OVOL2-Mediated ZEB1 Downregulation May Prevent Promotion of Actinic Keratosis to Cutaneous Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tissue Samples
2.2. Immunohistochemical Analysis
2.3. Cell Culture
2.4. A Gene-Specific Small Interfering RNA (siRNA) Transfection
2.5. Quantitative Reverse-Transcription Polymerase Chain Reaction (qRT-PCR) Analysis
2.6. Western Blotting
2.7. Matrigel Invasion Chamber Assay
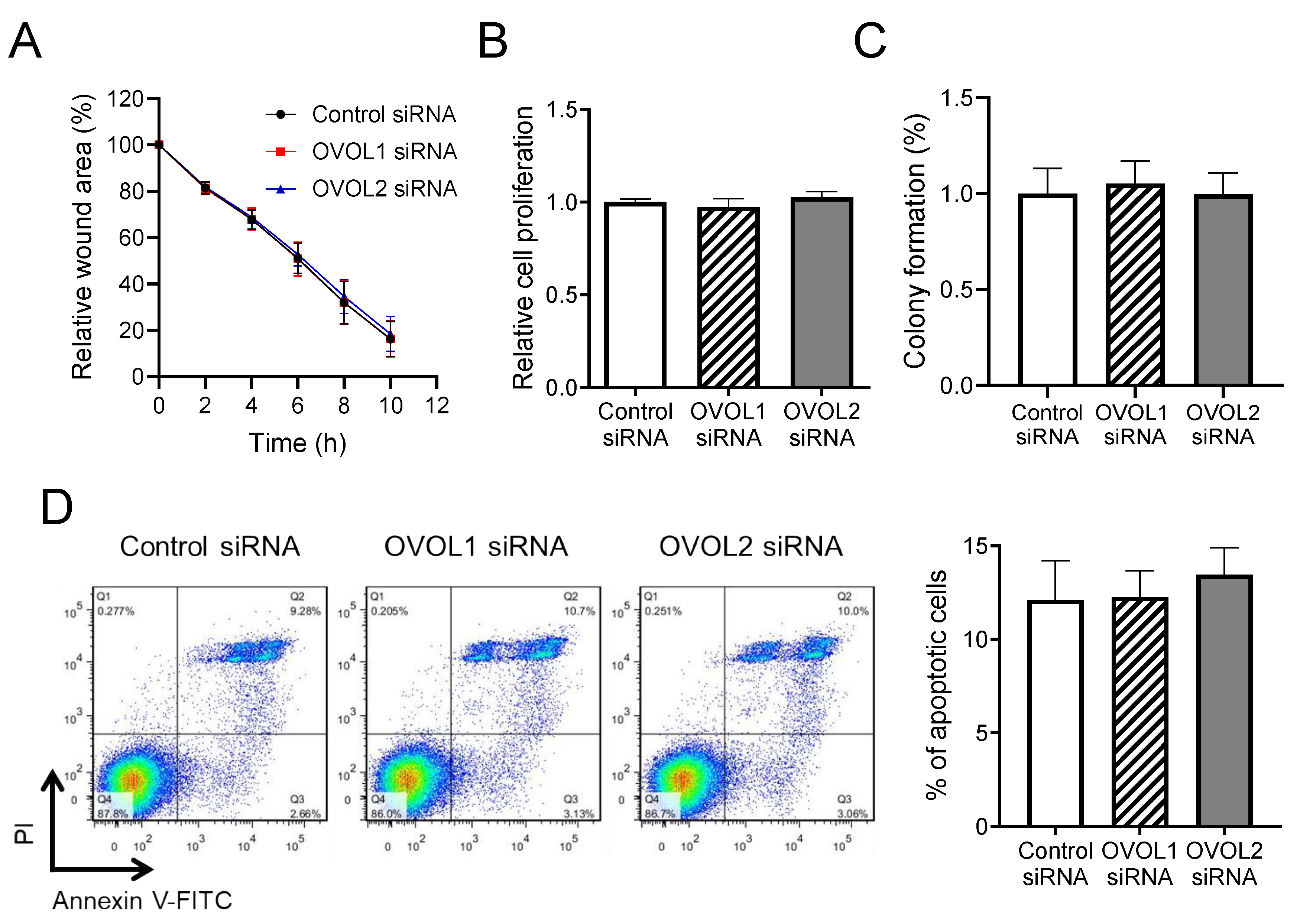
2.8. Wound Healing Assay
2.9. Proliferation Assay
2.10. Colony Formation Assay
2.11. Apoptosis Assay
2.12. Statistical Analysis
3. Results
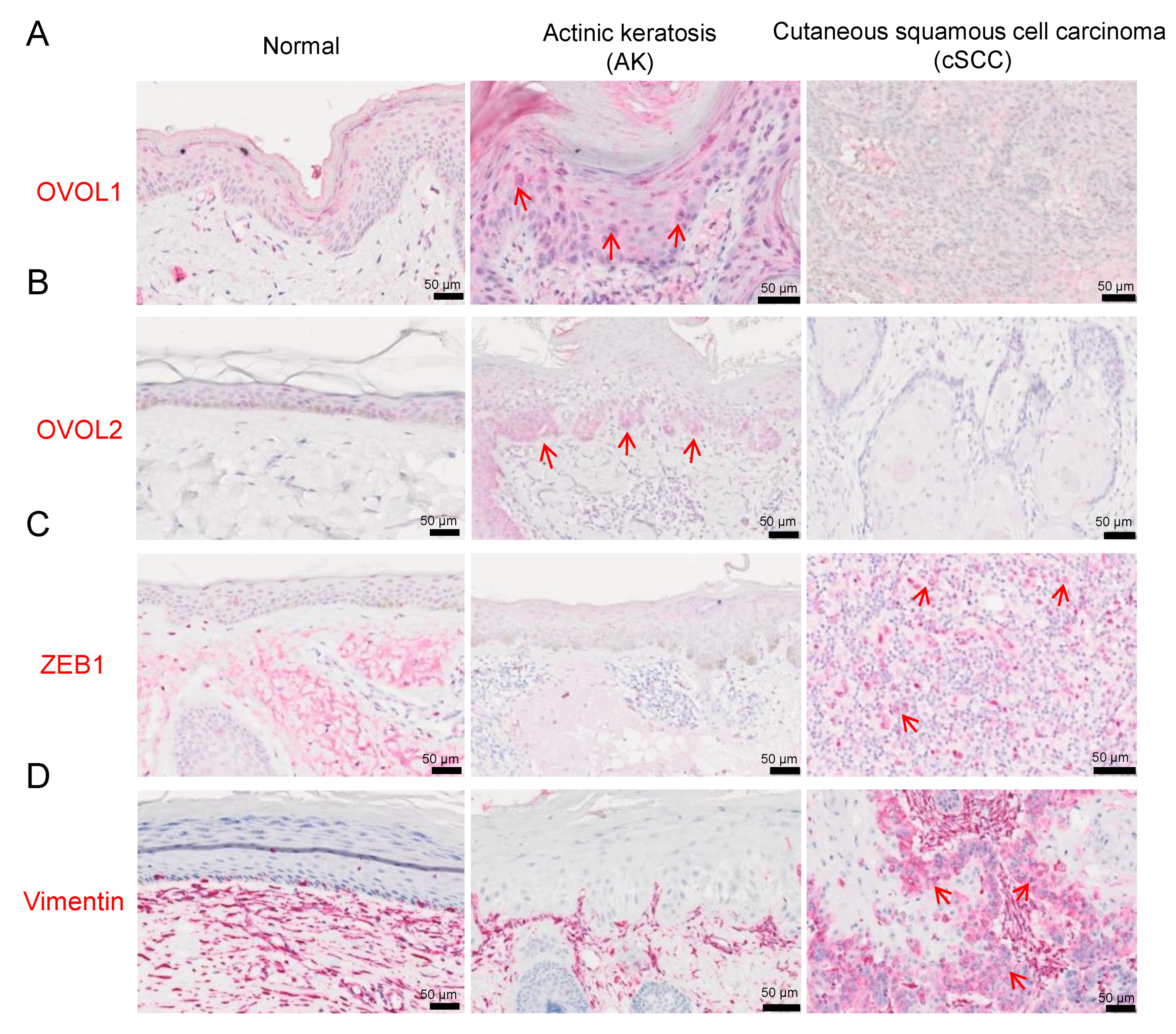
3.1. Immunohistochemistry
3.1.1. OVOL1/2 Expression in AK and cSCC
3.1.2. ZEB1 and Vimentin Expression in AK and cSCC
3.1.3. Associations between OVOL1/2 and ZEB1 or Vimentin in AK and cSCC
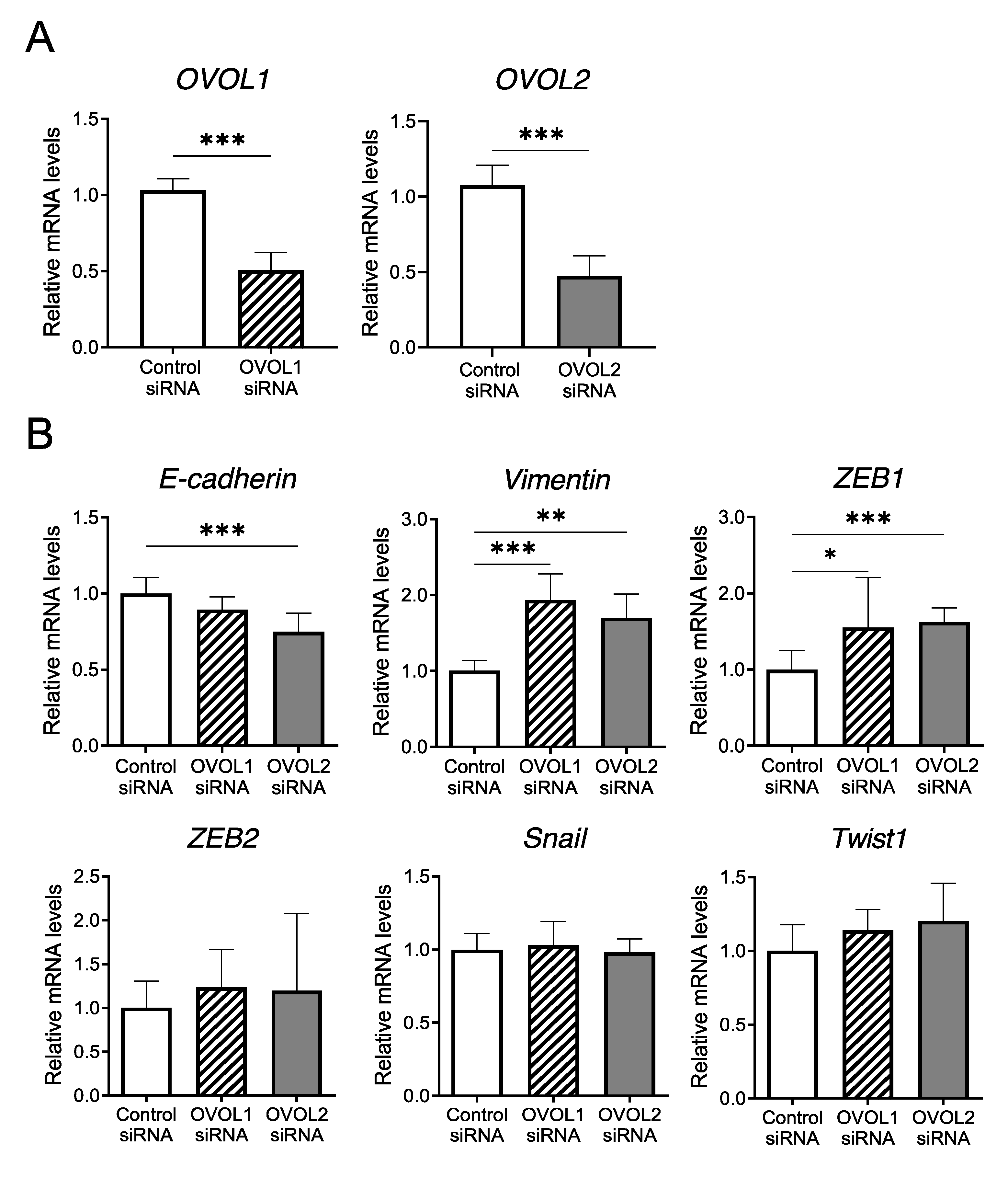
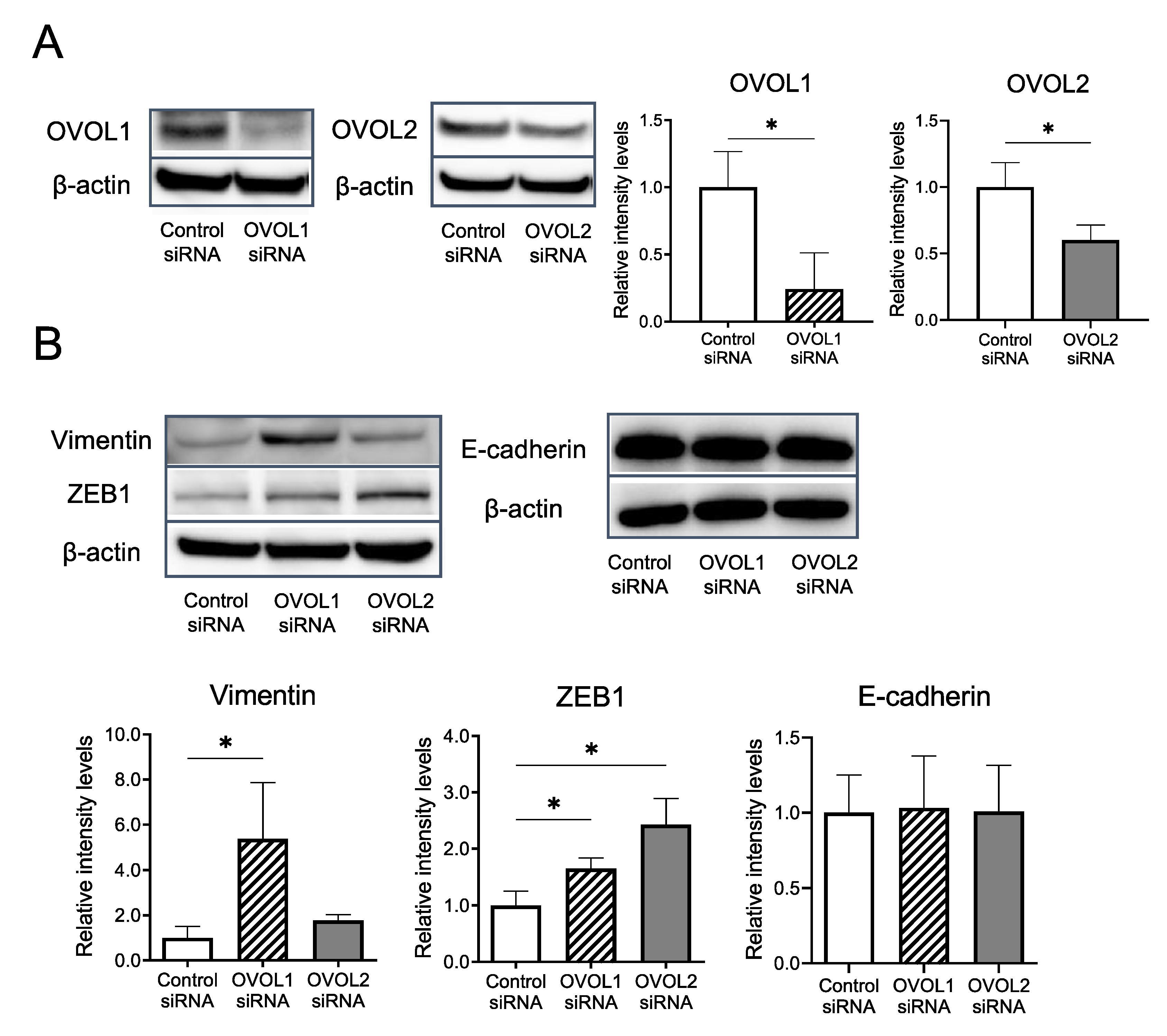
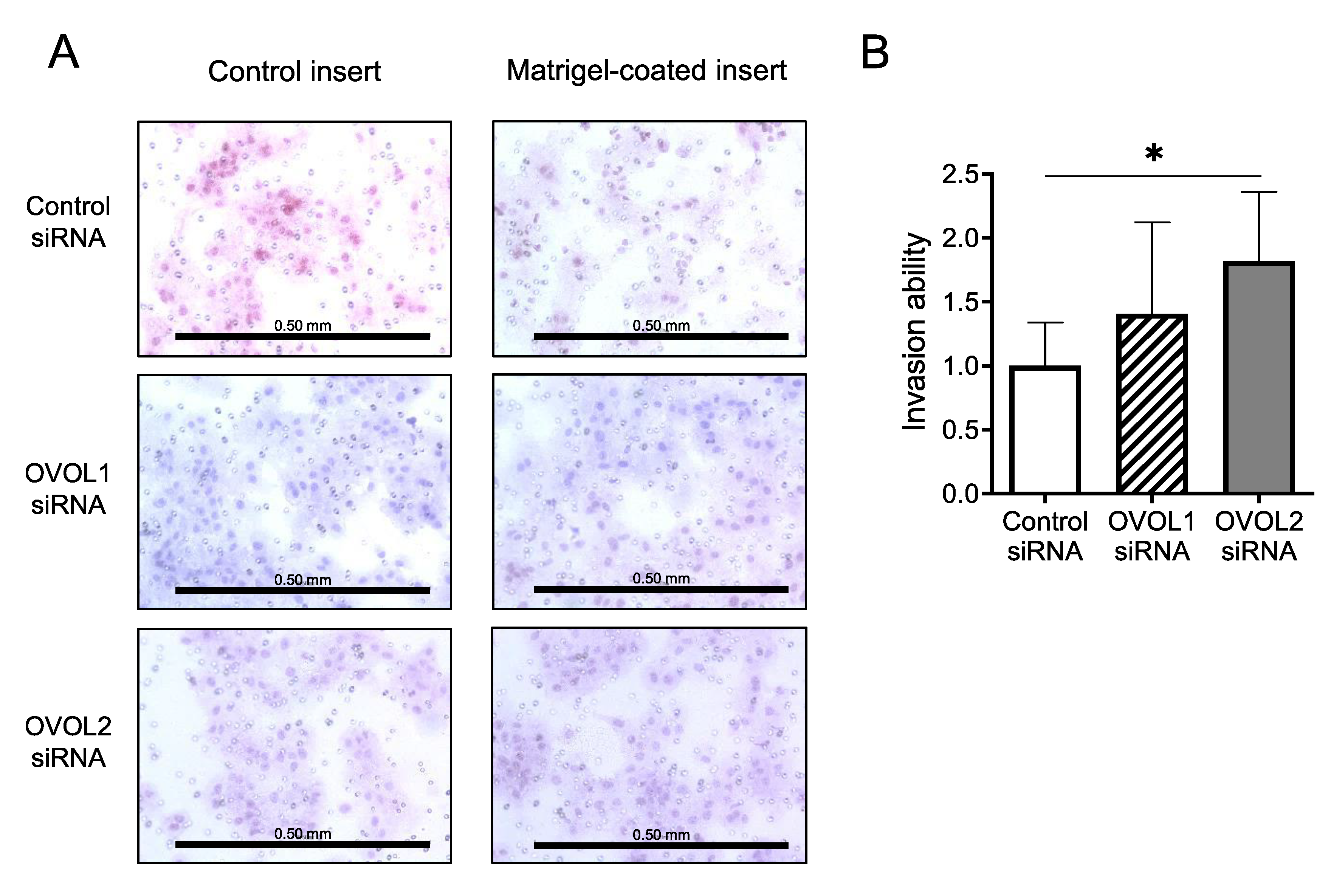
3.2. In Vitro Assays Using an A431 Cell Line
3.2.1. Screening of EMT-Related Factors Influenced by OVOL1/2 Knockdown
3.2.2. Impacts of OVOL1/2 Knockdown on Vimentin, ZEB1, and E-cadherin Protein Levels
3.2.3. Increased Invasion upon OVOL2 Knockdown
