Clinical Implications and Gender Differences of KCNQ1 p.Gly168Arg Pathogenic Variant in Long QT Syndrome
Abstract
1. Introduction
2. Methods
2.1. Study Population
2.2. Genetic Testing
2.3. Statistical Analysis
3. Results
4. Discussion
5. Limitations
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mazzanti, A.; Maragna, R.; Vacanti, G.; Monteforte, N.; Bloise, R.; Marino, M.; Braghieri, L.; Gambelli, P.; Memmi, M.; Pagan, E.; et al. Interplay Between Genetic Substrate, QTc Duration, and Arrhythmia Risk in Patients with Long QT Syndrome. J. Am. Coll. Cardiol. 2018, 71, 1663–1671. [Google Scholar] [CrossRef] [PubMed]
- Kapplinger, J.D.; Tester, D.J.; Salisbury, B.A.; Carr, J.L.; Harris-Kerr, C.; Pollevick, G.D.; Wilde, A.A.M.; Ackerman, M.J. Spectrum and prevalence of mutations from the first 2500 consecutive unrelated patients referred for the FAMILION long QT syndrome genetic test. HeartRhythm 2009, 6, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Napolitano, C.; Priori, S.G.; Schwartz, P.J.; Bloise, R.; Ronchetti, E.; Nastoli, J.; Bottelli, G.; Cerrone, M.; Leonardi, S. Genetic testing in the long QT syndrome: Development and validation of an efficient approach to genotyping in clinical practice. JAMA 2005, 294, 2975–2980. [Google Scholar] [CrossRef]
- Locati, E.T. QT interval duration remains a major risk factor in long QT syndrome patients. J. Am. Coll. Cardiol. 2006, 48, 1053–1055. [Google Scholar] [CrossRef]
- Goldenberg, I.; Horr, S.; Moss, A.J.; Lopes, C.M.; Barsheshet, A.; McNitt, S.; Zareba, W.; Andrews, M.L.; Robinson, J.L.; Locati, E.H.; et al. Risk for life-threatening cardiac events in patients with genotype-confirmed long-QT syndrome and normal-range corrected QT intervals. J. Am. Coll. Cardiol. 2011, 57, 51–59. [Google Scholar] [CrossRef] [PubMed]
