Can Additional Patient Information Improve the Diagnostic Performance of Deep Learning for the Interpretation of Knee Osteoarthritis Severity
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset
2.2. Labeling
2.3. DL Algorithm
3. Statistical Analysis
3.1. Results
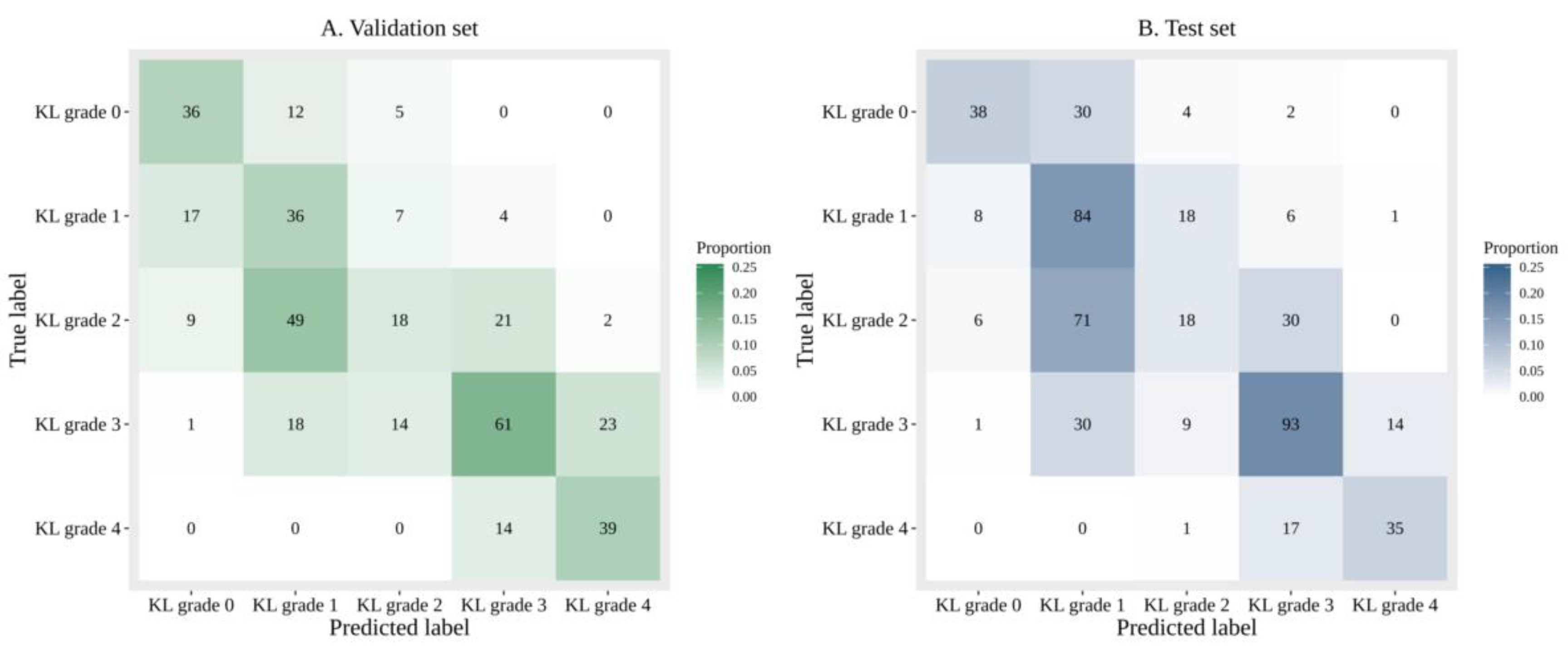
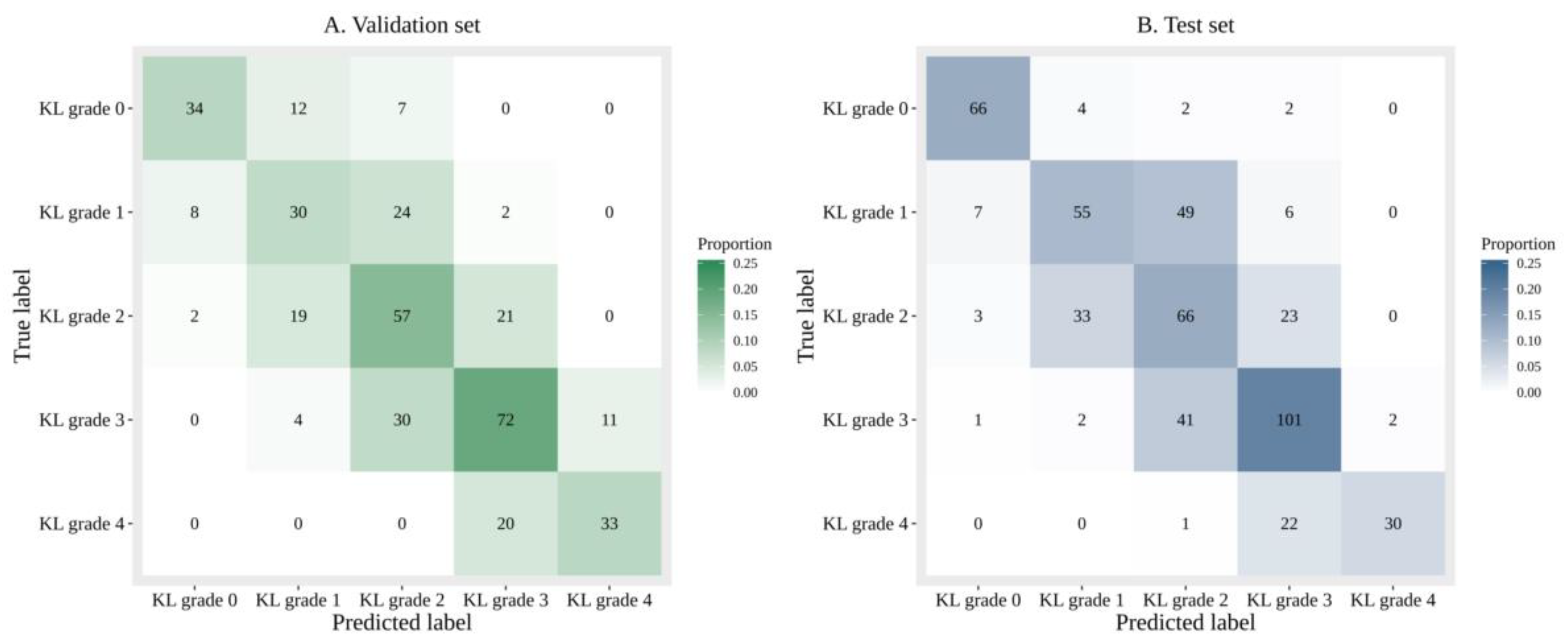
3.1.1. Performance of DL with Sole Image Data
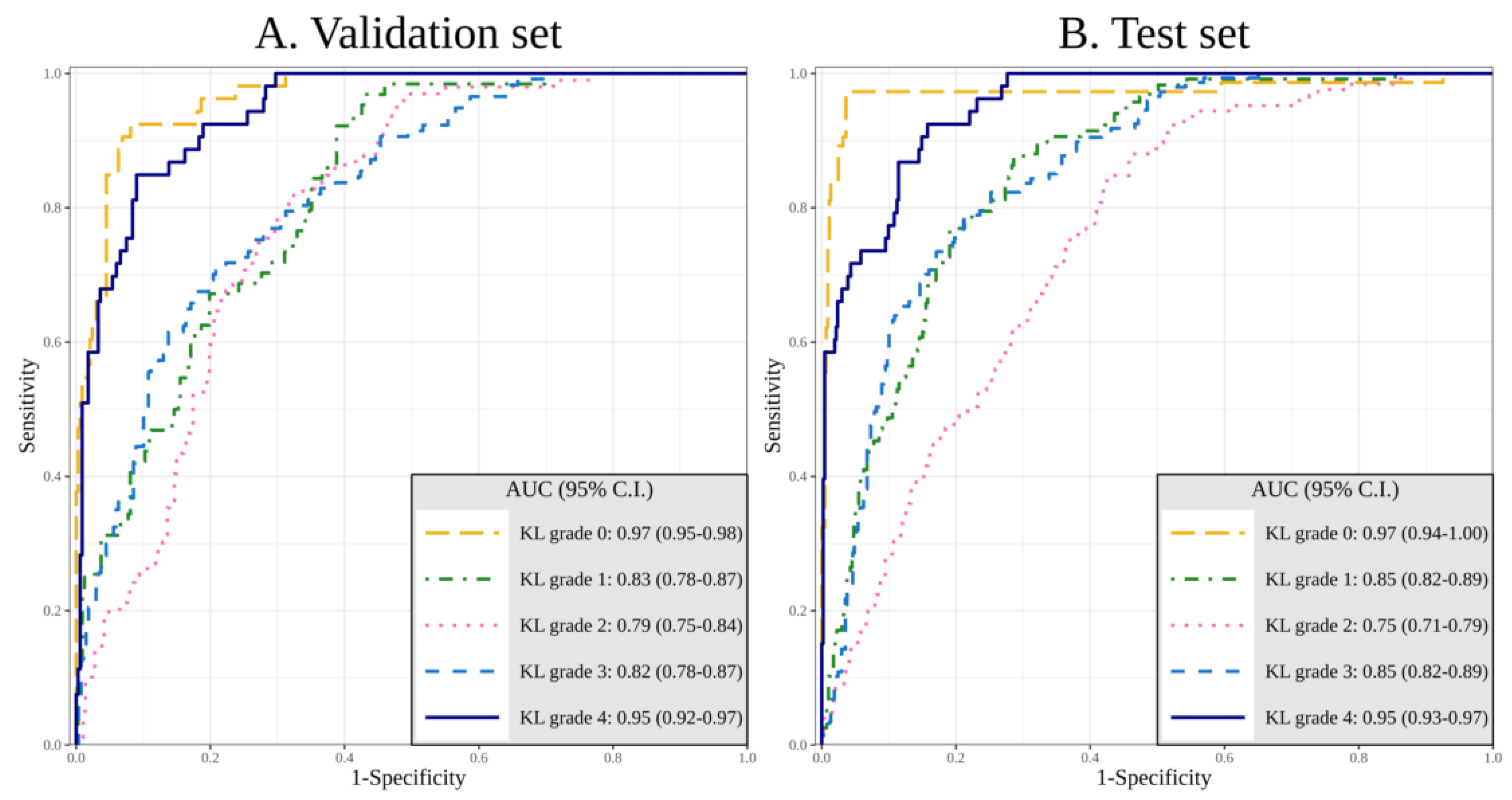
3.1.2. Performance of DL with Image Data and Additional Patient Information
3.1.3. Comparison of the Accuracy and Diagnosing Performance Between Sole Image Data and Image Data with Additional Patient Information
3.1.4. Gradient-Weighted Class Activation Mapping (Grad-CAM)
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Cross, M.; Smith, E.; Hoy, D.; Nolte, S.; Ackerman, I.; Fransen, M.; Bridgett, L.; Williams, S.; Guillemin, F.; Hill, C.L.; et al. The global burden of hip and knee osteoarthritis: Estimates from the Global Burden of Disease 2010 study. Ann. Rheum. Dis. 2014, 73, 1323–1330. [Google Scholar] [CrossRef]
- Eyre, D.R. Collagens and Cartilage Matrix Homeostasis. Clin. Orthop. Relat. Res. 2004, 427, S118–S122. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Morey, V.; Kang, J.Y.; Kim, K.W.; Kim, T.K. Prevalence and Risk Factors of Spine, Shoulder, Hand, Hip, and Knee Osteoarthritis in Community-dwelling Koreans Older Than Age 65 Years. Clin. Orthop. Relat. Res. 2015, 473, 3307–3314. [Google Scholar] [CrossRef] [PubMed]
- Heidari, B. Knee Osteoarthritis Prevalence, Risk Factors, Pathogenesis and Features: Part I. Casp. J. Intern. Med. 2011, 2, 205–212. [Google Scholar]