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ratushny, V.; Gober, M.D.; Hick, R.; Ridky, T.W.; Seykora, J.T. From keratinocyte to cancer: The pathogenesis and modeling of cutaneous squamous cell carcinoma. J. Clin. Investig. 2012, 122, 464–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karia, P.S.; Han, J.; Schmults, C.D. Cutaneous squamous cell carcinoma: Estimated incidence of disease, nodal metastasis, and deaths from disease in the United States, 2012. J. Am. Acad. Dermatol. 2013, 68, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Criscione, V.D.; Weinstock, M.A.; Naylor, M.F.; Luque, C.; Eide, M.J.; Bingham, S.F. Department of Veteran Affairs Topical Tretinoin Chemoprevention Trial Group. Actinic keratoses: Natural history and risk of malignant transformation in the Veterans Affairs Topical Tretinoin Chemoprevention Trial. Cancer 2009, 115, 2523–2530. [Google Scholar] [CrossRef]
- Werner, R.N.; Stockfleth, E.; Connolly, S.M.; Correia, O.; Erdmann, R.; Foley, P.; Gupta, A.K.; Jacobs, A.; Kerl, H.; Lim, H.W.; et al. Evidence- and consensus-based (S3) guidelines for the treatment of actinic keratosis - International League of Dermatological Societies in cooperation with the European Dermatology Forum-short version. J. Eur. Acad. Dermatol. Venereol. 