Artificial Intelligence Tools for Refining Lung Cancer Screening
Abstract
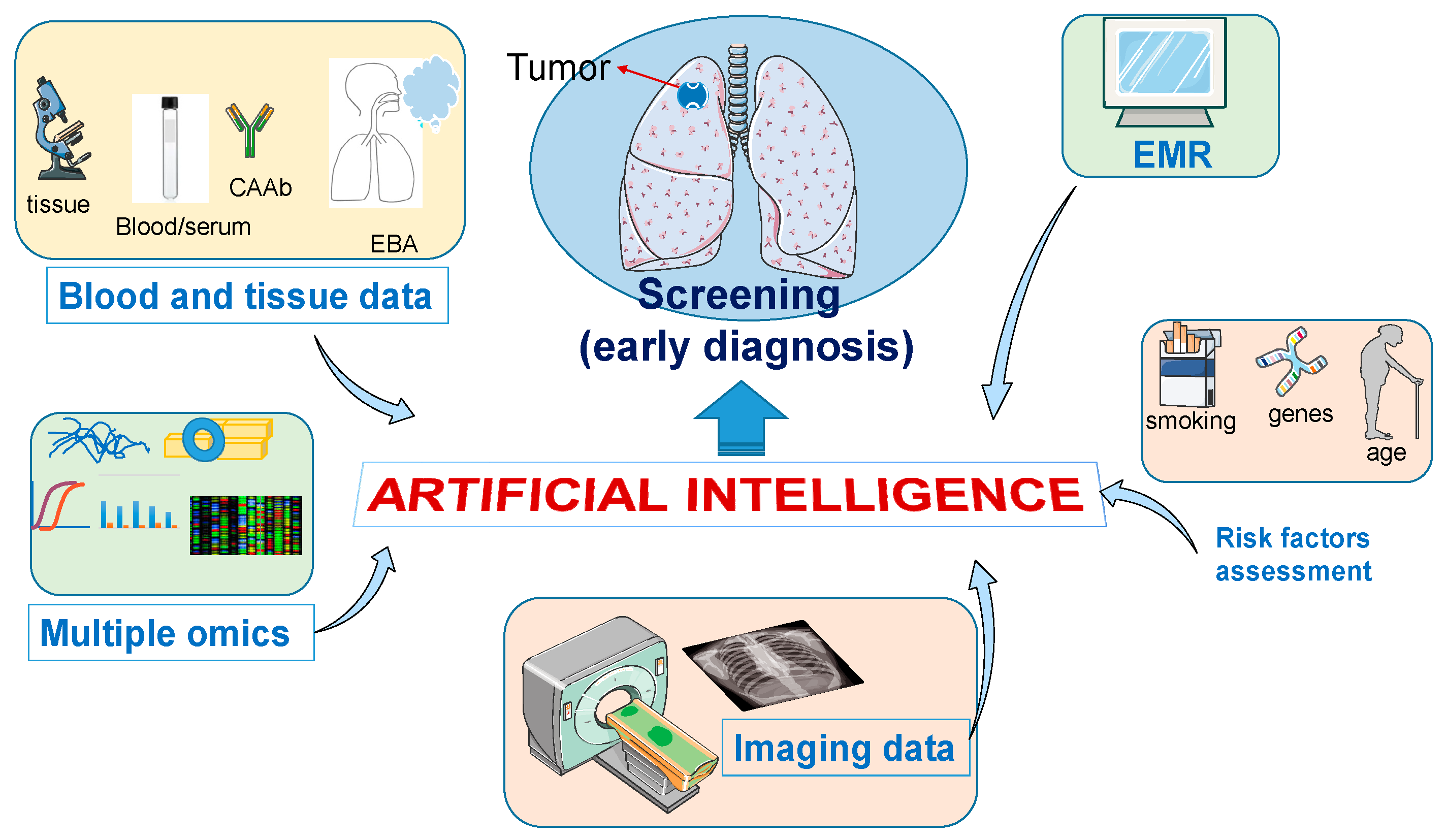
:1. Introduction
2. Lung Cancer: Epidemiological and Clinical Considerations
3. Lung Cancer Screening
4. AI and CT Scan Images
5. Concluding Remarks and Future Directions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bayle, G.L. Recherches sur la Phthisie Pulmonaire; Ouvrage lu à la Société de la Faculté de Médecine de Paris, Dans Diverses Séances, en 1809 et 1810; Gabon: Paris, French, 1810; 439p. [Google Scholar]
- Witschi, H. A short history of lung cancer. Toxicol. Sci. 2001, 64, 4–6. [Google Scholar] [CrossRef] [Green Version]
- Proctor, R.N. The history of the discovery of the cigarette-lung cancer link: Evidentiary traditions, corporate denial, global toll. Tob. Control 2012, 21, 87–91. [Google Scholar] [CrossRef]
- Barta, J.A.; Powell, C.A.; Wisnivesky, J.P. Global Epidemiology of Lung Cancer. Ann. Glob. Health 2019, 85. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [Green Version]
- de Groot, P.M.; Wu, C.C.; Carter, B.W.; Munden, R.F. The epidemiology of lung cancer. Transl. Lung Cancer Res. 2018, 7, 220–233. [Google Scholar] [CrossRef]
- Goebel, C.; Louden, C.L.; McKenna, R.; Onugha, O.; Wachtel, A.; Long, T. Diagnosis of Non-small Cell Lung Cancer for Early Stage Asymptomatic Patients. Cancer Genom. Proteom. 2019, 16, 229–244. [Google Scholar] [CrossRef] [Green Version]
- Blandin Knight, S.; Crosbie, P.A.; Balata, H.; Chudziak, J.; Hussell, T.; Dive, C. Progress and prospects of early detection in lung cancer. Open Biol. 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Sharma, D.; Newman, T.G.; Aronow, W.S. Lung cancer screening: History, current perspectives, and future directions. Arch. Med. Sci. 2015, 11, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Benzaquen, J.; Boutros, J.; Marquette, C.; Delingette, H.; Hofman, P. Lung Cancer Screening, Towards a Multidimensional Approach: Why and How? Cancers 2019, 11, 212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capuano, R.; Catini, A.; Paolesse, R.; Di Natale, C. Sensors for Lung Cancer Diagnosis. J. Clin. Med. 2019, 8, 235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onoi, K.; Chihara, Y.; Uchino, J.; Shimamoto, T.; Morimoto, Y.; Iwasaku, M.; Kaneko, Y.; Yamada, T.; Takayama, K. Immune Checkpoint Inhibitors for Lung Cancer Treatment: A Review. J. Clin. Med. 2020, 9, 1362. [Google Scholar] [CrossRef] [PubMed]
- Owais, M.; Arsalan, M.; Choi, J.; Park, K.R. Effective Diagnosis and Treatment through Content-Based Medical Image Retrieval (CBMIR) by Using Artificial Intelligence. J. Clin. Med. 2019, 8, 462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espinoza, J.L. Machine learning for tackling microbiota data and infection complications in immunocompromised patients with cancer. J. Intern. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Mobiny, A.; Singh, A.; Van Nguyen, H. Risk-Aware Machine Learning Classifier for Skin Lesion Diagnosis. J. Clin. Med. 2019, 8, 1241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azuaje, F.; Kim, S.Y.; Perez Hernandez, D.; Dittmar, G. Connecting Histopathology Imaging and Proteomics in Kidney Cancer through Machine Learning. J. Clin. Med. 2019, 8, 1535. [Google Scholar] [CrossRef] [Green Version]
- Bi, W.L.; Hosny, A.; Schabath, M.B.; Giger, M.L.; Birkbak, N.J.; Mehrtash, A.; Allison, T.; Arnaout, O.; Abbosh, C.; Dunn, I.F.; et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J. Clin. 2019. [Google Scholar] [CrossRef] [Green Version]
