Evaluation of Transfer Learning with Deep Convolutional Neural Networks for Screening Osteoporosis in Dental Panoramic Radiographs
Abstract
:1. Introduction
2. Patients and Methods
2.1. Patients
2.2. Data Preprocessing
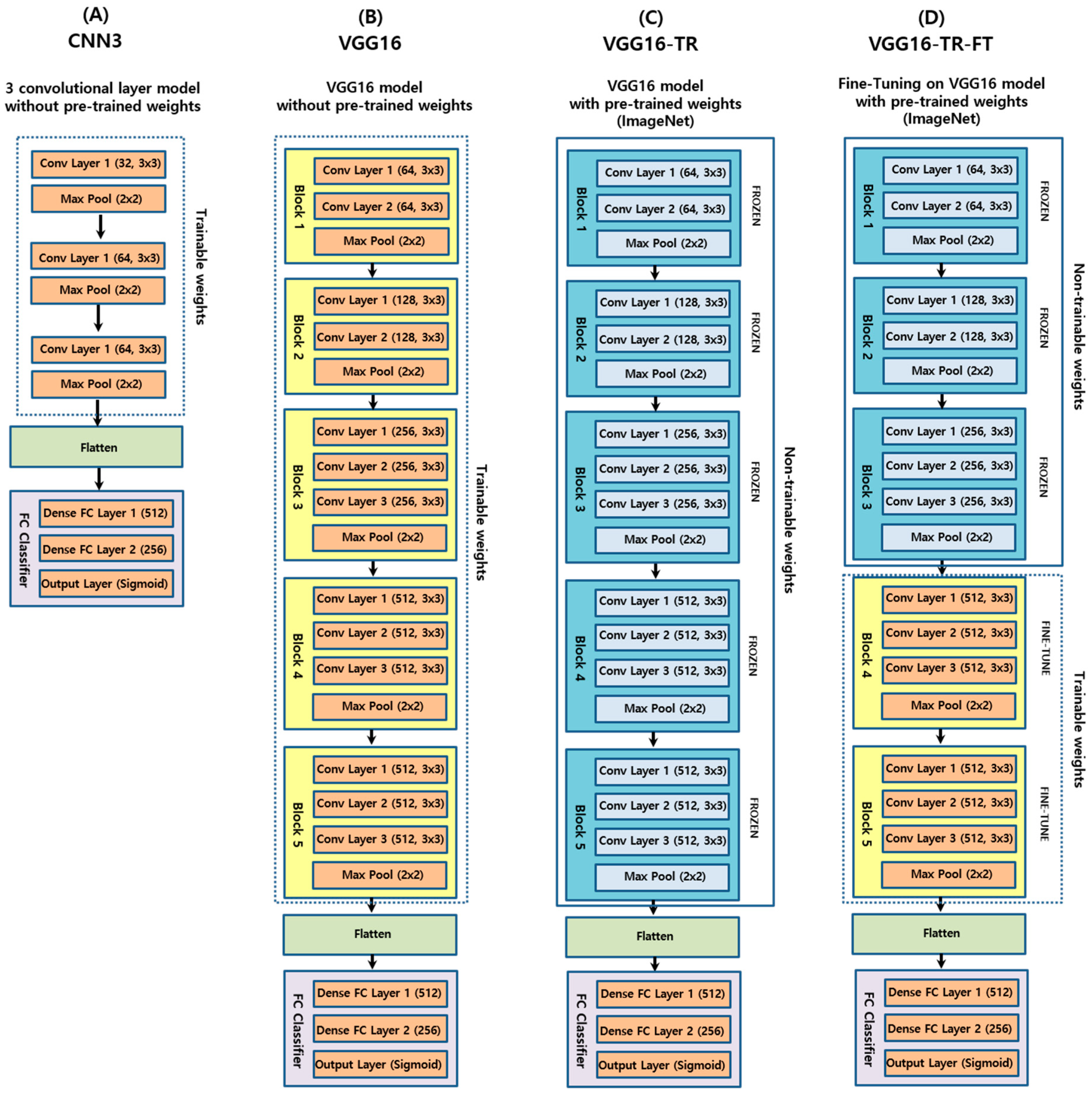