2015, 29, 2069–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Figueras, M.T. From actinic keratosis to squamous cell carcinoma: Pathophysiology revisited. J. Eur. Acad. Dermatol. Venereol. 2017, 31, 5–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartis, D.; Mise, N.; Mahida, R.Y.; Eickelberg, O.; Thickett, D.R. Epithelial-mesenchymal transition in lung development and disease: Does it exist and is it important? Thorax 2014, 69, 760–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef]
- Thiery, J.P.; Sleeman, J.P. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006, 7, 131–142. [Google Scholar] [CrossRef]
- Acloque, H.; Adams, M.S.; Fishwick, K.; Bronner-Fraser, M.; Nieto, M.A. Epithelial-mesenchymal transitions: The importance of changing cell state in development and disease. J. Clin. Investig. 2009, 119, 1438–1449. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Unternaehrer, J.J. Epithelial-mesenchymal Transition and Cancer Stem Cells: At the Crossroads of Differentiation and Dedifferentiation. Dev. Dyn. 2019, 248, 10–20. [Google Scholar] [CrossRef] [Green Version]
- Saenz-Sardà, X.; Carrato, C.; Pérez-Roca, L.; Puig, L.; Ferrándiz, C.; Ariza, A.; Fernández-Figueras, M.T. Epithelial-to-mesenchymal transition contributes to invasion in squamous cell carcinomas originated from actinic keratosis through the differentiated pathway, whereas proliferation plays a more significant role in the classical pathway. J. Eur. Acad. Dermatol. Venereol. 