- Chassagnon, G.; Vakalopoulou, M.; Paragios, N.; Revel, M.P. Artificial intelligence applications for thoracic imaging. Eur. J. Radiol. 2020, 123, 108774. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Jin, G.; Pang, Y.; Wang, W.; Zhang, H.; Tuo, G.; Wu, P.; Wang, Z.; Zhu, Z. The diagnostic accuracy of artificial intelligence in thoracic diseases: A protocol for systematic review and meta-analysis. Medicine (Baltimore) 2020, 99, e19114. [Google Scholar] [CrossRef]
- Blanchard, E.M.; Arnaoutakis, K.; Hesketh, P.J. Lung cancer in octogenarians. J. Thorac. Oncol. 2010, 5, 909–916. [Google Scholar] [CrossRef] [Green Version]
- Smolle, E.; Pichler, M. Non-Smoking-Associated Lung Cancer: A distinct Entity in Terms of Tumor Biology, Patient Characteristics and Impact of Hereditary Cancer Predisposition. Cancers 2019, 11, 204. [Google Scholar] [CrossRef] [Green Version]
- Kanwal, M.; Ding, X.J.; Cao, Y. Familial risk for lung cancer. Oncol. Lett. 2017, 13, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Torres-Durán, M.; Ruano-Ravina, A.; Parente-Lamelas, I.; Leiro-Fernández, V.; Abal-Arca, J.; Montero-Martínez, C.; Pena-Álvarez, C.; Castro-Añón, O.; Golpe-Gómez, A.; Martínez, C.; et al. Residential radon and lung cancer characteristics in never smokers. Int. J. Radiat. Biol. 2015, 91, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Lukas, R.V.; Gondi, V.; Kamson, D.O.; Kumthekar, P.; Salgia, R. State-of-the-art considerations in small cell lung cancer brain metastases. Oncotarget 2017, 8, 71223–71233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsoukalas, N.; Aravantinou-Fatorou, E.; Baxevanos, P.; Tolia, M.; Tsapakidis, K.; Galanopoulos, M.; Liontos, M.; Kyrgias, G. Advanced small cell lung cancer (SCLC): New challenges and new expectations. Ann. Transl. Med. 2018, 6, 145. [Google Scholar] [CrossRef] [PubMed]
- Gadgeel, S.M. Targeted Therapy and Immune Therapy for Small Cell Lung Cancer. Curr. Treat. Options Oncol. 2018, 19, 53. [Google Scholar] [CrossRef]
- Sgambato, A.; Casaluce, F.; Maione, P.; Gridelli, C. Targeted therapies in non-small cell lung cancer: A focus on ALK/ROS1 tyrosine kinase inhibitors. Expert Rev. Anticancer Ther. 2018, 18, 71–80. [Google Scholar] [CrossRef]
- Forde, P.M.; Ettinger, D.S. Targeted therapy for non-small-cell lung cancer: Past, present and future. Expert Rev. Anticancer Ther. 2013, 13, 745–758. [Google Scholar] [CrossRef] [Green Version]
- Farhat, F.S.; Houhou, W. Targeted therapies in non-small cell lung carcinoma: What have we achieved so far? Ther. Adv. Med. Oncol. 2013, 5, 249–270. [Google Scholar] [CrossRef] [Green Version]
- Calles, A.; Aguado, G.; Sandoval, C.; Álvarez, R. The role of immunotherapy in small cell lung cancer. Clin. Transl. Oncol. 2019. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Y.L. Immunotherapy for small-cell lung cancer. Lancet Oncol. 2016, 17, 846–847. [Google Scholar] [CrossRef] [Green Version]
- Shen, K.; Cui, J.; Wei, Y.; Chen, X.; Liu, G.; Gao, X.; Li, W.; Lu, H.; Zhan, P.; Lv, T.; et al. Effectiveness and safety of PD-1/PD-L1 or CTLA4 inhibitors combined with chemotherapy as a first-line treatment for lung cancer: A meta-analysis. J. Thorac. Dis. 2018, 10, 6636–6652. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yu, X.; Wang, W. A meta-analysis of efficacy and safety of antibodies targeting PD-1/PD-L1 in treatment of advanced nonsmall cell lung cancer. Medicine (Baltimore) 2016, 95, e5539. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.W.; Xiong, Y.; Chen, S.; Xia, F.; Li, Q.; Hu, J. Anti-PD-1/PD-L1 antibody therapy for pretreated advanced nonsmall-cell lung cancer: A meta-analysis of randomized clinical trials. Medicine (Baltimore) 2016, 95, e4611. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Xie, R.; Lin, S.; You, X.; Weng, X. Anti-PD-1/PD-L1 Antibody Therapy for Pretreated Advanced or Metastatic Nonsmall Cell Lung Carcinomas and the Correlation between PD-L1 Expression and Treatment Effectiveness: An Update Meta-Analysis of Randomized Clinical Trials. BioMed Res. Int. 2018, 2018, 3820956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saquib, N.; Saquib, J.; Ioannidis, J.P. Does screening for disease save lives in asymptomatic adults? Systematic review of meta-analyses and randomized trials. Int. J. Epidemiol. 2015, 44, 264–277. [Google Scholar] [CrossRef] [Green Version]
- Shieh, Y.; Eklund, M.; Sawaya, G.F.; Black, W.C.; Kramer, B.S.; Esserman, L.J. Population-based screening for cancer: Hope and hype. Nat. Rev. Clin. Oncol. 2016, 13, 550–565. [Google Scholar] [CrossRef]
- Brett, G.Z. The value of lung cancer detection by six-monthly chest radiographs. Thorax 1968, 23, 414–420. [Google Scholar] [CrossRef] [Green Version]
- Melamed, M.R.; Flehinger, B.J.; Zaman, M.B.; Heelan, R.T.; Perchick, W.A.; Martini, N. Screening for early lung cancer. Results of the Memorial Sloan-Kettering study in New York. Chest 1984, 86, 44–53. [Google Scholar] [CrossRef]
- Hocking, W.G.; Hu, P.; Oken, M.M.; Winslow, S.D.; Kvale, P.A.; Prorok, P.C.; Ragard, L.R.; Commins, J.; Lynch, D.A.; Andriole, G.L.; et al. Lung cancer screening in the randomized Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial. J. Natl. Cancer Inst. 2010, 102, 722–731. [Google Scholar] [CrossRef]
- Kaneko, M.; Eguchi, K.; Ohmatsu, H.; Kakinuma, R.; Naruke, T.; Suemasu, K.; Moriyama, N. Peripheral lung cancer: Screening and detection with low-dose spiral CT versus radiography. Radiology 1996, 201, 798–802. [Google Scholar] [CrossRef]
- Sone, S.; Takashima, S.; Li, F.; Yang, Z.; Honda, T.; Maruyama, Y.; Hasegawa, M.; Yamanda, T.; Kubo, K.; Hanamura, K.; et al. Mass screening for lung cancer with mobile spiral computed tomography scanner. Lancet 1998, 351, 1242–1245. [Google Scholar] [CrossRef]
- Henschke, C.I. Early lung cancer action project: Overall design and findings from baseline screening. Cancer 2000, 89, 2474–2482. [Google Scholar] [CrossRef]
- Henschke, C.I.; Yankelevitz, D.F.; Libby, D.M.; Pasmantier, M.W.; Smith, J.P.; Miettinen, O.S.; International Early Lung Cancer Action Program Investigators. Survival of patients with stage I lung cancer detected on CT screening. N. Engl. J. Med. 2006, 355, 1763–1771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swensen, S.J.; Jett, J.R.; Hartman, T.E.; Midthun, D.E.; Mandrekar, S.J.; Hillman, S.L.; Sykes, A.M.; Aughenbaugh, G.L.; Bungum, A.O.; Allen, K.L. CT screening for lung cancer: Five-year prospective experience. Radiology 2005, 235, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Aberle, D.R.; Adams, A.M.; Berg, C.D.; Black, W.C.; Clapp, J.D.; Fagerstrom, R.M.; Gareen, I.F.; Gatsonis, C.; Marcus, P.M.; Sicks, J.D.; et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saghir, Z.; Dirksen, A.; Ashraf, H.; Bach, K.S.; Brodersen, J.; Clementsen, P.F.; Døssing, M.; Hansen, H.; Kofoed, K.F.; Larsen, K.R.; et al. CT screening for lung cancer brings forward early disease. The randomised Danish Lung Cancer Screening Trial: Status after five annual screening rounds with low-dose CT. Thorax 2012, 67, 296–301. [Google Scholar] [CrossRef] [Green Version]