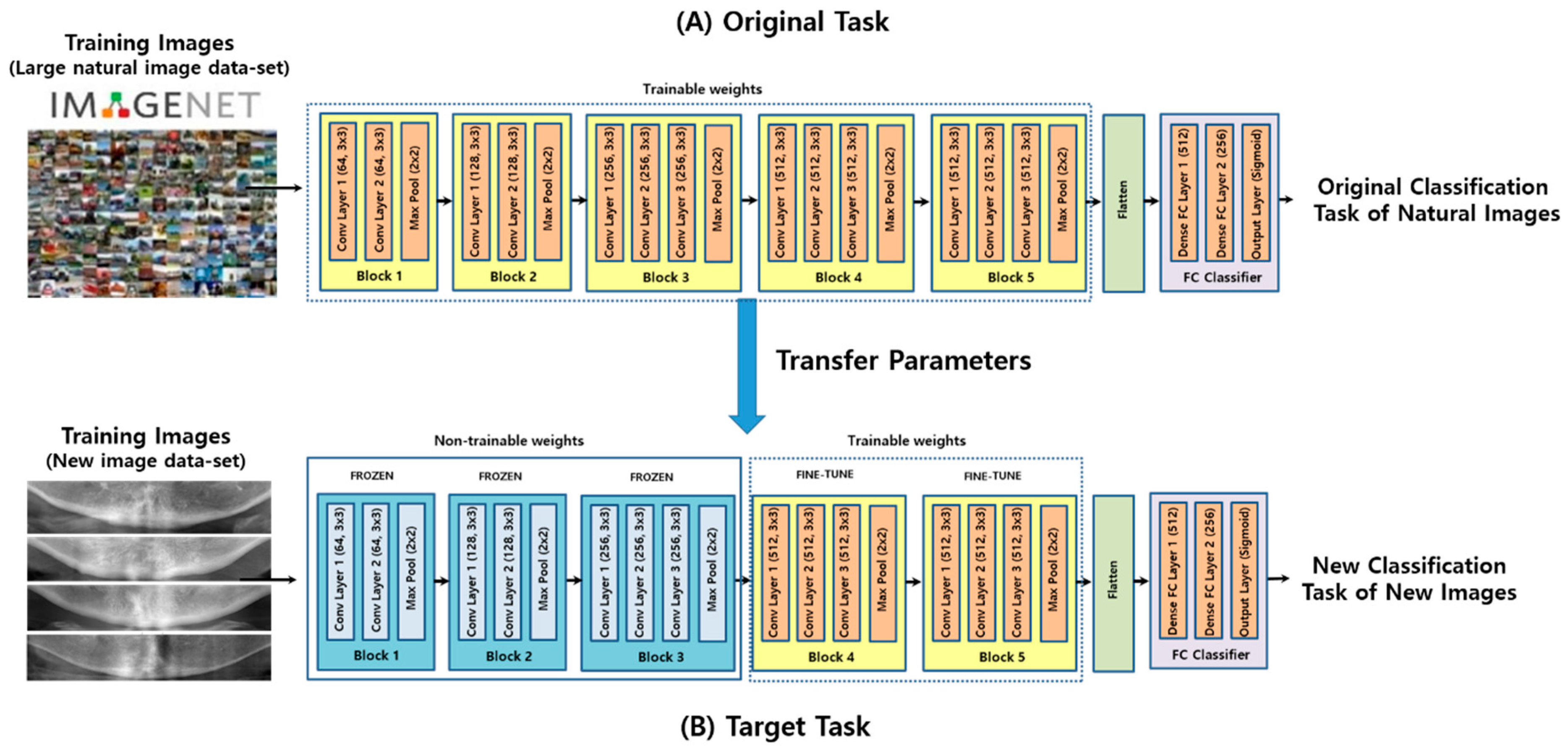
2.3. Convolutional Neural Networks
2.4. Model Training
2.5. Performance Evaluation
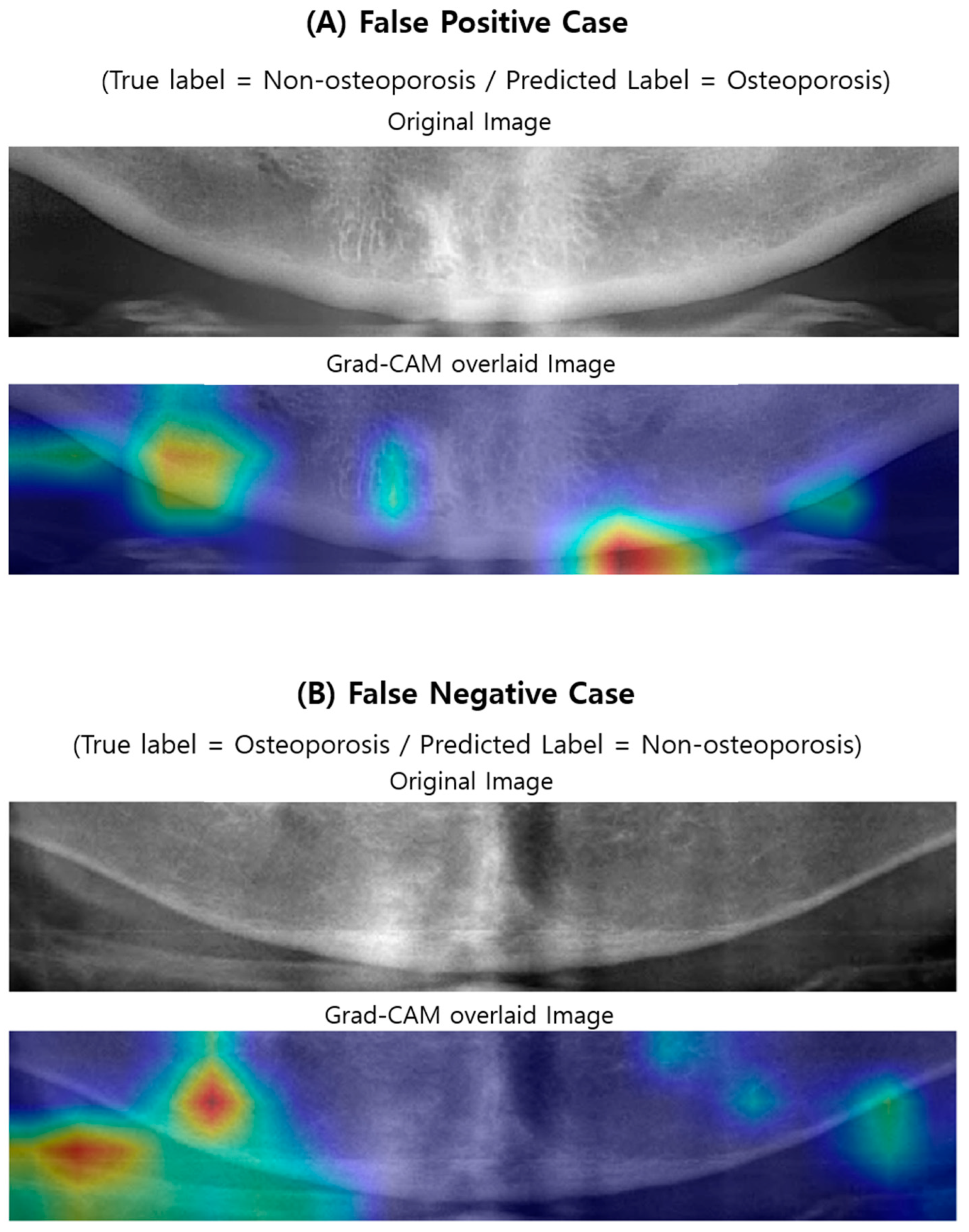
2.6. Visualizing Model Decisions
3. Results
3.1. Baseline Clinical and Demographic Characteristics of the Subjects
3.2. Prediction Performance
3.3. Visualizing Model Decisions
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- NIH Consensus Development Panel on Osteoporosis Prevention, Diagnosis, and Therapy, March 7–29, 2000: Highlights of the conference. South. Med. J. 2001, 94, 569–573.