2018, 32, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Moustakas, A.; Heldin, C.H. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci. 2007, 98, 1512–1520. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Sun, Y.; Ma, L. ZEB1: At the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle. 2015, 14, 481–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Wu, Q.; Wang, Y.; Wei, Y.; Wu, H.; Duan, L.; Zhang, Q.; Wu, Y. Ovol2 induces mesenchymal-epithelial transition via targeting ZEB1 in osteosarcoma. Onco Targets Ther. 2018, 11, 2963–2973. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Zheng, X.; Zhou, J.; Zhang, Y.; Chen, K. ZEB1 promotes the progression and metastasis of cervical squamous cell carcinoma via the promotion of epithelial-mesenchymal transition. Int. J. Clin. Exp. Pathol. 2015, 8, 11258–11267. [Google Scholar]
- Roca, H.; Hernandez, J.; Weidner, S.; McEachin, R.C.; Fuller, D.; Sud, S.; Schumann, T.; Wilkinson, J.E.; Zaslavsky, A.; Li, H.; et al. Transcription factors OVOL1 and OVOL2 induce the mesenchymal to epithelial transition in human cancer. PLoS ONE 2013, 8, e76773. [Google Scholar] [CrossRef]
- Li, S.; Yang, J. Ovol proteins: Guardians against EMT during epithelial differentiation. Dev. Cell 2014, 29, 1–2. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Schonbaum, C.; Degenstein, L.; Bai, W.; Mahowald, A.; Fuchs, E. The ovo gene required for cuticle formation and oogenesis in flies is involved in hair formation and spermatogenesis in mice. Genes Dev. 1998, 12, 3452–3463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, T.; Tsuji, G.; Ohno, F.; Uchi, H.; Nakahara, T.; Hashimoto-Hachiya, A.; Yoshida, Y.; Yamamoto, O.; Oda, Y.; Furue, M. Activation of the OVOL1-OVOL2 Axis in the Hair Bulb and in Pilomatricoma. Am. J. Pathol. 2016, 186, 1036–1043. [Google Scholar] [CrossRef] [Green Version]
- Nair, M.; Teng, A.; Bilanchone, V.; Agrawal, A.; Li, B.; Dai, X. Ovol1 regulates the growth arrest of embryonic epidermal progenitor cells and represses c-myc transcription. J. Cell Biol. 2006, 173, 253–264. [Google Scholar] [CrossRef]
- Johnson, A.D.; Fitzsimmons, D.; Hagman, J.; Chamberlin, H.M. EGL-38 Pax regulates the ovo-related gene lin-48 during Caenorhabditis elegans organ development. Development 2001, 128, 2857–2865. [Google Scholar] [PubMed]