- Infante, M.; Cavuto, S.; Lutman, F.R.; Passera, E.; Chiarenza, M.; Chiesa, G.; Brambilla, G.; Angeli, E.; Aranzulla, G.; Chiti, A.; et al. Long-Term Follow-up Results of the DANTE Trial, a Randomized Study of Lung Cancer Screening with Spiral Computed Tomography. Am. J. Respir. Crit. Care Med. 2015, 191, 1166–1175. [Google Scholar] [CrossRef]
- Paci, E.; Puliti, D.; Lopes Pegna, A.; Carrozzi, L.; Picozzi, G.; Falaschi, F.; Pistelli, F.; Aquilini, F.; Ocello, C.; Zappa, M.; et al. Mortality, survival and incidence rates in the ITALUNG randomised lung cancer screening trial. Thorax 2017, 72, 825–831. [Google Scholar] [CrossRef] [Green Version]
- Becker, N.; Motsch, E.; Trotter, A.; Heussel, C.P.; Dienemann, H.; Schnabel, P.A.; Kauczor, H.U.; Maldonado, S.G.; Miller, A.B.; Kaaks, R.; et al. Lung cancer mortality reduction by LDCT screening-Results from the randomized German LUSI trial. Int. J. Cancer 2020, 146, 1503–1513. [Google Scholar] [CrossRef]
- Pastorino, U.; Sverzellati, N.; Sestini, S.; Silva, M.; Sabia, F.; Boeri, M.; Cantarutti, A.; Sozzi, G.; Corrao, G.; Marchianò, A. Ten-year results of the Multicentric Italian Lung Detection trial demonstrate the safety and efficacy of biennial lung cancer screening. Eur. J. Cancer 2019, 118, 142–148. [Google Scholar] [CrossRef]
- Pastorino, U.; Silva, M.; Sestini, S.; Sabia, F.; Boeri, M.; Cantarutti, A.; Sverzellati, N.; Sozzi, G.; Corrao, G.; Marchianò, A. Prolonged lung cancer screening reduced 10-year mortality in the MILD trial: New confirmation of lung cancer screening efficacy. Ann. Oncol. 2019, 30, 1162–1169. [Google Scholar] [CrossRef] [PubMed]
- de Koning, H.J.; van der Aalst, C.M.; de Jong, P.A.; Scholten, E.T.; Nackaerts, K.; Heuvelmans, M.A.; Lammers, J.J.; Weenink, C.; Yousaf-Khan, U.; Horeweg, N.; et al. Reduced Lung-Cancer Mortality with Volume CT Screening in a Randomized Trial. N. Engl. J. Med. 2020, 382, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Moyer, V.A.; US Preventive Services Task Force. Screening for lung cancer: U.S. Preventive Services Task Force recommendation statement. Ann. Intern. Med. 2014, 160, 330–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oken, M.M.; Hocking, W.G.; Kvale, P.A.; Andriole, G.L.; Buys, S.S.; Church, T.R.; Crawford, E.D.; Fouad, M.N.; Isaacs, C.; Reding, D.J.; et al. Screening by chest radiograph and lung cancer mortality: The Prostate, Lung, Colorectal, and Ovarian (PLCO) randomized trial. JAMA 2011, 306, 1865–1873. [Google Scholar] [CrossRef]
- Pinsky, P.F.; Berg, C.D. Applying the National Lung Screening Trial eligibility criteria to the US population: What percent of the population and of incident lung cancers would be covered? J. Med. Screen 2012, 19, 154–156. [Google Scholar] [CrossRef]
- Kozubek, M. Challenges and Benchmarks in Bioimage Analysis. Adv. Anat. Embryol. Cell Biol. 2016, 219, 231–262. [Google Scholar] [CrossRef]
- Jacobs, C.; van Rikxoort, E.M.; Murphy, K.; Prokop, M.; Schaefer-Prokop, C.M.; van Ginneken, B. Computer-aided detection of pulmonary nodules: A comparative study using the public LIDC/IDRI database. Eur. Radiol. 2016, 26, 2139–2147. [Google Scholar] [CrossRef]
- Armato, S.G.; McLennan, G.; Bidaut, L.; McNitt-Gray, M.F.; Meyer, C.R.; Reeves, A.P.; Zhao, B.; Aberle, D.R.; Henschke, C.I.; Hoffman, E.A.; et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A completed reference database of lung nodules on CT scans. Med. Phys. 2011, 38, 915–931. [Google Scholar] [CrossRef]
- Data Science Bowl 2017, Can You Improve Lung Cancer Detection? Available online: https://www.kaggle.com/c/data-science-bowl-2017 (accessed on 14 May 2018).
