.; Cameron, J.L.; Dholakia, A.; Wild, A.T.; Shin, E.J.; Khashab, M.A.; O’Broin-Lennon, A.M.; et al. Reliable detection of somatic mutations in fine needle aspirates of pancreatic cancer with next-generation sequencing: Implications for surgical management. Ann. Surg. 2016, 263, 153–161. [Google Scholar] [CrossRef]
- Gleeson, F.C.; Kerr, S.E.; Kipp, B.R.; Voss, J.S.; Minot, D.M.; Tu, Z.J.; Henry, M.R.; Graham, R.P.; Vasmatzis, G.; Cheville, J.C.; et al. Targeted next generation sequencing of endoscopic ultrasound acquired cytology from ampullary and pancreatic adenocarcinoma has the potential to aid patient stratification for optimal therapy selection. Oncotarget 2016, 7, 54526–54536. [Google Scholar] [CrossRef] [Green Version]
- Larson, B.K.; Tuli, R.; Jamil, L.H.; Lo, S.K.; Deng, N.; Hendifar, A.E. Utility of endoscopic ultrasound-guided biopsy for next-generation sequencing of pancreatic exocrine malignancies. Pancreas 2018, 47, 990–995. [Google Scholar] [CrossRef]
- Elhanafi, S.; Mahmud, N.; Vergara, N.; Kochman, M.L.; Das, K.K.; Ginsberg, G.G.; Rajala, M.; Chandrasekhara, V. Comparison of endoscopic ultrasound tissue acquisition methods for genomic analysis of pancreatic cancer. J. Gastroenterol. Hepatol. 2019, 34, 907–913. [Google Scholar] [CrossRef]
- Young, G.; Wang, K.; He, J.; Otto, G.; Hawryluk, M.; Zwirco, Z.; Brennan, T.; Nahas, M.; Donahue, A.; Yelensky, R.; et al. Clinical next-generation sequencing successfully applied to fine-needle aspirations of pulmonary and pancreatic neoplasms. Cancer Cytopathol. 2013, 121, 688–694. [Google Scholar] [CrossRef]
- Gleeson, F.C.; Voss, J.S.; Kipp, B.R.; Kerr, S.E.; Van Arnam, J.S.; Mills, J.R.; Marcou, C.A.; Schneider, A.R.; Tu, Z.J.; Henry, M.R.; et al. Assessment of pancreatic neuroendocrine tumor cytologic genotype diversity to guide personalized medicine using a custom gastroenteropancreatic next-generation sequencing panel. Oncotarget 2017, 8, 93464–93475. [Google Scholar] [CrossRef] [Green Version]
- Kubota, Y.; Kawakami, H.; Natsuizaka, M.; Kawakubo, K.; Marukawa, K.; Kudo, T.; Abe, Y.; Kubo, K.; Kuwatani, M.; Hatanaka, Y.; et al. CTNNB1 mutational analysis of solid-pseudopapillary neoplasms of the pancreas using endoscopic ultrasound-guided fine-needle aspiration and next-generation deep sequencing. J. Gastroenterol. 2015, 50, 203–210. [Google Scholar] [CrossRef]
- Springer, S.; Wang, Y.; Dal Molin, M.; Masica, D.L.; Jiao, Y.; Kinde, I.; Blackford, A.; Raman, S.P.; Wolfgang, C.L.; Tomita, T.; et al. A combination of molecular markers and clinical features improve the classification of pancreatic cysts. Gastroenterology 2015, 149, 1501–1510. [Google Scholar] [CrossRef]
- Laffan, T.A.; Horton, K.M.; Klein, A.P.; Berlanstein, B.; Siegelman, S.S.; Kawamoto, S.; Johnson, P.T.; Fishman, E.K.; Hruban, R.H. Prevalence of unsuspected pancreatic cysts on MDCT. Am. J. Roentgenol. 2008, 191, 802–807. [Google Scholar] [CrossRef]
- Lee, K.S.; Sekhar, A.; Rofsky, N.M.; Pedrosa, I. Prevalence of incidental pancreatic cysts in the adult population on MR imaging. Am. J. Gastroenterol. 2010, 105, 2079–2084. [Google Scholar] [CrossRef]