- Altman, R.; Asch, E.; Bloch, D.; Bole, G.; Borenstein, D.; Brandt, K.; Christy, W.; Cooke, T.D.; Greenwald, R.; Hochberg, M.; et al. Development of Criteria for the Classification and Reporting of Osteoarthritis: Classification of Osteoarthritis of the Knee. Arthritis Rheum. 1986, 29, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Tiulpin, A.; Thevenot, J.; Rahtu, E.; Lehenkari, P.; Saarakkala, S. Automatic Knee Osteoarthritis Diagnosis from Plain Radiographs: A Deep Learning-Based Approach. Sci. Rep. 2018, 8, 1727. [Google Scholar] [CrossRef]
- Kellgren, J.H.; Lawrence, J.S. Radiological Assessment of Osteo-Arthrosis. Ann. Rheum. Dis. 1957, 16, 494–502. [Google Scholar] [CrossRef]
- Gossec, L.; Jordan, J.M.; Mazzuca, S.A.; Lam, M.A.; Suarez-Almazor, M.E.; Renner, J.B.; Lopez-Olivo, M.A.; Hawker, G.; Dougados, M.; Maillefert, J.F. Comparative Evaluation of Three Semi-Quantitative Radiographic Grading Techniques for Knee Osteoarthritis in Terms of Validity and Reproducibility in 1759 X-Rays: Report of the Oarsi–Omeract Task Force. Osteoarthr. Cartil. 2008, 16, 742–748. [Google Scholar] [CrossRef]
- Sheehy, L.; Culham, E.; McLean, L.; Niu, J.; Lynch, J.; Segal, N.A.; Singh, J.A.; Nevitt, M.; Cooke, T.D. Validity and Sensitivity to Change of Three Scales for the Radiographic Assessment of Knee Osteoarthritis Using Images from the Multicenter Osteoarthritis Study (Most). Osteoarthr. Cartil. 2015, 23, 1491–1498. [Google Scholar] [CrossRef]
- Culvenor, A.G.; Engen, C.N.; Øiestad, B.E.; Engebretsen, L.; Risberg, M.A. Defining the Presence of Radiographic Knee Osteoarthritis: A Comparison Between the Kellgren and Lawrence System and Oarsi Atlas Criteria. Knee Surg. Sports Traumatol. Arthrosc. 2015, 23, 3532–3539. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, K.J.; Sunwoo, L.; Choi, D.; Nam, C.M.; Cho, J.; Kim, J.; Bae, Y.J.; Yoo, R.E.; Choi, B.S.; et al. Deep Learning in Diagnosis of Maxillary Sinusitis Using Conventional Radiography. Investig. Radiol. 