- Maier-Hein, L.; Eisenmann, M.; Reinke, A.; Onogur, S.; Stankovic, M.; Scholz, P.; Arbel, T.; Bogunovic, H.; Bradley, A.P.; Carass, A.; et al. Why rankings of biomedical image analysis competitions should be interpreted with care. Nat. Commun. 2018, 9, 5217. [Google Scholar] [CrossRef] [Green Version]
- Maier-Hein, L.; Reinke, A.; Kozubek, M.; Martel, A.L.; Arbel, T.; Eisenmann, M.; Hanbury, A.; Jannin, P.; Müller, H.; Onogur, S.; et al. BIAS: Transparent reporting of biomedical image analysis challenges. Med. Image Anal. 2020, 66, 101796. [Google Scholar] [CrossRef]
- McWilliams, A.; Tammemagi, M.C.; Mayo, J.R.; Roberts, H.; Liu, G.; Soghrati, K.; Yasufuku, K.; Martel, S.; Laberge, F.; Gingras, M.; et al. Probability of cancer in pulmonary nodules detected on first screening CT. N. Engl. J. Med. 2013, 369, 910–919. [Google Scholar] [CrossRef] [Green Version]
- Pinsky, P.F.; Gierada, D.S.; Black, W.; Munden, R.; Nath, H.; Aberle, D.; Kazerooni, E. Performance of Lung-RADS in the National Lung Screening Trial: A retrospective assessment. Ann. Intern. Med. 2015, 162, 485–491. [Google Scholar] [CrossRef] [Green Version]
- McKee, B.J.; Regis, S.M.; McKee, A.B.; Flacke, S.; Wald, C. Performance of ACR Lung-RADS in a clinical CT lung screening program. J. Am. Coll. Radiol. 2015, 12, 273–276. [Google Scholar] [CrossRef] [Green Version]
- Ciompi, F.; Chung, K.; van Riel, S.J.; Setio, A.A.A.; Gerke, P.K.; Jacobs, C.; Scholten, E.T.; Schaefer-Prokop, C.; Wille, M.M.W.; Marchianò, A.; et al. Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci. Rep. 2017, 7, 46479. [Google Scholar] [CrossRef]
- Petousis, P.; Han, S.X.; Aberle, D.; Bui, A.A. Prediction of lung cancer incidence on the low-dose computed tomography arm of the National Lung Screening Trial: A dynamic Bayesian network. Artif. Intell. Med. 2016, 72, 42–55. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Sun, X.; Dang, K.; Li, K.; Guo, X.W.; Chang, J.; Yu, Z.Q.; Huang, F.Y.; Wu, Y.S.; Liang, Z.; et al. Toward an Expert Level of Lung Cancer Detection and Classification Using a Deep Convolutional Neural Network. Oncologist 2019, 24, 1159–1165. [Google Scholar] [CrossRef] [Green Version]
- Petousis, P.; Winter, A.; Speier, W.; Aberle, D.R.; Hsu, W.; Bui, A.A.T. Using Sequential Decision Making to Improve Lung Cancer Screening Performance. IEEE Access 2019, 7, 119403–119419. [Google Scholar] [CrossRef]
- Huang, P.; Lin, C.T.; Li, Y.; Tammemagi, M.C.; Brock, M.V.; Atkar-Khattra, S.; Xu, Y.; Hu, P.; Mayo, J.R.; Schmidt, H.; et al. Prediction of lung cancer risk at follow-up screening with low-dose CT: A training and validation study of a deep learning method. Lancet Digit. Health 2019, 1, e353–e362. [Google Scholar] [CrossRef] [Green Version]
- Ardila, D.; Kiraly, A.P.; Bharadwaj, S.; Choi, B.; Reicher, J.J.; Peng, L.; Tse, D.; Etemadi, M.; Ye, W.; Corrado, G.; et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 2019, 25, 954–961. [Google Scholar] [CrossRef]
- Cui, S.; Ming, S.; Lin, Y.; Chen, F.; Shen, Q.; Li, H.; Chen, G.; Gong, X.; Wang, H. Development and clinical application of deep learning model for lung nodules screening on CT images. Sci. Rep. 2020, 10, 13657. [Google Scholar] [CrossRef]
- Fornacon-Wood, I.; Faivre-Finn, C.; O’Connor, J.P.B.; Price, G.J. Radiomics as a personalized medicine tool in lung cancer: Separating the hope from the hype. Lung Cancer 2020, 146, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Koçak, B.; Durmaz, E.; Ateş, E.; Kılıçkesmez, Ö. Radiomics with artificial intelligence: A practical guide for beginners. Diagn. Interv. Radiol. 2019, 25, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, S.; Wang, H.; Liu, Y.; Garcia, A.; Stringfield, O.; Krewer, H.; Li, Q.; Cherezov, D.; Gatenby, R.A.; Balagurunathan, Y.; et al. Predicting Malignant Nodules from Screening CT Scans. J. Thorac. Oncol. 2016, 11, 2120–2128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, S.J.; Wang, C.W.; Pan, K.T.; Wu, Y.C.; Wu, C.T. Localized thin-section CT with radiomics feature extraction and machine learning to classify early-detected pulmonary nodules from lung cancer screening. Phys. Med. Biol. 2018, 63, 065005. [Google Scholar] [CrossRef]