- De Oliveira, P.B.; Puchnick, A.; Szejnfeld, J.; Goldman, S.M. Prevalence of incidental pancreatic cysts on 3 tesla magnetic resonance. PLoS ONE 2015, 10, e0121317. [Google Scholar] [CrossRef]
- Tanaka, M.; Fernandez-Del Castillo, C.; Kamisawa, T.; Jang, J.Y.; Levy, P.; Ohtsuka, T.; Salvia, R.; Shimizu, Y.; Tada, M.; Wolfgang, C.L. Revisions of international consensus Fukuoka guidelines for the management of IPMN of the pancreas. Pancreatology 2017, 17, 738–753. [Google Scholar] [CrossRef]
- Pergolini, I.; Sahora, K.; Ferrone, C.R.; Morales-Oyarvide, V.; Wolpin, B.M.; Mucci, L.A.; Brugge, W.R.; Mino-Kenudson, M.; Patino, M.; Sahani, D.V.; et al. Long-term risk of pancreatic malignancy in patients with branch duct intraductal papillary mucinous neoplasm in a referral center. Gastroenterology 2017, 153, 1284–1294.e1. [Google Scholar] [CrossRef]
- Thornton, G.D.; McPhail, M.J.; Nayagam, S.; Hewitt, M.J.; Vlavianos, P.; Monahan, K.J. Endoscopic ultrasound guided fine needle aspiration for the diagnosis of pancreatic cystic neoplasms: A meta-analysis. Pancreatology 2013, 13, 48–57. [Google Scholar] [CrossRef]
- Suzuki, R.; Thosani, N.; Annangi, S.; Guha, S.; Bhutani, M.S. Diagnostic yield of EUS-FNA-based cytology distinguishing malignant and benign IPMNs: A systematic review and meta-analysis. Pancreatology 2014, 14, 380–384. [Google Scholar] [CrossRef]
- Singhi, A.D.; Nikiforova, M.N.; Fasanella, K.E.; McGrath, K.M.; Pai, R.K.; Ohori, N.P.; Bartholow, T.L.; Brand, R.E.; Chennat, J.S.; Lu, X. Preoperative GNAS and KRAS testing in the diagnosis of pancreatic mucinous cysts. Clin. Cancer Res. 2014, 20, 4381–4389. [Google Scholar] [CrossRef]
- Wu, J.; Matthaei, H.; Maitra, A.; Dal Molin, M.; Wood, L.D.; Eshleman, J.R.; Goggins, M.; Canto, M.I.; Schulick, R.D.; Edil, B.H.; et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci. Transl. Med. 2011, 3, 92ra66. [Google Scholar] [CrossRef]
- Nikiforova, M.N.; Khalid, A.; Fasanella, K.E.; McGrath, K.M.; Brand, R.E.; Chennat, J.S.; Slivka, A.; Zeh, H.J.; Zureikat, A.H.; Krasinskas, A.M.; et al. Integration of KRAS testing in the diagnosis of pancreatic cystic lesions: a clinical experience of 618 pancreatic cysts. Mod. Pathol. 2013, 26, 1478–1487. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Jiao, Y.; Dal Molin, M.; Maitra, A.; de Wilde, R.F.; Wood, L.D.; Eshleman, J.R.; Goggins, M.G.; Wolfgang, C.L.; Canto, M.I.; et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc. Natl. Acad. Sci. USA 2011, 108, 21188–21193. [Google Scholar] [CrossRef] [Green Version]
- Jones, M.; Zheng, Z.; Wang, J.; Dudley, J.; Albanese, E.; Kadayifci, A.; Dias-Santagata, D.; Le, L.; Brugge, W.R.; Fernandez-del Castillo, C.; et al. Impact of next-generation sequencing on the clinical diagnosis of pancreatic cysts. Gastrointest. Endosc. 2016, 83, 140–148. [Google Scholar] [CrossRef]
- Singhi, A.D.; McGrath, K.; Brand, R.E.; Khalid, A.; Zeh, H.J.; Chennat, J.S.; Fasanella, K.E.; Papachristou, G.I.; Slivka, A.; Bartlett, D.L.; et al. Preoperative next-generation sequencing of pancreatic cyst fluid is highly accurate in cyst classification and detection of advanced neoplasia. Gut 2018, 67, 2131–2141. [Google Scholar] [CrossRef]