2019, 54, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Chee, C.G.; Kim, Y.; Kang, Y.; Lee, K.J.; Chae, H.D.; Cho, J.; Nam, C.M.; Choi, D.; Lee, E.; Lee, J.W.; et al. Performance of a Deep Learning Algorithm in Detecting Osteonecrosis of the Femoral Head on Digital Radiography: A Comparison With Assessments by Radiologists. Am. J. Roentgenol. 2019, 213, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Heo, J.; Jang, D.K.; Sunwoo, L.; Kim, J.; Lee, K.J.; Kang, S.H.; Park, S.J.; Kwon, O.K.; Oh, C.W. Machine Learning for Detecting Moyamoya Disease in Plain Skull Radiography Using a Convolutional Neural Network. EBioMedicine 2019, 40, 636–642. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Gao, L.; Shi, X.; Allen, K.; Yang, L. Fully Automatic Knee Osteoarthritis Severity Grading Using Deep Neural Networks with a Novel Ordinal Loss. Comput. Med. Imaging Graph. 2019, 75, 84–92. [Google Scholar] [CrossRef]
- Shahrokh Esfahani, M.; Dougherty, E.R. Effect of Separate Sampling on Classification Accuracy. Bioinformatics 2014, 30, 242–250. [Google Scholar] [CrossRef]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef] [PubMed]
- Kohn, M.D.; Sassoon, A.A.; Fernando, N.D. Classifications in Brief: Kellgren-Lawrence Classification of Osteoarthritis. Clin. Orthop. Relat. Res. 2016, 474, 1886–1893. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Albanie, S.; Sun, G.; Wu, E. Squeeze-and-Excitation Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 42, 2011–2023. [Google Scholar] [CrossRef] [PubMed]
- Delong, E.R.; Delong, D.M.; Clarke-Pearson, D.L. Comparing the Areas under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Iizuka, T.; Fukasawa, M.; Kameyama, M. Deep-Learning-Based Imaging-Classification Identified Cingulate Island Sign in Dementia with Lewy Bodies. Sci. Rep. 2019, 9, 8944. [Google Scholar] [CrossRef] [PubMed]