- Delzell, D.A.P.; Magnuson, S.; Peter, T.; Smith, M.; Smith, B.J. Machine Learning and Feature Selection Methods for Disease Classification with Application to Lung Cancer Screening Image Data. Front. Oncol. 2019, 9, 1393. [Google Scholar] [CrossRef] [Green Version]
- Grossmann, P.; Stringfield, O.; El-Hachem, N.; Bui, M.M.; Rios Velazquez, E.; Parmar, C.; Leijenaar, R.T.; Haibe-Kains, B.; Lambin, P.; Gillies, R.J.; et al. Defining the biological basis of radiomic phenotypes in lung cancer. eLife 2017, 6. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Irvin, J.; Ball, R.L.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.P.; et al. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. 2018, 15, e1002686. [Google Scholar] [CrossRef]
- Meziane, M.; Mazzone, P.; Novak, E.; Lieber, M.L.; Lababede, O.; Phillips, M.; Obuchowski, N.A. A comparison of four versions of a computer-aided detection system for pulmonary nodules on chest radiographs. J. Thorac. Imaging 2012, 27, 58–64. [Google Scholar] [CrossRef]
- Schalekamp, S.; van Ginneken, B.; Koedam, E.; Snoeren, M.M.; Tiehuis, A.M.; Wittenberg, R.; Karssemeijer, N.; Schaefer-Prokop, C.M. Computer-aided detection improves detection of pulmonary nodules in chest radiographs beyond the support by bone-suppressed images. Radiology 2014, 272, 252–261. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.H.; Goo, J.M.; Park, C.M.; Lee, H.J.; Jin, K.N. Computer-aided detection of malignant lung nodules on chest radiographs: Effect on observers’ performance. Korean J. Radiol. 2012, 13, 564–571. [Google Scholar] [CrossRef]
- Nam, J.G.; Park, S.; Hwang, E.J.; Lee, J.H.; Jin, K.N.; Lim, K.Y.; Vu, T.H.; Sohn, J.H.; Hwang, S.; Goo, J.M.; et al. Development and Validation of Deep Learning-based Automatic Detection Algorithm for Malignant Pulmonary Nodules on Chest Radiographs. Radiology 2019, 290, 218–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cha, M.J.; Chung, M.J.; Lee, J.H.; Lee, K.S. Performance of Deep Learning Model in Detecting Operable Lung Cancer with Chest Radiographs. J. Thorac. Imaging 2019, 34, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.Y.; Lin, J.T.; Huang, H.H.; Gao, Y.; Yan, M.R.; Sun, M.; Xu, W.P.; Yan, H.H.; Zhong, W.Z.; Yang, X.N. Development and Validation of a. Clin. Lung Cancer 2020, 21, 47–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deppen, S.A.; Blume, J.D.; Kensinger, C.D.; Morgan, A.M.; Aldrich, M.C.; Massion, P.P.; Walker, R.C.; McPheeters, M.L.; Putnam, J.B.; Grogan, E.L. Accuracy of FDG-PET to diagnose lung cancer in areas with infectious lung disease: A meta-analysis. JAMA 2014, 312, 1227–1236. [Google Scholar] [CrossRef] [PubMed]
- Schwyzer, M.; Ferraro, D.A.; Muehlematter, U.J.; Curioni-Fontecedro, A.; Huellner, M.W.; von Schulthess, G.K.; Kaufmann, P.A.; Burger, I.A.; Messerli, M. Automated detection of lung cancer at ultralow dose PET/CT by deep neural networks—Initial results. Lung Cancer 2018, 126, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Biederer, J.; Ohno, Y.; Hatabu, H.; Schiebler, M.L.; van Beek, E.J.R.; Vogel-Claussen, J.; Kauczor, H.U. Screening for lung cancer: Does MRI have a role? Eur. J. Radiol. 2017, 86, 353–360. [Google Scholar] [CrossRef]
- Allen, B.D.; Schiebler, M.L.; Sommer, G.; Kauczor, H.U.; Biederer, J.; Kruser, T.J.; Carr, J.C.; Hazen, G. Cost-effectiveness of lung MRI in lung cancer screening. Eur. Radiol. 2020, 30, 1738–1746. [Google Scholar] [CrossRef]
- Meier-Schroers, M.; Homsi, R.; Gieseke, J.; Schild, H.H.; Thomas, D. Lung cancer screening with MRI: Evaluation of MRI for lung cancer screening by comparison of LDCT- and MRI-derived Lung-RADS categories in the first two screening rounds. Eur. Radiol. 2019, 29, 898–905. [Google Scholar] [CrossRef]