- Zhang, M.L.; Arpin, R.N.; Brugge, W.R.; Forcione, D.G.; Basar, O.; Pitman, M.B. Moray micro forceps biopsy improves the diagnosis of specific pancreatic cysts. Cancer Cytopathol. 2018, 126, 414–420. [Google Scholar] [CrossRef] [Green Version]
- Shakhatreh, M.H.; Naini, S.R.; Brijbassie, A.A.; Grider, D.J.; Shen, P.; Yeaton, P. Use of a novel through-the-needle biopsy forceps in endoscopic ultrasound. Endosc. Int. Open 2016, 4, E439–E442. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA-cancer connection: The beginning of a new tale. Cancer Res. 2006, 66, 7390–7394. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007, 21, 1025–1030. [Google Scholar] [CrossRef]
- He, L.; Thomson, J.M.; Hemann, M.T.; Hernando-Monge, E.; Mu, D.; Goodson, S.; Powers, S.; Cordon-Cardo, C.; Lowe, S.W.; Hannon, G.J.; et al. A microRNA polycistron as a potential human oncogene. Nature 2005, 435, 828–833. [Google Scholar] [CrossRef]
- Voorhoeve, P.M.; le Sage, C.; Schrier, M.; Gillis, A.J.; Stoop, H.; Nagel, R.; Liu, Y.P.; van Duijse, J.; Drost, J.; Griekspoor, A.; et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 2006, 124, 1169–1181. [Google Scholar] [CrossRef]
- Gironella, M.; Seux, M.; Xie, M.J.; Cano, C.; Tomasini, R.; Gommeaux, J.; Garcia, S.; Nowak, J.; Yeung, M.L.; Jeang, K.T.; et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc. Natl. Acad. Sci. USA 2007, 104, 16170–16175. [Google Scholar] [CrossRef] [Green Version]
- Szafranska, A.E.; Doleshal, M.; Edmunds, H.S.; Gordon, S.; Luttges, J.; Munding, J.B.; Barth, R.J.; Gutmann, E.J.; Suriawinata, A.A.; Marc Pipas, J.; et al. Analysis of microRNAs in pancreatic fine-needle aspirates can classify benign and malignant tissues. Clin. Chem. 2008, 54, 1716–1724. [Google Scholar] [CrossRef]
- Giovannetti, E.; Funel, N.; Peters, G.J.; Del Chiaro, M.; Erozenci, L.A.; Vasile, E.; Leon, L.G.; Pollina, L.E.; Groen, A.; Falcone, A.; et al. MicroRNA-21 in pancreatic cancer: Correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010, 70, 4528–4538. [Google Scholar] [CrossRef]
- Jamieson, N.B.; Morran, D.C.; Morton, J.P.; Ali, A.; Dickson, E.J.; Carter, C.R.; Sansom, O.J.; Evans, T.R.J.; McKay, C.J.; Oien, K.A. MicroRNA molecular profiles associated with diagnosis, clinicopathologic criteria, and overall survival in patients with resectable pancreatic ductal adenocarcinoma. Clin. Cancer Res. 2012, 18, 534–545. [Google Scholar] [CrossRef]
- Matthaei, H.; Wylie, D.; Lloyd, M.B.; Dal Molin, M.; Kemppainen, J.; Mayo, S.C.; Wolfgang, C.L.; Schulick, R.D.; Langfield, L.; Andruss, B.F.; et al. miRNA biomarkers in cyst fluid augment the diagnosis and management of pancreatic cysts. Clin. Cancer Res. 2012, 18, 4713–4724. [Google Scholar] [CrossRef]
- Wang, J.; Paris, P.L.; Chen, J.; Ngo, V.; Yao, H.; Frazier, M.L.; Killary, A.M.; Liu, C.G.; Liang, H.; Mathy, C.; et al. Next generation sequencing of pancreatic cyst fluid microRNAs from low grade-benign and high grade-invasive lesions. Cancer Lett. 2015, 356, 404–409. [Google Scholar] [CrossRef]