- Abedin, J.; Antony, J.; McGuinness, K.; Moran, K.; O’Connor, N.E.; Rebholz-Schuhmann, D.; Newell, J. Predicting Knee Osteoarthritis Severity: Comparative Modeling Based on Patient’s Data and Plain X-Ray Images. Sci. Rep. 2019, 9, 5761. [Google Scholar] [CrossRef] [PubMed]


| Characteristic | Training Set | Validation Set | Test Set | Total | p-Value |
|---|---|---|---|---|---|
| Age (Year) | 62.3 ± 12.5 | 63.2 ± 12.6 | 61.5 ± 14.9 | 62.3 ± 2.8 | 0.359 |
| Gender (M/F) | 701/2763 | 74/312 | 146/370 | 921/3445 | <0.001* |
| BMI (kg/m2) | 25.5 ± 3.19 | 25.4 ± 2.96 | 25.9 ± 3.42 | 25.5 ± 3.20 | 0.025 * |
| WBL Ratio | 0.31 ± 0.17 | 0.32 ± 0.15 | 0.36 ± 0.14 | 0.32 ± 0.16 | <0.001 * |
| DM/HTN | 629/1552 | 63/181 | 64/184 | 756/1917 | 0.005/<0.001 * |
| K-L 0 | 473 | 53 | 74 | 600 | |
| K-L 1 | 574 | 64 | 117 | 755 | |
| K-L 2 | 889 | 99 | 125 | 1113 | |
| K-L 3 | 1055 | 117 | 147 | 1319 | |
| K-L 4 | 473 | 53 | 53 | 579 | |
| Total | 3464 | 386 | 516 | 4366 |
| DL with Sole Image Data | DL with Image Data and Patient’s Information | p-Value | |
|---|---|---|---|
| KL Grade 0 | 0.91 (0.88–0.95) | 0.97 (0.94–1.00) | 0.008 * |
| KL Grade 1 | 0.80 (0.76–0.84) | 0.85 (0.82–0.89) | 0.020 * |
| KL Grade 2 | 0.69 (0.64–0.73) | 0.75 (0.71–0.79) | 0.027 * |
| KL Grade 3 | 0.86 (0.83–0.89) | 0.86 (0.82–0.89) | 0.553 |
| KL Grade 4 | 0.96 (0.94–0.98) | 0.95 (0.93–0.97) | 0.580 |
| Optimal Cutoff | Sensitivity of 90% | Specificity of 90% | |||
|---|---|---|---|---|---|
| DL With Sole Image Data | KL Grade 0 | Sensitivity | 64.9 (48/74) (52.9–75.6) | 83.8 (2/74) (73.4–91.3) | 64.9 (48/74) (52.9–75.6) |
| Specificity | 94.8 (419/442) (92.3–96.7) | 84.2 (372/442) (80.4–87.4) | 94.8 (419/442) (92.3–96.7) | ||