- European Heart Rhythm Association; Heart Rhythm Society; Zipes, D.P.; Camm, A.J.; Borggrefe, M.; Buxton, A.E.; Chaitman, B.; Fromer, M.; Gregoratos, G.; Klein, G.; et al. ACC/AHA/ESC 2006 guidelines for management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: A report of the American College of Cardiology/American Heart Association Task Force and the European Society of Cardiology Committee for Practice Guidelines (Writing Committee to Develop Guidelines for Management of Patients with Ventricular Arrhythmias and the Prevention of Sudden Cardiac Death). J. Am. Coll. Cardiol. 2006, 48, e247–e346. [Google Scholar] [CrossRef]
- Priori, S.G.; Blomström-Lundqvist, C.; Mazzanti, A.; Blom, N.; Borggrefe, M.; Camm, J.; Elliott, P.M.; Fitzsimons, D.; Hatala, R.; Hindricks, G.; et al. 2015 ESC Guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: The Task Force for the Management of Patients with Ventricular Arrhythmias and the Prevention of Sudden Cardiac Death of the European Society of Cardiology (ESC). Endorsed by: Association for European Paediatric and Congenital Cardiology (AEPC). Eur. Heart J. 2015, 36, 2793–2867. [Google Scholar] [CrossRef] [PubMed]
- Priori, S.G.; Wilde, A.A.; Horie, M.; Cho, Y.; Behr, E.R.; Berul, C.; Blom, N.; Brugada, J.; Chiang, C.-E.; Huikuri, H.; et al. HRS/EHRA/APHRS expert consensus statement on the diagnosis and management of patients with inherited primary arrhythmia syndromes: Document endorsed by HRS, EHRA, and APHRS in May 2013 and by ACCF, AHA, PACES, and AEPC in June 2013. HeartRhythm 2013, 10, 1932–1963. [Google Scholar] [CrossRef]
- Priori, S.G.; Schwartz, P.J.; Napolitano, C.; Bloise, R.; Ronchetti, E.; Grillo, M.; Vicentini, A.; Spazzolini, C.; Nastoli, J.; Bottelli, G.; et al. Risk stratification in the long-QT syndrome. N. Engl. J. Med. 2003, 348, 1866–1874. [Google Scholar] [CrossRef]
- Schwartz, P.J.; Crotti, L.; Insolia, R. Long-QT syndrome: From genetics to management. Circ. Arrhythm. Electrophysiol. 2012, 5, 868–877. [Google Scholar] [CrossRef]