- Roy-Chowdhuri, S.; Stewart, J. Preanalytic variables in cytology: Lessons learned from next-generation sequencing-The MD Anderson experience. Arch. Pathol. Lab. Med. 2016, 140, 1191–1199. [Google Scholar] [CrossRef]
- Lunardi, S.; Muschel, R.J.; Brunner, T.B. The stromal compartments in pancreatic cancer: Are there any therapeutic targets? Cancer Lett. 2014, 343, 147–155. [Google Scholar] [CrossRef]
- Torphy, R.J.; Wang, Z.; True-Yasaki, A.; Volmar, K.E.; Rashid, N.; Yeh, B.; Anderson, J.M.; Johansen, J.S.; Hollingsworth, M.A.; Yeh, J.J.; et al. Stromal content is correlated with tissue site, contrast retention, and survival in pancreatic adenocarcinoma. JCO Precis. Oncol. 2018, 2018. [Google Scholar] [CrossRef]
- Oh, D.; Seo, D.W.; Hong, S.M.; Jun, J.H.; Song, T.J.; Park, D.H.; Son, B.K.; Lee, S.S.; Lee, S.K.; Kim, M.H. The usefulness of contrast-enhanced harmonic EUS-guided fine-needle aspiration for evaluation of hepatic lesions (with video). Gastrointest. Endosc. 2018, 88, 495–501. [Google Scholar] [CrossRef]
- Erickson, R.A.; Sayage-Rabie, L.; Beissner, R.S. Factors predicting the number of EUS-guided fine-needle passes for diagnosis of pancreatic malignancies. Gastrointest. Endosc. 2000, 51, 184–190. [Google Scholar] [CrossRef]
- Klapman, J.B.; Logrono, R.; Dye, C.E.; Waxman, I. Clinical impact of on-site cytopathology interpretation on endoscopic ultrasound-guided fine needle aspiration. Am. J. Gastroenterol. 2003, 98, 1289–1294. [Google Scholar] [CrossRef]
- Wani, S.; Muthusamy, V.R.; Komanduri, S. EUS-guided tissue acquisition: An evidence-based approach (with videos). Gastrointest. Endosc. 2014, 80, 939–959.e7. [Google Scholar] [CrossRef]
- Da Cunha Santos, G.; Ko, H.M.; Saieg, M.A.; Geddie, W.R. “The petals and thorns” of ROSE (rapid on-site evaluation). Cancer Cytopathol. 2013, 121, 4–8. [Google Scholar] [CrossRef]
- Siddiqui, A.A.; Brown, L.J.; Hong, S.K.; Draganova-Tacheva, R.A.; Korenblit, J.; Loren, D.E.; Kowalski, T.E.; Solomides, C. Relationship of pancreatic mass size and diagnostic yield of endoscopic ultrasound-guided fine needle aspiration. Dig. Dis. Sci. 2011, 56, 3370–3375. [Google Scholar] [CrossRef]
- Cheng, B.; Zhang, Y.; Chen, Q.; Sun, B.; Deng, Z.; Shan, H.; Dou, L.; Wang, J.; Li, Y.; Yang, X.; et al. Analysis of fine-needle biopsy vs fine-needle aspiration in diagnosis of pancreatic and abdominal masses: A prospective, multicenter, randomized controlled trial. Clin. Gastroenterol. Hepatol. 2018, 16, 1314–1321. [Google Scholar] [CrossRef]
- Vanbiervliet, G.; Napoleon, B.; Saint Paul, M.C.; Sakarovitch, C.; Wangermez, M.; Bichard, P.; Subtil, C.; Koch, S.; Grandval, P.; Gincul, R.; et al. Core needle versus standard needle for endoscopic ultrasound-guided biopsy of solid pancreatic masses: A randomized crossover study. Endoscopy 2014, 46, 1063–1070. [Google Scholar] [CrossRef]
- Aadam, A.A.; Wani, S.; Amick, A.; Shah, J.N.; Bhat, Y.M.; Hamerski, C.M.; Klapman, J.B.; Muthusamy, V.R.; Watson, R.R.; Rademaker, A.W.; et al. A randomized controlled cross-over trial and cost analysis comparing endoscopic ultrasound fine needle aspiration and fine needle biopsy. Endosc. Int. Open 2016, 4, E497–E505. [Google Scholar] [CrossRef] [Green Version]