- Wang, X.; Wan, Q.; Chen, H.; Li, Y.; Li, X. Classification of pulmonary lesion based on multiparametric MRI: Utility of radiomics and comparison of machine learning methods. Eur. Radiol. 2020, 30, 4595–4605. [Google Scholar] [CrossRef]
- Janowczyk, A.; Madabhushi, A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J. Pathol. Inform. 2016, 7, 29. [Google Scholar] [CrossRef]
- Ding, H.; Xia, W.; Zhang, L.; Mao, Q.; Cao, B.; Zhao, Y.; Xu, L.; Jiang, F.; Dong, G. CT-Based Deep Learning Model for Invasiveness Classification and Micropapillary Pattern Prediction within Lung Adenocarcinoma. Front. Oncol. 2020, 10, 1186. [Google Scholar] [CrossRef] [PubMed]
- Kanavati, F.; Toyokawa, G.; Momosaki, S.; Rambeau, M.; Kozuma, Y.; Shoji, F.; Yamazaki, K.; Takeo, S.; Iizuka, O.; Tsuneki, M. Weakly-supervised learning for lung carcinoma classification using deep learning. Sci. Rep. 2020, 10, 9297. [Google Scholar] [CrossRef] [PubMed]
- Marquette, C.H.; Boutros, J.; Benzaquen, J.; Ferreira, M.; Pastre, J.; Pison, C.; Padovani, B.; Bettayeb, F.; Fallet, V.; Guibert, N.; et al. Circulating tumour cells as a potential biomarker for lung cancer screening: A prospective cohort study. Lancet Respir. Med. 2020, 8, 709–716. [Google Scholar] [CrossRef]
- Wang, T.; Liu, H.; Pei, L.; Wang, K.; Song, C.; Wang, P.; Ye, H.; Zhang, J.; Ji, Z.; Ouyang, S.; et al. Screening of tumor-associated antigens based on Oncomine database and evaluation of diagnostic value of autoantibodies in lung cancer. Clin. Immunol. 2020, 210, 108262. [Google Scholar] [CrossRef]
- Fehlmann, T.; Kahraman, M.; Ludwig, N.; Backes, C.; Galata, V.; Keller, V.; Geffers, L.; Mercaldo, N.; Hornung, D.; Weis, T.; et al. Evaluating the Use of Circulating MicroRNA Profiles for Lung Cancer Detection in Symptomatic Patients. JAMA Oncol. 2020, 6, 714–723. [Google Scholar] [CrossRef]
- Cai, D.; Xu, Y.; Ding, R.; Qiu, K.; Zhang, R.; Wang, H.; Huang, L.; Xie, X.; Yan, H.; Deng, Y.; et al. Extensive serum biomarker analysis in patients with non-small-cell lung carcinoma. Cytokine 2020, 126, 154868. [Google Scholar] [CrossRef]
- Aasi, A.; Aghaei, S.M.; Panchapakesan, B. A density functional theory study on the interaction of toluene with transition metal decorated carbon nanotubes: A promising platform for early detection of lung cancer from human breath. Nanotechnology 2020, 31, 415707. [Google Scholar] [CrossRef]
- Robles, A.I.; Harris, C.C. Integration of multiple “OMIC” biomarkers: A precision medicine strategy for lung cancer. Lung Cancer 2017, 107, 50–58. [Google Scholar] [CrossRef] [Green Version]
- Nicora, G.; Vitali, F.; Dagliati, A.; Geifman, N.; Bellazzi, R. Integrated Multi-Omics Analyses in Oncology: A Review of Machine Learning Methods and Tools. Front. Oncol. 2020, 10, 1030. [Google Scholar] [CrossRef]
- Holzinger, A.; Haibe-Kains, B.; Jurisica, I. Why imaging data alone is not enough: AI-based integration of imaging, omics, and clinical data. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2722–2730. [Google Scholar] [CrossRef]
- Lo Gullo, R.; Daimiel, I.; Morris, E.A.; Pinker, K. Combining molecular and imaging metrics in cancer: Radiogenomics. Insights Imaging 2020, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Zhu, Z.; Wu, H.; Han, W.; Cheng, X.; Li, J.; Du, H.; Lei, J.; Sui, X.; Song, W.; et al. Individualized nomogram for predicting ALK rearrangement status in lung adenocarcinoma patients. Eur. Radiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, S.; Petrella, F.; Buscarino, V.; De Maria, F.; Raimondi, S.; Barberis, M.; Fumagalli, C.; Spitaleri, G.; Rampinelli, C.; De Marinis, F.; et al. CT Radiogenomic Characterization of EGFR, K-RAS, and ALK Mutations in Non-Small Cell Lung Cancer. Eur. Radiol. 2016, 26, 32–42. [Google Scholar] [CrossRef] [PubMed]