| KL Grade 1 | Sensitivity | 94.0 (110/117) (88.1–97.6) | 99.1 (116/117) (95.3–100.0) | 58.1 (68/117) (48.6–67.2) | |
| Specificity | 49.6 (198/399) (44.6–54.6) | 38.6 (154/399) (33.8–43.6) | 80.5 (321/399) (76.2–84.2) | ||
| KL Grade 2 | Sensitivity | 80.0 (100/125) (71.9–86.6) | 78.4 (98/125) (70.2–85.3) | 29.6 (37/125) (21.8–38.4) | |
| Specificity | 51.2 (200/391) (46.1–56.2) | 51.9 (203/391) (46.8–57.0) | 82.4 (322/391) (78.2–86.0) | ||
| KL Grade 3 | Sensitivity | 75.5 (111/147) (67.7–82.2) | 93.9 (138/147) (88.7–97.2) | 56.5 (83/147) (48.0–64.6) | |
| Specificity | 78.3 (289/369) (73.8–82.4) | 61.5 (227/369) (56.3–66.5) | 89.2 (329/369) (85.5–92.1) | ||
| KL grade 4 | Sensitivity | 77.4 (41/53) (63.8–87.7) | 84.9 (45/53) (72.4–93.3) | 75.5 (40/53) (61.7–86.2) | |
| Specificity | 90.7 (420/463) (87.7–93.2) | 87.3 (404/463) (83.9–90.2) | 92.0 (426/463) (89.2–94.3) | ||
| DL With Image Data and Patient’s Information | KL Grade 0 | Sensitivity | 97.3 (72/74) (90.6–99.7) | 97.3 (72/74) (90.6–99.7) | 97.3 (72/74) (90.6–99.7) |
| Specificity | 92.3 (408/442) (89.4–94.6) | 93.9 (415/442) (91.2–95.9) | 88.5 (391/442) (85.1–91.3) | ||
| KL Grade 1 | Sensitivity | 92.3 (108/117) (85.9–96.4) | 90.6 (106/117) (83.8–95.2) | 48.7 (57/117) (39.4–58.1) | |
| Specificity | 56.9 (227/399) (51.9–61.8) | 62.2 (248/399) (57.2–66.9) | 90.2 (360/399) (86.9–93.0) | ||
| KL Grade 2 | Sensitivity | 76.0 (95/125) (67.5–83.2) | 88.8 (111/125) (8.19–93.7) | 27.2 (34/125) (19.6–35.9) | |
| Specificity | 61.6 (241/391) (56.6–66.5) | 51.4 (201/391) (46.3–56.5) | 90.5 (354/391) (87.2–93.2) | ||
| KL Grade 3 | Sensitivity | 73.5 (108/147) (65.6–80.4) | 89.8 (132/147) (83.7–94.2) | 51.0 (75/147) (42.7–59.3) | |
| Specificity | 81.8 (302/369) (77.5–85.6) | 61.5 (227/369) (56.3–66.5) | 91.1 (336/369) (87.7–93.8) | ||