- Bang, J.Y.; Hebert-Magee, S.; Trevino, J.; Ramesh, J.; Varadarajulu, S. Randomized trial comparing the 22-gauge aspiration and 22-gauge biopsy needles for EUS-guided sampling of solid pancreatic mass lesions. Gastrointest. Endosc. 2012, 76, 321–327. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.N.; Moon, J.H.; Kim, H.K.; Choi, H.J.; Choi, M.H.; Kim, D.C.; Lee, T.H.; Cha, S.W.; Cho, Y.D.; Park, S.H. Core biopsy needle versus standard aspiration needle for endoscopic ultrasound-guided sampling of solid pancreatic masses: A randomized parallel-group study. Endoscopy 2014, 46, 1056–1062. [Google Scholar] [CrossRef]
- Bang, J.Y.; Hawes, R.; Varadarajulu, S. A meta-analysis comparing ProCore and standard fine-needle aspiration needles for endoscopic ultrasound-guided tissue acquisition. Endoscopy 2016, 48, 339–349. [Google Scholar] [CrossRef]
- Kandel, P.; Tranesh, G.; Nassar, A.; Bingham, R.; Raimondo, M.; Woodward, T.A.; Gomez, V.; Wallace, M.B. EUS-guided fine needle biopsy sampling using a novel fork-tip needle: A case-control study. Gastrointest. Endosc. 2016, 84, 1034–1039. [Google Scholar] [CrossRef]
- Armellini, E.; Manfrin, E.; Trisolini, E.; Andorno, S.; Ballare, M.; Bernardoni, L.; Boldorini, R.L.; Gabbrielli, A.; Frulloni, L.; Larghi, A.; et al. Histologic retrieval rate of a newly designed side-bevelled 20G needle for EUS-guided tissue acquisition of solid pancreatic lesions. United European Gastroenterol. J. 2019, 7, 96–104. [Google Scholar] [CrossRef]
- Naveed, M.; Siddiqui, A.A.; Kowalski, T.E.; Loren, D.E.; Khalid, A.; Soomro, A.; Mazhar, S.M.; Yoo, J.; Hasan, R.; Yalamanchili, S. A multicenter comparative trial of a novel EUS-guided core biopsy needle (SharkCore()) with the 22-gauge needle in patients with solid pancreatic mass lesions. Endosc. Ultrasound 2018, 7, 34–40. [Google Scholar]
- Nayar, M.K.; Paranandi, B.; Dawwas, M.F.; Leeds, J.S.; Darne, A.; Haugk, B.; Majumdar, D.; Ahmed, M.M.; Oppong, K.W. Comparison of the diagnostic performance of 2 core biopsy needles for EUS-guided tissue acquisition from solid pancreatic lesions. Gastrointest. Endosc. 2017, 85, 1017–1024. [Google Scholar] [CrossRef]
- Bang, J.Y.; Hebert-Magee, S.; Navaneethan, U.; Hasan, M.K.; Hawes, R.; Varadarajulu, S. Randomized trial comparing the Franseen and Fork-tip needles for EUS-guided fine-needle biopsy sampling of solid pancreatic mass lesions. Gastrointest. Endosc. 2018, 87, 1432–1438. [Google Scholar] [CrossRef]
- De Biase, D.; Visani, M.; Malapelle, U.; Simonato, F.; Cesari, V.; Bellevicine, C.; Pession, A.; Troncone, G.; Fassina, A.; Tallini, G. Next-generation sequencing of lung cancer EGFR exons 18-21 allows effective molecular diagnosis of small routine samples (cytology and biopsy). PLoS ONE 2013, 8, e83607. [Google Scholar] [CrossRef]
- Roy-Chowdhuri, S.; Goswami, R.S.; Chen, H.; Patel, K.P.; Routbort, M.J.; Singh, R.R.; Broaddus, R.R.; Barkoh, B.A.; Manekia, J.; Yao, H.; et al. Factors affecting the success of next-generation sequencing in cytology specimens. Cancer Cytopathol. 2015, 123, 659–668. [Google Scholar] [CrossRef]