- Cauley, J.A. Public health impact of osteoporosis. J. Gerontol. A Biol. Sci. Med. Sci. 2013, 68, 1243–1251. [Google Scholar] [CrossRef] [Green Version]
- Bliuc, D.; Nguyen, N.D.; Nguyen, T.V.; Eisman, J.A.; Center, J.R. Compound risk of high mortality following osteoporotic fracture and refracture in elderly women and men. J. Bone Miner. Res. 2013, 28, 2317–2324. [Google Scholar] [CrossRef] [PubMed]
- Sozen, T.; Ozisik, L.; Basaran, N.C. An overview and management of osteoporosis. Eur. J. Rheumatol. 2017, 4, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Melton, L.J., 3rd; Chrischilles, E.A.; Cooper, C.; Lane, A.W.; Riggs, B.L. Perspective. How many women have osteoporosis? J. Bone Miner. Res. 1992, 7, 1005–1010. [Google Scholar] [CrossRef]
- Melton, L.J., 3rd; Atkinson, E.J.; O’Connor, M.K.; O’Fallon, W.M.; Riggs, B.L. Bone density and fracture risk in men. J. Bone Miner. Res. 1998, 13, 1915–1923. [Google Scholar] [CrossRef]
- Kanis, J.A.; Johnell, O.; Oden, A.; Sembo, I.; Redlund-Johnell, I.; Dawson, A.; De Laet, C.; Jonsson, B. Long-term risk of osteoporotic fracture in Malmo. Osteoporos. Int. 2000, 11, 669–674. [Google Scholar] [CrossRef]
- Kalinowski, P.; Rozylo-Kalinowska, I.; Piskorz, M.; Bojakowska-Komsta, U. Correlations between periodontal disease, mandibular inferior cortex index and the osteoporotic fracture probability assessed by means of the fracture risk assessment body mass index tool. BMC Med. Imaging 2019, 19, 41. [Google Scholar] [CrossRef]
- Marcucci, G.; Brandi, M.L. Rare causes of osteoporosis. Clin. Cases Miner. Bone Metab. 2015, 12, 151–156. [Google Scholar] [CrossRef]
- Kanis, J.A.; Johnell, O. Requirements for DXA for the management of osteoporosis in Europe. Osteoporos. Int. 2005, 16, 229–238. [Google Scholar] [CrossRef]
- Kanis, J.A. Diagnosis of osteoporosis and assessment of fracture risk. Lancet 2002, 359, 1929–1936. [Google Scholar] [CrossRef]
- Mithal, A.; Bansal, B.; Kyer, C.S.; Ebeling, P. The Asia-Pacific Regional Audit-Epidemiology, Costs, and Burden of Osteoporosis in India 2013: A report of International Osteoporosis Foundation. Indian J. Endocrinol. Metab. 2014, 18, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, A.; Suei, Y.; Ohtsuka, M.; Otani, K.; Tanimoto, K.; Ohtaki, M. Usefulness of panoramic radiography in the diagnosis of postmenopausal osteoporosis in women. Width and morphology of inferior cortex of the mandible. Dentomaxillofac. Radiol. 1996, 25, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Ledgerton, D.; Horner, K.; Devlin, H.; Worthington, H. Radiomorphometric indices of the mandible in a British female population. Dentomaxillofac. Radiol. 1999, 28, 173–181. [Google Scholar] [CrossRef]
- White, S.C.; Taguchi, A.; Kao, D.; Wu, S.; Service, S.K.; Yoon, D.; Suei, Y.; Nakamoto, T.; Tanimoto, K. Clinical and panoramic predictors of femur bone mineral density. Osteoporos. Int. 2005, 16, 339–346. [Google Scholar] [CrossRef]
- Yasar, F.; Akgunlu, F. The differences in panoramic mandibular indices and fractal dimension between patients with and without spinal osteoporosis. Dentomaxillofac. Radiol. 2006, 35, 1–9. [Google Scholar] [CrossRef]
- Taguchi, A.; Ohtsuka, M.; Tsuda, M.; Nakamoto, T.; Kodama, I.; Inagaki, K.; Noguchi, T.; Kudo, Y.; Suei, Y.; Tanimoto, K. Risk of vertebral osteoporosis in post-menopausal women with alterations of the mandible. Dentomaxillofac. Radiol. 2007, 36, 143–148. [Google Scholar] [CrossRef]
- Devlin, H.; Karayianni, K.; Mitsea, A.; Jacobs, R.; Lindh, C.; van der Stelt, P.; Marjanovic, E.; Adams, J.; Pavitt, S.; Horner, K. Diagnosing osteoporosis by using dental panoramic radiographs: The OSTEODENT project. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2007, 104, 821–828. [Google Scholar] [CrossRef]