- Wang, Z.H.; Li, Z.; Hu, M.; Yang, Q.J.; Yan, S.; Wu, R.S.; Li, BA.; Guo, M. Ovol2 gene inhibits the Epithelial-to-Mesenchymal Transition in lung adenocarcinoma by transcriptionally repressing Twist1. Gene 2017, 600, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.S.; Hong, J.J.; Wu, J.F.; Yan, S.; Wu, D.; Liu, N.; Liu, Q.F.; Wu, Q.W.; Xie, Y.Y.; Liu, Y.J.; et al. OVOL2 antagonizes TGF-β signaling to regulate epithelial to mesenchymal transition during mammary tumor metastasis. Oncotarget 2017, 8, 39401–39416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuji, G.; Hashimoto-Hachiya, A.; Kiyomatsu-Oda, M.; Takemura, M.; Ohno, F.; Ito, T.; Morino-Koga, S.; Mitoma, C.; Nakahara, T.; Uchi, H.; et al. Aryl hydrocarbon receptor activation restores filaggrin expression via OVOL1 in atopic dermatitis. Cell Death Dis. 2017, 8, e2931. [Google Scholar] [CrossRef] [Green Version]
- Ito, T.; Tsuji, G.; Ohno, F.; Nakahara, T.; Uchi, H.; Furue, M. Potential role of the OVOL1-OVOL2 axis and c-Myc in the progression of cutaneous squamous cell carcinoma. Mod. Pathol. 2017, 30, 919–927. [Google Scholar] [CrossRef]
- Toll, A.; Masferrer, E.; Hernández-Ruiz, M.E.; Ferrandiz-Pulido, C.; Yébenes, M.; Jaka, A.; Tuneu, A.; Jucglà, A.; Gimeno, J.; Baró, T.; et al. Epithelial to mesenchymal transition markers are associated with an increased metastatic risk in primary cutaneous squamous cell carcinomas but are attenuated in lymph node metastases. J. Dermatol. Sci. 2013, 72, 93–102. [Google Scholar] [CrossRef]