- Lindeman, N.I.; Cagle, P.T.; Beasley, M.B.; Chitale, D.A.; Dacic, S.; Giaccone, G.; Jenkins, R.B.; Kwiatkowski, D.J.; Saldivar, J.S.; Squire, J.; et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: Guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J. Mol. Diagn. 2013, 15, 415–453. [Google Scholar]
- Ellison, G.; Zhu, G.; Moulis, A.; Dearden, S.; Speake, G.; McCormack, R. EGFR mutation testing in lung cancer: A review of available methods and their use for analysis of tumour tissue and cytology samples. J. Clin. Pathol. 2013, 66, 79–89. [Google Scholar] [CrossRef]
- Williams, C.; Ponten, F.; Moberg, C.; Soderkvist, P.; Uhlen, M.; Ponten, J.; Sitbon, G.; Lundeberg, J. A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am. J. Pathol. 1999, 155, 1467–1471. [Google Scholar] [CrossRef]
- Hartley, C.P.; Mahajan, A.M.; Selvaggi, S.M.; Rehrauer, W.M. FNA smears of pancreatic ductal adenocarcinoma are superior to formalin-fixed paraffin-embedded tissue as a source of DNA: Comparison of targeted KRAS amplification and genotyping in matched preresection and postresection samples. Cancer Cytopathol. 2017, 125, 838–847. [Google Scholar] [CrossRef]
- Wei, S.; Lieberman, D.; Morrissette, J.J.; Baloch, Z.W.; Roth, D.B.; McGrath, C. Using “residual” FNA rinse and body fluid specimens for next-generation sequencing: An institutional experience. Cancer Cytopathol. 2016, 124, 324–349. [Google Scholar] [CrossRef]
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, M.B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Kerr, K.; et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: Guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J. Mol. Diagn. 2018, 20, 129–159. [Google Scholar]
- Laquière, A.; Lefort, C.; Maire, F.; Aubert, A.; Gincul, R.; Prat, F.; Grandval, P.; Croizet, O.; Boulant, J.; Vanbiervliet, G.; et al. 19 G nitinol needle versus 22 G needle for transduodenal endoscopic ultrasound-guided sampling of pancreatic solid masses: A randomized study. Endoscopy 2018, 51, 436–443. [Google Scholar] [CrossRef]
- Song, T.J.; Kim, J.H.; Lee, S.S.; Eum, J.B.; Moon, S.H.; Park, D.Y.; Seo, D.W.; Lee, S.K.; Jang, S.J.; Yun, S.C.; et al. The prospective randomized, controlled trial of endoscopic ultrasound-guided fine-needle aspiration using 22G and 19G aspiration needles for solid pancreatic or peripancreatic masses. Am. J. Gastroenterol. 2010, 105, 1739–1745. [Google Scholar] [CrossRef]
- Itoi, T.; Itokawa, F.; Sofuni, A.; Nakamura, K.; Tsuchida, A.; Yamao, K.; Kawai, T.; Moriyasu, F. Puncture of solid pancreatic tumors guided by endoscopic ultrasonography: A pilot study series comparing Trucut and 19-gauge and 22-gauge aspiration needles. Endoscopy 2005, 37, 362–366. [Google Scholar] [CrossRef]
- Affolter, K.E.; Schmidt, R.L.; Matynia, A.P.; Adler, D.G.; Factor, R.E. Needle size has only a limited effect on outcomes in EUS-guided fine needle aspiration: A systematic review and meta-analysis. Dig. Dis. Sci. 2013, 58, 1026–1034. [Google Scholar] [CrossRef]