- Brink, P.A.; Crotti, L.; Corfield, V.; Goosen, A.; Durrheim, G.; Hedley, P.; Heradien, M.; Geldenhuys, G.; Vanoli, E.; Bacchini, S.; et al. Phenotypic variability and unusual clinical severity of congenital long-QT syndrome in a founder population. Circulation 2005, 112, 2602–2610. [Google Scholar] [CrossRef] [PubMed]
- Crotti, L.; Spazzolini, C.; Schwartz, P.J.; Shimizu, W.; Denjoy, I.; Schulze-Bahr, E.; Zaklyazminskaya, E.V.; Swan, H.; Ackerman, M.J.; Moss, A.J.; et al. The common long-QT syndrome mutation KCNQ1/A341V causes unusually severe clinical manifestations in patients with different ethnic backgrounds: Toward a mutation-specific risk stratification. Circulation 2007, 116, 2366–2375. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, P.J.; Priori, S.G.; Spazzolini, C.; Moss, A.J.; Vincent, G.M.; Napolitano, C.; Denjoy, I.; Guicheney, P.; Breithardt, G.; Keating, M.T.; et al. Genotype-phenotype correlation in the long-QT syndrome: Gene-specific triggers for life-threatening arrhythmias. Circulation 2001, 103, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Rautaharju, P.M.; Surawicz, B.; Gettes, L.S.; Bailey, J.J.; Childers, R.; Deal, B.J.; Gorgels, A.; Hancock, E.W.; Josephson, M.; Kligfield, P.; et al. AHA/ACCF/HRS recommendations for the standardization and interpretation of the electrocardiogram: Part IV: The ST segment, T and U waves, and the QT interval: A scientific statement from the American Heart Association Electrocardiography and Arrhythmias Committee, Council on Clinical Cardiology; the American College of Cardiology Foundation; and the Heart Rhythm Society. Endorsed by the International Society for Computerized Electrocardiology. J. Am. Coll. Cardiol. 2009, 53, 982–991. [Google Scholar] [CrossRef] [PubMed]
- Adler, F.H. Memoir of H. C. Bazett (1885–1950). Trans. Stud. Coll. Physicians Phila. 1952, 19, 121. [Google Scholar]