- Jia, D.; Jolly, M.K.; Boareto, M.; Parsana, P.; Mooney, S.M.; Pienta, K.J.; Levine, H.; Ben-Jacob, E. OVOL guides the epithelial-hybrid-mesenchymal transition. Oncotarget 2015, 6, 15436–15448. [Google Scholar] [CrossRef] [Green Version]
- Fu, H.; Qi, L.; Chen, L.; He, Y.; Zhang, N.; Guo, H. Expression of Ovol2 is related to epithelial characteristics and shows a favorable clinical outcome in hepatocellular carcinoma. Onco Targets Ther. 2016, 9, 5963–5973. [Google Scholar]
- Haensel, D.; Sun, P.; MacLean, A.L.; Ma, X.; Zhou, Y.; Stemmler, M.P.; Brabletz, S.; Berx, G.; Plikus, M.V.; Nie, Q.; et al. An Ovol2-Zeb1 transcriptional circuit regulates epithelial directional migration and proliferation. EMBO Rep. 2018, 20, e46273. [Google Scholar] [CrossRef]
- Lo, H.C.; Zhang, X.H. EMT in Metastasis: Finding the Right Balance. Dev. Cell 2018, 45, 663–665. [Google Scholar] [CrossRef] [Green Version]

| Total | OVOL1 | p-Value | ||
|---|---|---|---|---|
| High | Low | |||
| ZEB1 | 0.245 | |||
| Positive | 19 | 10 | 9 | |
| Negative | 41 | 29 | 12 | |
| Total | OVOL2 | p-value | ||