| KL Grade 4 | Sensitivity | 73.6 (39/53) (59.7–84.7) | 92.5 (49/53) (81.8–97.9) | 73.6 (39/53) (59.7–84.7) | |
| Specificity | 91.4 (423/463) (88.4–93.8) | 84.2 (390/463) (80.6–87.4) | 91.1 (422/463) (88.2–93.6) | ||
| DL With Sole Image Data | KL Grade 0 | PPV | 67.6 (48/71) (55.5–78.2) | 47.6 (62/132) (38.2–55.8) | 67.6 (48/71) (55.5–78.2) |
| NPV | 94.2 (419/445) (91.6–96.1) | 96.9 (372/384) (94.6–98.4) | 94.2 (419/445) (91.6–96.1) | ||
| KL Grade 1 | PPV | 35.4 (110/311) (30.1–41.0) | 32.1 (116/361) (27.3–37.2) | 46.6 (68/146) (38.3–55.0) | |
| NPV | 96.6 (198/205) (93.1–98.6) | 99.4 (154/155) (96.5–100.0) | 86.8 (321/370) (82.9–90.0) | ||
| KL Grade 2 | PPV | 34.4 (100/291) (28.9–40.1) | 34.3 (98/286) (28.8–40.1) | 34.9 (37/106) (25.9–44.8) | |
| NPV | 88.9 (200/225) (84.0–92.7) | 88.3 (203/230) (83.4–92.1) | 78.5 (322/410) (74.2–82.4) | ||
| KL Grade 3 | PPV | 58.1 (111/191) (50.8–65.2) | 49.3 (138/280) (43.3–55.3) | 67.5 (83/123) (58.4–75.6) | |
| NPV | 88.9 (289/325) (85.0–92.1) | 96.2 (227/236) (92.9–98.2) | 83.7 (329/393) (79.7–87.2) | ||
| KL Grade 4 | PPV | 48.8 (41/84) (37.7–60.0) | 43.3 (45/104) (33.6–53.3) | 51.9 (40/77) (40.3–63.5) | |
| NPV | 97.2 (420/432) (95.2–98.6) | 98.1 (404/412) (96.2–99.2) | 97.0 (426/439) (95.0–98.4) | ||
| Accuracy | 51.9 (268/516) (47.5–56.3) | ||||
| DL With Image Data and Patient’s Information | KL grade 0 | PPV | 67.9 (72/106) (58.2–76.7) | 72.7 (72/99) (62.9–81.2) | 58.5 (72/123) (49.3–67.3) |
| NPV | 99.5 (408/410) (98.2–99.9) | 99.5 (415/417) (98.3–99.9) | 99.5 (391/393) (98.2–99.9) | ||
| KL grade 1 | PPV | 38.6 (108/280) (32.8–44.5) | 41.2 (106/257) (35.2–47.5) | 59.4 (57/96) (48.9–69.3) | |
| NPV | 96.2 (227/236) (92.9–98.2) | 95.8 (248/259) (92.5–97.9) | 85.7 (360/420) (82.0–88.9) | ||
| KL Grade 2 | PPV | 38.8 (95.245) (32.6–45.2) | 36.9 (111/301) (31.4–42.6) | 47.9 (34/71) (35.9–60.1) | |