- Gómez, J.; Reguero, J.R.; Morís, C.; Martín, M.; Alvarez, V.; Alonso, B.; Iglesias, S.; Coto, E. Mutation analysis of the main hypertrophic cardiomyopathy genes using multiplex amplification and semiconductor next-generation sequencing. Circ. J. Off. J. Jpn. Circ. Soc. 2014, 78, 2963–2971. [Google Scholar] [CrossRef]
- Gómez, J.; Lorca, R.; Reguero, J.R.; Morís, C.; Martín, M.; Tranche, S.; Alonso, B.; Iglesias, S.; Alvarez, V.; Díaz-Molina, B.; et al. Screening of the Filamin C Gene in a Large Cohort of Hypertrophic Cardiomyopathy Patients. Circ. Cardiovasc. Genet. 2017, 10. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. Off. J. Am. Coll. Med. Genet. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Moss, A.J.; Shimizu, W.; Wilde, A.A.M.; Towbin, J.A.; Zareba, W.; Robinson, J.L.; Qi, M.; Vincent, G.M.; Ackerman, M.J.; Kaufman, E.S.; et al. Clinical aspects of type-1 long-QT syndrome by location, coding type, and biophysical function of mutations involving the KCNQ1 gene. Circulation 2007, 115, 2481–2489. [Google Scholar] [CrossRef]
- Shimizu, W.; Horie, M.; Ohno, S.; Takenaka, K.; Yamaguchi, M.; Shimizu, M.; Washizuka, T.; Aizawa, Y.; Nakamura, K.; Ohe, T.; et al. Mutation site-specific differences in arrhythmic risk and sensitivity to sympathetic stimulation in the LQT1 form of congenital long QT syndrome: Multicenter study in Japan. J. Am. Coll. Cardiol. 2004, 44, 117–125. [Google Scholar] [CrossRef]
- Barsheshet, A.; Goldenberg, I.; O-Uchi, J.; Moss, A.J.; Jons, C.; Shimizu, W.; Wilde, A.A.; McNitt, S.; Peterson, D.R.; Zareba, W.; et al. Mutations in cytoplasmic loops of the KCNQ1 channel and the risk of life-threatening events: Implications for mutation-specific response to β-blocker therapy in type 1 long-QT syndrome. Circulation 2012, 125, 1988–1996. [Google Scholar] [CrossRef] [PubMed]
- Zareba, W.; Moss, A.J.; Sheu, G.; Kaufman, E.S.; Priori, S.; Vincent, G.M.; Towbin, J.A.; Benhorin, J.; Schwartz, P.J.; Napolitano, C.; et al. Location of mutation in the KCNQ1 and phenotypic presentation of long QT syndrome. J. Cardiovasc. Electrophysiol. 2003, 14, 1149–1153. [Google Scholar] [CrossRef] [PubMed]