- Madhoun, M.F.; Wani, S.B.; Rastogi, A.; Early, D.; Gaddam, S.; Tierney, W.M.; Maple, J.T. The diagnostic accuracy of 22-gauge and 25-gauge needles in endoscopic ultrasound-guided fine needle aspiration of solid pancreatic lesions: A meta-analysis. Endoscopy 2013, 45, 86–92. [Google Scholar] [CrossRef]
- Siddiqui, U.D.; Rossi, F.; Rosenthal, L.S.; Padda, M.S.; Murali-Dharan, V.; Aslanian, H.R. EUS-guided FNA of solid pancreatic masses: A prospective, randomized trial comparing 22-gauge and 25-gauge needles. Gastrointest. Endosc. 2009, 70, 1093–1097. [Google Scholar] [CrossRef]
- Varadarajulu, S.; Fockens, P.; Hawes, R.H. Best practices in endoscopic ultrasound-guided fine-needle aspiration. Clin. Gastroenterol. Hepatol. 2012, 10, 697–703. [Google Scholar] [CrossRef]
- Wani, S. Basic techniques in endoscopic ultrasound-guided fine-needle aspiration: Role of a stylet and suction. Endosc. Ultrasound 2014, 3, 17–21. [Google Scholar] [CrossRef] [Green Version]
- Nakai, Y.; Isayama, H.; Chang, K.J.; Yamamoto, N.; Hamada, T.; Uchino, R.; Mizuno, S.; Miyabayashi, K.; Yamamoto, K.; Kawakubo, K.; et al. Slow pull versus suction in endoscopic ultrasound-guided fine-needle aspiration of pancreatic solid masses. Dig. Dis. Sci. 2014, 59, 1578–1585. [Google Scholar] [CrossRef]
- Chen, J.Y.; Ding, Q.Y.; Lv, Y.; Guo, W.; Zhi, F.C.; Liu, S.D.; Cheng, T.M. Slow-pull and different conventional suction techniques in endoscopic ultrasound-guided fine-needle aspiration of pancreatic solid lesions using 22-gauge needles. World J. Gastroenterol. 2016, 22, 8790–8797. [Google Scholar] [CrossRef]
- Saxena, P.; El Zein, M.; Stevens, T.; Abdelgelil, A.; Besharati, S.; Messallam, A.; Kumbhari, V.; Azola, A.; Brainard, J.; Shin, E.J.; et al. Stylet slow-pull versus standard suction for endoscopic ultrasound-guided fine-needle aspiration of solid pancreatic lesions: A multicenter randomized trial. Endoscopy 2018, 50, 497–504. [Google Scholar] [CrossRef]
- Lee, K.Y.; Cho, H.D.; Hwangbo, Y.; Yang, J.K.; Han, S.J.; Choi, H.J.; Lee, Y.N.; Cha, S.W.; Moon, J.H.; Cho, Y.D.; et al. Efficacy of 3 fine-needle biopsy techniques for suspected pancreatic malignancies in the absence of an on-site cytopathologist. Gastrointest. Endosc. 2019, 89, 825–831.e1. [Google Scholar] [CrossRef]
- Roy-Chowdhuri, S.; Chen, H.; Singh, R.R.; Krishnamurthy, S.; Patel, K.P.; Routbort, M.J.; Manekia, J.; Barkoh, B.A.; Yao, H.; Sabir, S.; et al. Concurrent fine needle aspirations and core needle biopsies: a comparative study of substrates for next-generation sequencing in solid organ malignancies. Mod. Pathol. 2017, 30, 499–508. [Google Scholar] [CrossRef] [Green Version]
| Author (Year) | Study Type | No. of Patients | Type of Tumor | Biopsy Type | Needle | Adequacy Rate for NGS | P-Value | Required Tumor Fraction | Genes Targeted | Frequency of Genomic Alterations |
|---|---|---|---|---|---|---|---|---|---|---|
| Elhanafi S, et al. (2018) | Retrospective cohort study | 167 | PDAC | EUS-FNA/B | 70.1% | ≥10% | Custom panel (47 genes) | KRAS (88%), TP53 (68%), SMAD4 (16%) | ||
| 145 | EUS-FNA | EUSN-3 (22-gauge) | 66.9% | 0.02 | ||||||