- Okabe, S.; Morimoto, Y.; Ansai, T.; Yoshioka, I.; Tanaka, T.; Taguchi, A.; Kito, S.; Wakasugi-Sato, N.; Oda, M.; Kuroiwa, H.; et al. Assessment of the relationship between the mandibular cortex on panoramic radiographs and the risk of bone fracture and vascular disease in 80-year-olds. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2008, 106, 433–442. [Google Scholar] [CrossRef]
- Taguchi, A. Triage screening for osteoporosis in dental clinics using panoramic radiographs. Oral Dis. 2010, 16, 316–327. [Google Scholar] [CrossRef]
- Al-Dam, A.; Blake, F.; Atac, A.; Amling, M.; Blessmann, M.; Assaf, A.; Hanken, H.; Smeets, R.; Heiland, M. Mandibular cortical shape index in non-standardised panoramic radiographs for identifying patients with osteoporosis as defined by the German Osteology Organization. J. Craniomaxillofac. Surg. 2013, 41, e165–e169. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, M.S.; Asano, A.; Taguchi, A.; Kurita, T.; Sanada, M. Diagnosis of osteoporosis from dental panoramic radiographs using the support vector machine method in a computer-aided system. BMC Med. Imaging 2012, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, M.S.; Ganesh Kumar, P.; Park, S.Y.; Huh, K.H.; Heo, M.S.; Kurita, T.; Asano, A.; An, S.Y.; Chien, S.I. Automatic detection of osteoporosis based on hybrid genetic swarm fuzzy classifier approaches. Dentomaxillofac. Radiol. 2016, 45, 20160076. [Google Scholar] [CrossRef] [Green Version]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.; van Ginneken, B.; Sanchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, C.; Took, C.C.; Seong, J.K. Machine learning in biomedical engineering. Biomed. Eng. Lett. 2018, 8, 1–3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Baker, B.; Gupta, O.; Naik, N.; Raskar, R. Designing neural network architectures using reinforcement learning. arXiv 2016, arXiv:1611.02167. [Google Scholar]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? arXiv 2014, arXiv:1411.1792. [Google Scholar]
- Pan, S.J.; Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2009, 22, 1345–1359. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems, Lake Tahoe, Nevada, 3–8 December 2012; Curran Associates Inc.: Red Hook, NY, USA, 2012; Volume 1, pp. 1097–1105. [Google Scholar]
- Han, Z.; Wei, B.; Zheng, Y.; Yin, Y.; Li, K.; Li, S. Breast cancer multi-classification from histopathological images with structured deep learning model. Sci. Rep. 2017, 7, 4172. [Google Scholar] [CrossRef] [PubMed]
- Christopher, M.; Belghith, A.; Bowd, C.; Proudfoot, J.A.; Goldbaum, M.H.; Weinreb, R.N.; Girkin, C.A.; Liebmann, J.M.; Zangwill, L.M. Performance of Deep Learning Architectures and Transfer Learning for Detecting Glaucomatous Optic Neuropathy in Fundus Photographs. Sci. Rep. 2018, 8, 16685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, H.-C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravishankar, H.; Sudhakar, P.; Venkataramani, R.; Thiruvenkadam, S.; Annangi, P.; Babu, N.; Vaidya, V. Understanding the mechanisms of deep transfer learning for medical images. arXiv 2017, arXiv:1704.06040. [Google Scholar]
- Kanis, J.A. Assessment of fracture risk and its application to screening for postmenopausal osteoporosis: Synopsis of a WHO report. WHO Study Group. Osteoporos. Int. 1994, 4, 368–381. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M. Imagenet large scale visual recognition challenge. Int. J. Compute. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef] [Green Version]
- Stone, M. Cross-validatory choice and assessment of statistical predictions. J. R. Stat. Soc. Ser. B Methodol. 1974, 36, 111–133. [Google Scholar] [CrossRef]
- Chollet, F. Keras: Deep Learning Library for Theano and Tensorflow. 2015, 7, p. T1. Available online: https://keras.io (accessed on 30 January 2020).