| NPV | 88.9 (241/271) (84.6–92.4) | 93.5 (201/215) (89/3–96.4) | 79.6 (354/445) (75.5–83.2) | ||
| KL Grade 3 | PPV | 61.7 (108/175) (54.1–68.9) | 48.2 (132/274) (42.1–54.3) | 69.4 (75/108) (59.8–77.9) | |
| NPV | 88.6 (302/341) (84.7–91.7) | 93.8 (227/242) (90.0–96.5) | 82.4 (336/408) (78.3–85.9) | ||
| KL Grade 4 | PPV | 49.4 (39/79) | 40.2 (49/122) (31.4–49.4) | 48.8 (39/80) (37.4–60.2) | |
| NPV | 96.8 (423/437) (94.7–98.2) | 99.0 (390/394) (97.4–99.7) | 96.8 (422/436) (94.7–98.2) | ||
| Accuracy | 61.6 (318/516) (57.3–65.8) | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.H.; Lee, K.J.; Choi, D.; Lee, J.I.; Choi, H.G.; Lee, Y.S. Can Additional Patient Information Improve the Diagnostic Performance of Deep Learning for the Interpretation of Knee Osteoarthritis Severity. J. Clin. Med. 2020, 9, 3341. https://doi.org/10.3390/jcm9103341
Kim DH, Lee KJ, Choi D, Lee JI, Choi HG, Lee YS. Can Additional Patient Information Improve the Diagnostic Performance of Deep Learning for the Interpretation of Knee Osteoarthritis Severity. Journal of Clinical Medicine. 2020; 9(10):3341. https://doi.org/10.3390/jcm9103341
Chicago/Turabian StyleKim, Dong Hyun, Kyong Joon Lee, Dongjun Choi, Jae Ik Lee, Han Gyeol Choi, and Yong Seuk Lee. 2020. "Can Additional Patient Information Improve the Diagnostic Performance of Deep Learning for the Interpretation of Knee Osteoarthritis Severity" Journal of Clinical Medicine 9, no. 10: 3341. https://doi.org/10.3390/jcm9103341
APA StyleKim, D. H., Lee, K. J., Choi, D., Lee, J. I., Choi, H. G., & Lee, Y. S. (2020). Can Additional Patient Information Improve the Diagnostic Performance of Deep Learning for the Interpretation of Knee Osteoarthritis Severity. Journal of Clinical Medicine, 9(10), 3341. https://doi.org/10.3390/jcm9103341