| 22 | EUS-FNB | SharkCore/ProCore (22-gauge) | 90.9% | |||||||
| Larson BK, et al. (2018) | Retrospective study | 61 | Pancreatic exocrine malignancy | EUS-FNA/B | 67.2% | ≥20% | FoundationOne (315 genes) | NA | ||
| 7 | EUS-FNA | NA | 42.9% | 0.1494 | ||||||
| 54 | EUS-FNB | SharkCore/ProCore | 70.4% | |||||||
| Gleeson FC, et al. (2017) | Retrospective study | 156 | PanNET | EUS-FNA | NA | 58% | ≥20% | Custom GeneRead DNAseq Targeted Panel V2 (15 genes) | MEN1 (42%), DAXX (11%), ATRX (10%), TSC2 (8%) | |
| Young G, et al. (2013) | Retrospective study | 23 | PDAC, Mucinous adenocarcinoma, adenocarcinoma NOS, PanNET | EUS-FNA | NA | 100% | ≥20% | Custom panel (287 genes) | KRAS (78%), TP53 (74%), CDKN2A/B (35%), SMAD4 (17%), PTEN (13%) | |
| Gleeson FC, et al. (2016) | Retrospective study | 47 | PDAC, Ampullary adenocarcinoma, IPMN, Lynch syndrome associated PDAC | EUS-FNA | NA | 61.7% | ≥20% | Human Comprehensive Cancer GeneRead DNAseq Targeted Panel V2 (160 genes) | KRAS (93.1%), TP53 (72.4%), SMAD4 (31%), GNAS (10.3%) |
| Author (Year) | Study Type | No. of Patients | Type of Lesion | Biopsy Type | Needle | Adequacy Rate for NGS | Genes Targeted | Genomic Alteration Detected | Frequency of Genomic Alterations |
|---|---|---|---|---|---|---|---|---|---|
| Singhi AD, et al. (2018) | Prospective study | 673 | IPMN, MCN, SCA, Cystic PanNET, Acinar cell cystadenoma, Pseudocyst | EUS-FNA | NA | 93% | PancreaSeq (10 genes) | 57% | KRAS (42%), GNAS (26%), BRAF (1%), SMAD4 (17%), CTNNB1 (1%) |
| Jones M, et al. (2016) | Prospective study | 99 | IPMN, MCN, SCA, Cystic PanNET, NOS | EUS-FNA | NA | 97% | Custom panel (39 genes) | 57% | KRAS (47%), GNAS (24%) |
| Springer S, et al. | Retrospective study | 24 | 17 IPMN, 3 MCN, 2 SCA, 1 SPN, 1 ITPN | EUS-FNA | NA | NA | Custom panel (11 genes) | 87.5% | NA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imaoka, H.; Sasaki, M.; Hashimoto, Y.; Watanabe, K.; Ikeda, M. New Era of Endoscopic Ultrasound-Guided Tissue Acquisition: Next-Generation Sequencing by Endoscopic Ultrasound-Guided Sampling for Pancreatic Cancer. J. Clin. Med. 2019, 8, 1173. https://doi.org/10.3390/jcm8081173
Imaoka H, Sasaki M, Hashimoto Y, Watanabe K, Ikeda M. New Era of Endoscopic Ultrasound-Guided Tissue Acquisition: Next-Generation Sequencing by Endoscopic Ultrasound-Guided Sampling for Pancreatic Cancer. Journal of Clinical Medicine. 2019; 8(8):1173. https://doi.org/10.3390/jcm8081173
Chicago/Turabian StyleImaoka, Hiroshi, Mitsuhito Sasaki, Yusuke Hashimoto, Kazuo Watanabe, and Masafumi Ikeda. 2019. "New Era of Endoscopic Ultrasound-Guided Tissue Acquisition: Next-Generation Sequencing by Endoscopic Ultrasound-Guided Sampling for Pancreatic Cancer" Journal of Clinical Medicine 8, no. 8: 1173. https://doi.org/10.3390/jcm8081173
APA StyleImaoka, H., Sasaki, M., Hashimoto, Y., Watanabe, K., & Ikeda, M. (2019). New Era of Endoscopic Ultrasound-Guided Tissue Acquisition: Next-Generation Sequencing by Endoscopic Ultrasound-Guided Sampling for Pancreatic Cancer. Journal of Clinical Medicine, 8(8), 1173. https://doi.org/10.3390/jcm8081173