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv 2016, arXiv:1603.04467. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual explanations from deep networks via gradient-based localization. arXiv 2016, arXiv:1610.02391. [Google Scholar]
- Sawagashira, T.; Hayashi, T.; Hara, T.; Katsumata, A.; Muramatsu, C.; Zhou, X.; Iida, Y.; Katagi, K.; Fujita, H. An automatic detection method for carotid artery calcifications using top-hat filter on dental panoramic radiographs. IEICE Trans. Inf. Syst. 2013, 96, 1878–1881. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-S.; Adhikari, S.; Liu, L.; Jeong, H.-G.; Kim, H.; Yoon, S.-J. Osteoporosis detection in panoramic radiographs using a deep convolutional neural network-based computer-assisted diagnosis system: A preliminary study. Dentomaxillofac. Radiol. 2019, 48, 20170344. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, K.; Penatti, O.A.; Dos Santos, J.A. Towards better exploiting convolutional neural networks for remote sensing scene classification. Pattern Recognit. 2017, 61, 539–556. [Google Scholar] [CrossRef] [Green Version]





| Parameter | Without Osteoporosis (T-Score ≥ −2.5) | With Osteoporosis (T-Score < −2.5) | Total |
|---|---|---|---|
| Number of patients | 380 | 300 | 680 |
| Number of female/male | 332/48 | 233/67 | 565/115 |
| Mean age (±SD) | 58.5 (±11.8) | 68.4 (±8.4) | 63.0 (±11.6) |
| Model | AUC (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | Accuracy (95% CI) |
|---|---|---|---|---|
| CNN3 | 0.667 (±0.041) | 0.684 (±0.204) | 0.649 (±0.164) | 0.660 (±0.066) |
| VGG16 | 0.742 (±0.018) | 0.674 (±0.048) | 0.811 (±0.034) | 0.771 (±0.018) |
| VGG16-TR | 0.782 (±0.006) | 0.737 (±0.046) | 0.828 (±0.052) | 0.802 (±0.024) |
| VGG16-TR-TF | 0.858 (±0.008) | 0.900 (±0.019) | 0.815 (±0.032) | 0.840 (±0.018) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, K.-S.; Jung, S.-K.; Ryu, J.-J.; Shin, S.-W.; Choi, J. Evaluation of Transfer Learning with Deep Convolutional Neural Networks for Screening Osteoporosis in Dental Panoramic Radiographs. J. Clin. Med. 2020, 9, 392. https://doi.org/10.3390/jcm9020392
Lee K-S, Jung S-K, Ryu J-J, Shin S-W, Choi J. Evaluation of Transfer Learning with Deep Convolutional Neural Networks for Screening Osteoporosis in Dental Panoramic Radiographs. Journal of Clinical Medicine. 2020; 9(2):392. https://doi.org/10.3390/jcm9020392
Chicago/Turabian StyleLee, Ki-Sun, Seok-Ki Jung, Jae-Jun Ryu, Sang-Wan Shin, and Jinwook Choi. 2020. "Evaluation of Transfer Learning with Deep Convolutional Neural Networks for Screening Osteoporosis in Dental Panoramic Radiographs" Journal of Clinical Medicine 9, no. 2: 392. https://doi.org/10.3390/jcm9020392
APA StyleLee, K.-S., Jung, S.-K., Ryu, J.-J., Shin, S.-W., & Choi, J. (2020). Evaluation of Transfer Learning with Deep Convolutional Neural Networks for Screening Osteoporosis in Dental Panoramic Radiographs. Journal of Clinical Medicine, 9(2), 392. https://doi.org/10.3390/jcm9020392