| High | Low | |||
| ZEB1 | 0.0105* | |||
| Positive | 19 | 7 | 12 | |
| Negative | 41 | 30 | 11 | |
| Total | OVOL1 | p-Value | ||
|---|---|---|---|---|
| High | Low | |||
| Vimentin | 0.352 | |||
| Positive | 15 | 8 | 7 | |
| Negative | 45 | 31 | 14 | |
| Total | OVOL2 | p-value | ||
| High | Low | |||
| Vimentin | 0.543 | |||
| Positive | 15 | 8 | 7 | |
| Negative | 45 | 29 | 16 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murata, M.; Ito, T.; Tanaka, Y.; Yamamura, K.; Furue, K.; Furue, M. OVOL2-Mediated ZEB1 Downregulation May Prevent Promotion of Actinic Keratosis to Cutaneous Squamous Cell Carcinoma. J. Clin. Med. 2020, 9, 618. https://doi.org/10.3390/jcm9030618
Murata M, Ito T, Tanaka Y, Yamamura K, Furue K, Furue M. OVOL2-Mediated ZEB1 Downregulation May Prevent Promotion of Actinic Keratosis to Cutaneous Squamous Cell Carcinoma. Journal of Clinical Medicine. 2020; 9(3):618. https://doi.org/10.3390/jcm9030618
Chicago/Turabian StyleMurata, Maho, Takamichi Ito, Yuka Tanaka, Kazuhiko Yamamura, Kazuhisa Furue, and Masutaka Furue. 2020. "OVOL2-Mediated ZEB1 Downregulation May Prevent Promotion of Actinic Keratosis to Cutaneous Squamous Cell Carcinoma" Journal of Clinical Medicine 9, no. 3: 618. https://doi.org/10.3390/jcm9030618