- Valente, F.M.; Sparago, A.; Freschi, A.; Hill-Harfe, K.; Maas, S.M.; Frints, S.G.M.; Alders, M.; Pignata, L.; Franzese, M.; Angelini, C.; et al. Transcription alterations of KCNQ1 associated with imprinted methylation defects in the Beckwith-Wiedemann locus. Genet. Med. Off. J. Am. Coll. Med. Genet. 2019, 21, 1808–1820. [Google Scholar] [CrossRef] [PubMed]
- Eßinger, C.; Karch, S.; Moog, U.; Fekete, G.; Lengyel, A.; Pinti, E.; Eggermann, T.; Begemann, M. Frequency of KCNQ1 variants causing loss of methylation of Imprinting Centre 2 in Beckwith-Wiedemann syndrome. Clin. Epigenet. 2020, 12, 63. [Google Scholar] [CrossRef]
- Choufani, S.; Shuman, C.; Weksberg, R. Molecular findings in Beckwith-Wiedemann syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 2013, 163, 131–140. [Google Scholar] [CrossRef]
- Imboden, M.; Swan, H.; Denjoy, I.; Van Langen, I.M.; Latinen-Forsblom, P.J.; Napolitano, C.; Fressart, V.; Breithardt, G.; Berthet, M.; Priori, S.; et al. Female predominance and transmission distortion in the long-QT syndrome. N. Engl. J. Med. 2006, 355, 2744–2751. [Google Scholar] [CrossRef]
- Itoh, H.; Berthet, M.; Fressart, V.; Denjoy, I.; Maugenre, S.; Klug, D.; Mizusawa, Y.; Makiyama, T.; Hofman, N.; Stallmeyer, B.; et al. Asymmetry of parental origin in long QT syndrome: Preferential maternal transmission of KCNQ1 variants linked to channel dysfunction. Eur. J. Hum. Genet. 2016, 24, 1160–1166. [Google Scholar] [CrossRef]
- Huang, L.O.; Labbe, A.; Infante-Rivard, C. Transmission ratio distortion: Review of concept and implications for genetic association studies. Hum. Genet. 2013, 132, 245–263. [Google Scholar] [CrossRef]
- Mistry, H.D.; McCallum, L.A.; Kurlak, L.O.; Greenwood, I.A.; Broughton Pipkin, F.; Tribe, R.M. Novel expression and regulation of voltage-dependent potassium channels in placentas from women with preeclampsia. Hypertension 2011, 58, 497–504. [Google Scholar] [CrossRef]