| First Author/Year | Algorithm | Source of Data | No of Cases | Type of Validation | Main Finding |
|---|---|---|---|---|---|
| Ciompi F. [66], 2017 | CNN: Support vector machines | Data from the MILD trial and DLCST trial | 943 patients (1352 nodules) from MILD trial. | 468 patients (639 nodules) from DLCST trial. Accuracy: comparing with experienced radiologists (assessing 162 nodules from the test set) | The model outperformed classical patch classification approaches classifying lung nodules and its performance was comparable to that of experienced radiologists |
| Petousi P [67] | DBNs. Including three expert-driven DBNs and two DBNs derived from structure learning methods. | Retrospective clinical trial data from the NLST Trial. | Trained 10 times. Each time, 400 NLST cases (200 cancer and 200 non-cancer cases) randomly selected. | Internal validation using the complete LDCT arm of the NLST dataset (N = 25,486) | High discrimination and predictive power with the majority of cancer and non-cancer cases. Average AUC of the ROC was >0.75 for all DBN models outperforming logistic regression and naïve Bayes comparison models. |
| Zhang C [68] | CNN | Chest CT images from 3 sets of data. | 888 CT images from the LUNA16 dataset and 1397 CT images from the Kaggle dataset | CT images from three university hospitals in China. 25 experts graded prospectively collected CT images and compared with the CNN model | Performance of the model: Sensitivity 84.4% and specificity 83.0%. Subgroup analysis of smaller nodules (<10 mm) showed high sensitivity and specificity, similar to that of larger nodules (10–30 mm). |
| Petousi P [69] | Machine learning and DBN | NLST data | 5402 cases from the NLST LDCT trial arm with single indeterminate pulmonary nodules | -A five-fold stratified cross validation -Model’s decisions further compared with experts’ decisions | The model lowered the false-positive rate for most screenings in the NLST, while maintaining true positive detection rates; and improved early prediction of cancer cases with indeterminate pulmonary nodules |
| Huang [70] | Deep learning | Retrospective clinical trial data from the NLST Trial and from PanCan study. | The training cohort: 25,097 NLST cases | Double-blinded validation with 2294 PanCan cases | Compared with Lung-RADS, the model identified a high-risk group that was smaller and had a higher proportion of cancers, and more accurately identified people at very low-risk of lung cancer within 2 years. |
| Ardila [71], 2019 | Deep learning (Three-dimensional deep convolutional neural networks) | Retrospective clinical trial data from the NLST Trial. | 6716 NLST cases, | Independent 1139 cases. | AUC, 0.94 Better than radiologists when prior CT was not available. Equal as radiologists when prior CT was available |
| Cui [72], 2020 | Deep learning | Retrospective analysis of lung cancer screening data from three hospitals in China | Training test: 39,014 chest LDCT screening cases | Validation set (600 cases). External validation: the LUNA public database (888 studies) | Higher sensitivity than all radiologists. Low FPR. Better ROC-AUC and higher specificity than radiologists for classifying true positive cases. Good for differentiating nodule dimensions and nodule sub-types. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Espinoza, J.L.; Dong, L.T. Artificial Intelligence Tools for Refining Lung Cancer Screening. J. Clin. Med. 2020, 9, 3860. https://doi.org/10.3390/jcm9123860
Espinoza JL, Dong LT. Artificial Intelligence Tools for Refining Lung Cancer Screening. Journal of Clinical Medicine. 2020; 9(12):3860. https://doi.org/10.3390/jcm9123860
Chicago/Turabian StyleEspinoza, J. Luis, and Le Thanh Dong. 2020. "Artificial Intelligence Tools for Refining Lung Cancer Screening" Journal of Clinical Medicine 9, no. 12: 3860. https://doi.org/10.3390/jcm9123860