- Luo, Y.; Kumar, P.; Mendelson, C.R. Estrogen-related receptor γ (ERRγ) regulates oxygen-dependent expression of voltage-gated potassium (K+) channels and tissue kallikrein during human trophoblast differentiation. Mol. Endocrinol. 2013, 27, 940–952. [Google Scholar] [CrossRef]
- Shukla, K.K.; Mahdi, A.A.; Rajender, S. Ion channels in sperm physiology and male fertility and infertility. J. Androl. 2012, 33, 777–788. [Google Scholar] [CrossRef]
- Locati, E.H.; Zareba, W.; Moss, A.J.; Schwartz, P.J.; Vincent, G.M.; Lehmann, M.H.; Towbin, J.A.; Priori, S.G.; Napolitano, C.; Robinson, J.L.; et al. Age- and sex-related differences in clinical manifestations in patients with congenital long-QT syndrome: Findings from the International LQTS Registry. Circulation 1998, 97, 2237–2244. [Google Scholar] [CrossRef] [PubMed]
- Costa, J.; Lopes, C.M.; Barsheshet, A.; Moss, A.J.; Migdalovich, D.; Ouellet, G.; McNitt, S.; Polonsky, S.; Robinson, J.L.; Zareba, W.; et al. Combined assessment of sex- and mutation-specific information for risk stratification in type 1 long QT syndrome. HeartRhythm 2012, 9, 892–898. [Google Scholar] [CrossRef] [PubMed]
- Hara, M.; Danilo, P.; Rosen, M.R. Effects of gonadal steroids on ventricular repolarization and on the response to E4031. J. Pharmacol. Exp. Ther. 1998, 285, 1068–1072. [Google Scholar]
- Ridley, J.M.; Shuba, Y.M.; James, A.F.; Hancox, J.C. Modulation by testosterone of an endogenous hERG potassium channel current. J. Physiol. Pharmacol. Off. J. Pol. Physiol. Soc. 2008, 59, 395–407. [Google Scholar]
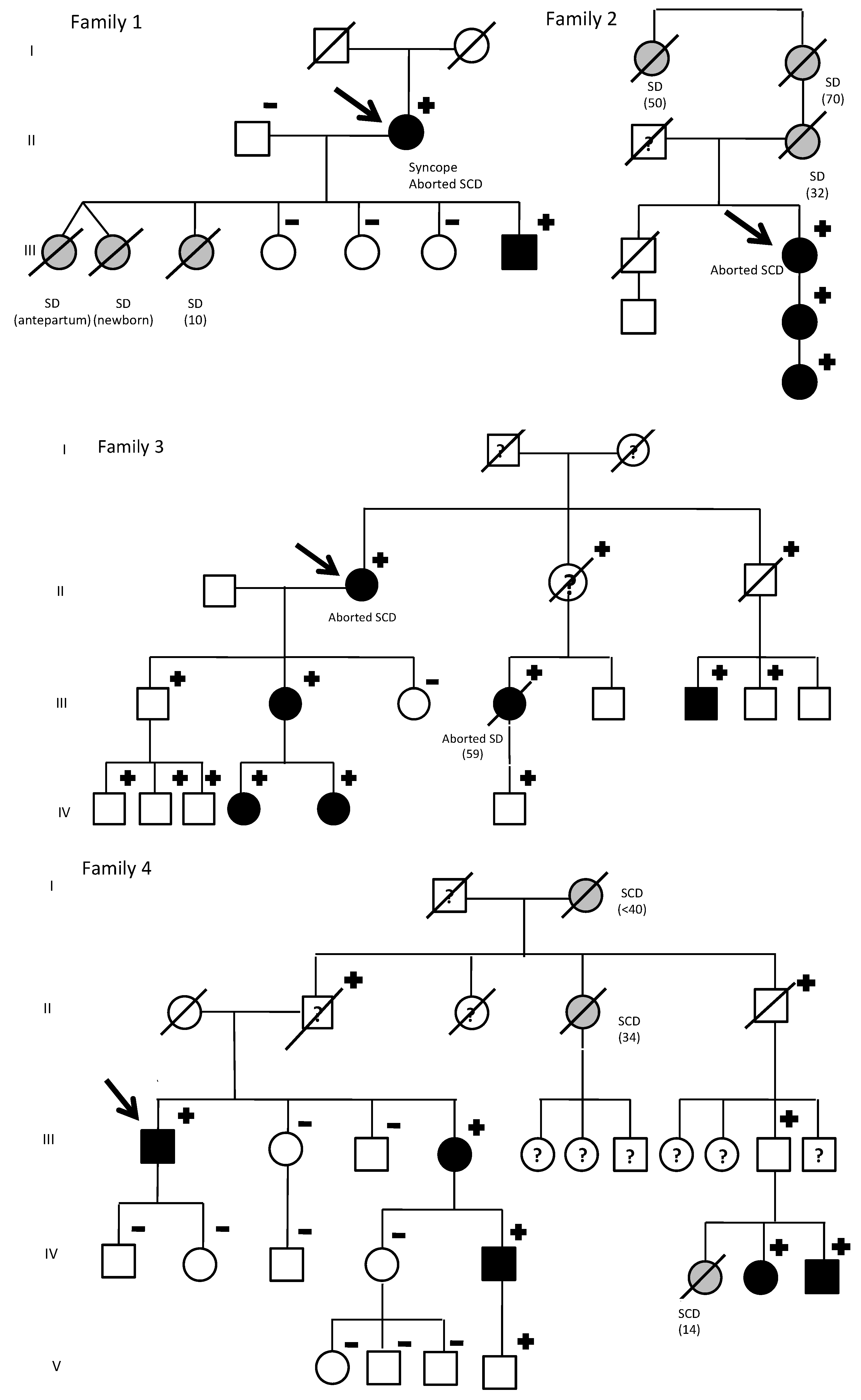
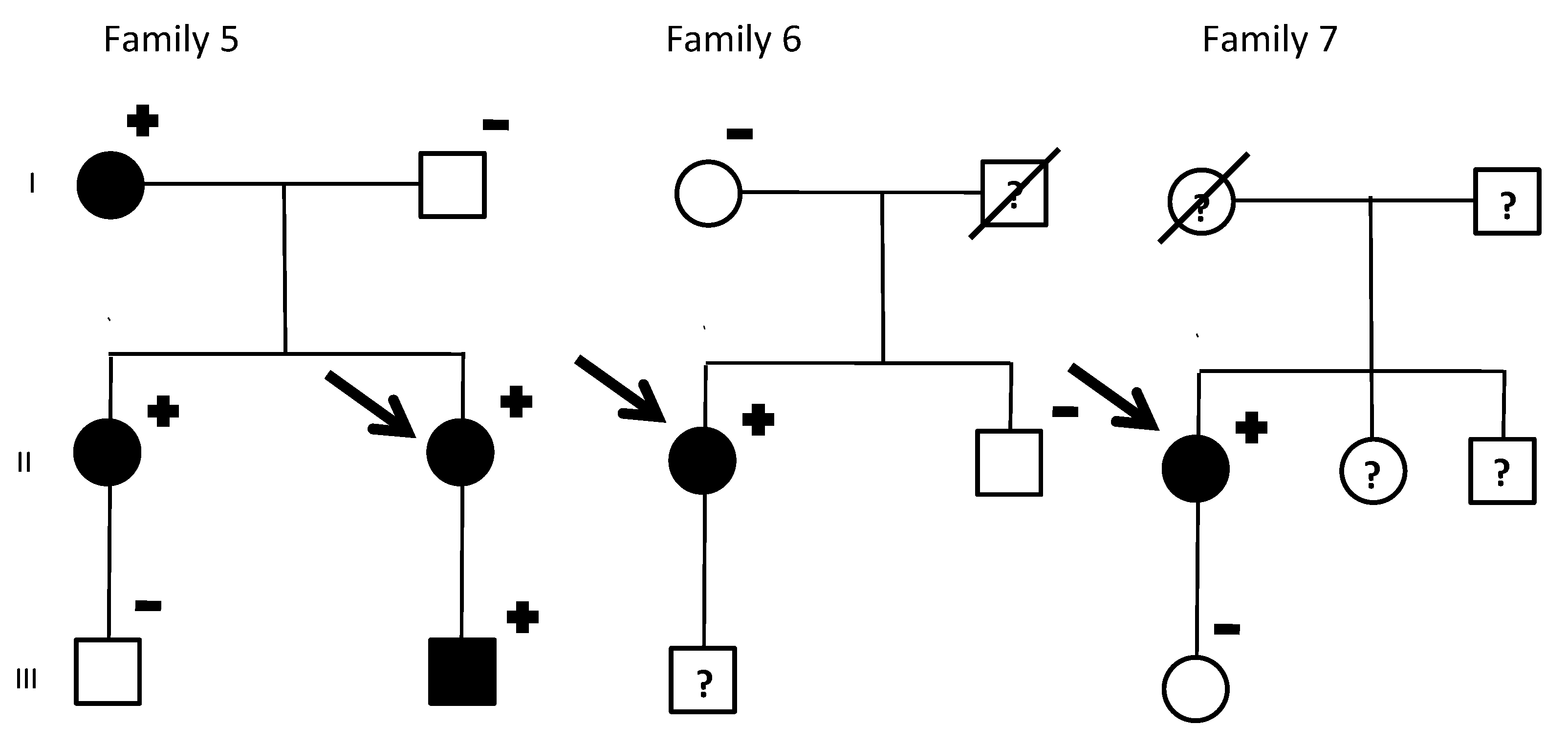
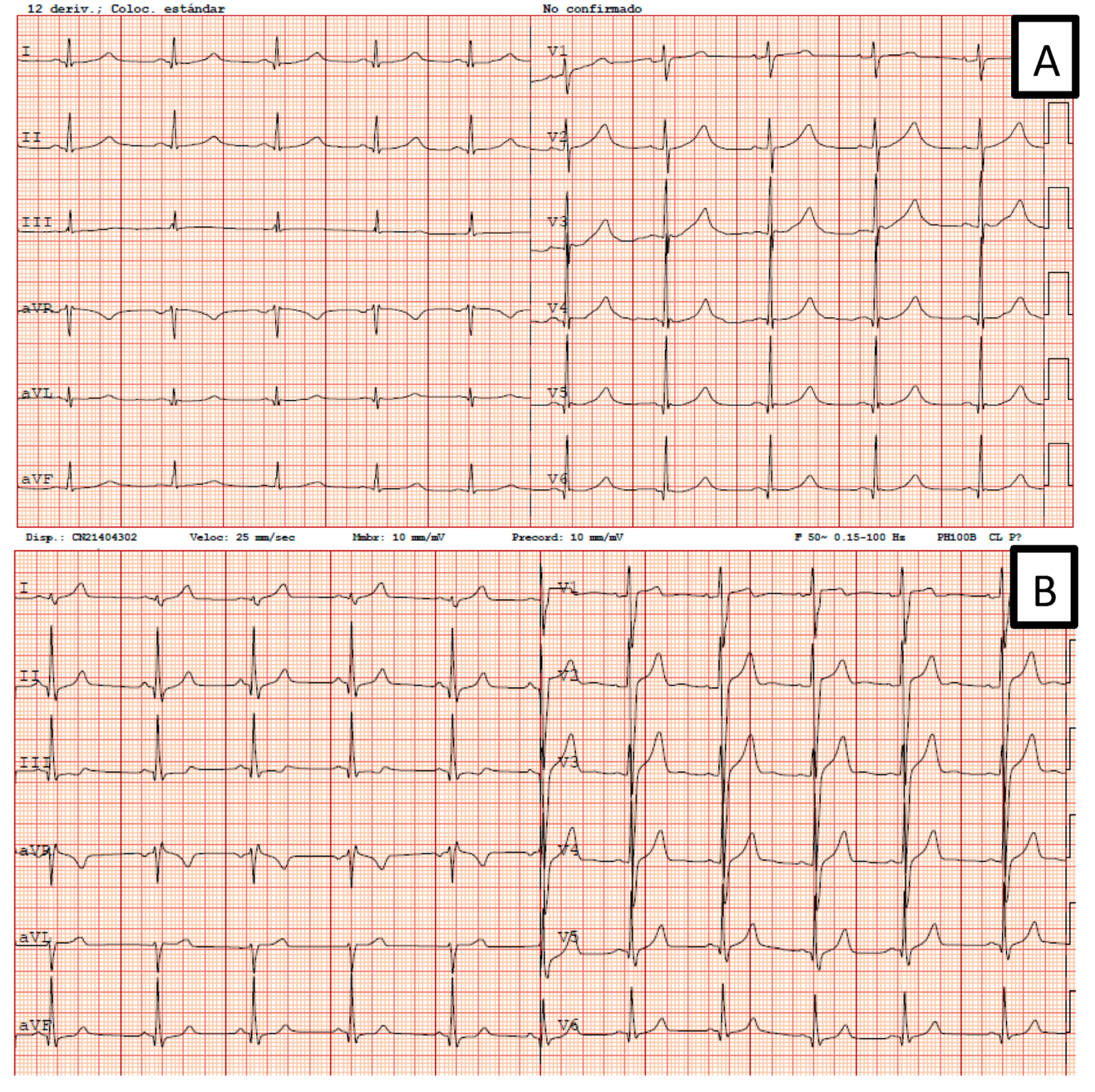
| Index Patient | Gender | Warning Symptoms | Aborted Sudden Deaths in the Family (Gender) | Sudden Deaths in the Family (Gender) | Male Carriers with LQTS Phenotype | Female Carriers with LQTS Phenotype |
|---|---|---|---|---|---|---|
| 1 | Female | Syncope | 1 (Female) | 3 (Female) | 100% (1/1) | 100% (1/1) |
| 2 | Female | Aborted SD | 1 (Female) | 3 (Female) | None | 100% (3/3) |
| 3 | Female | Aborted SD | 2 (Female) | 0 | 12.5% (1/8) | 100% (5/5) * |
| 4 | Male | None | 0 | 3 (Female) | 50% (3/6) * | 100% (2/2) |
| 5 | Female | None | 0 | 0 | 100% (1/1) | 100% (3/3) |
| 6 | Female | None | 0 | 0 | None | 100% (1/1) |
| 7 | Female | None | 0 | 0 | None | 100% (1/1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lorca, R.; Junco-Vicente, A.; Martin-Fernandez, M.; Pascual, I.; Aparicio, A.; Barja, N.; Cuesta-LLavona, E.; Roces, L.; Avanzas, P.; Moris, C.; et al. Clinical Implications and Gender Differences of KCNQ1 p.Gly168Arg Pathogenic Variant in Long QT Syndrome. J. Clin. Med. 2020, 9, 3846. https://doi.org/10.3390/jcm9123846
Lorca R, Junco-Vicente A, Martin-Fernandez M, Pascual I, Aparicio A, Barja N, Cuesta-LLavona E, Roces L, Avanzas P, Moris C, et al. Clinical Implications and Gender Differences of KCNQ1 p.Gly168Arg Pathogenic Variant in Long QT Syndrome. Journal of Clinical Medicine. 2020; 9(12):3846. https://doi.org/10.3390/jcm9123846
Chicago/Turabian StyleLorca, Rebeca, Alejandro Junco-Vicente, Maria Martin-Fernandez, Isaac Pascual, Andrea Aparicio, Noemi Barja, Elias Cuesta-LLavona, Luis Roces, Pablo Avanzas, Cesar Moris, and et al. 2020. "Clinical Implications and Gender Differences of KCNQ1 p.Gly168Arg Pathogenic Variant in Long QT Syndrome" Journal of Clinical Medicine 9, no. 12: 3846. https://doi.org/10.3390/jcm9123846
APA StyleLorca, R., Junco-Vicente, A., Martin-Fernandez, M., Pascual, I., Aparicio, A., Barja, N., Cuesta-LLavona, E., Roces, L., Avanzas, P., Moris, C., Coto, E., Rodríguez Reguero, J. J., & Gómez, J. (2020). Clinical Implications and Gender Differences of KCNQ1 p.Gly168Arg Pathogenic Variant in Long QT Syndrome. Journal of Clinical Medicine, 9(12), 3846. https://doi.org/10.3390/jcm9123846







