Association of SNP–SNP Interactions of Surfactant Protein Genes with Pediatric Acute Respiratory Failure
Abstract
1. Introduction
2. Subjects and Methods
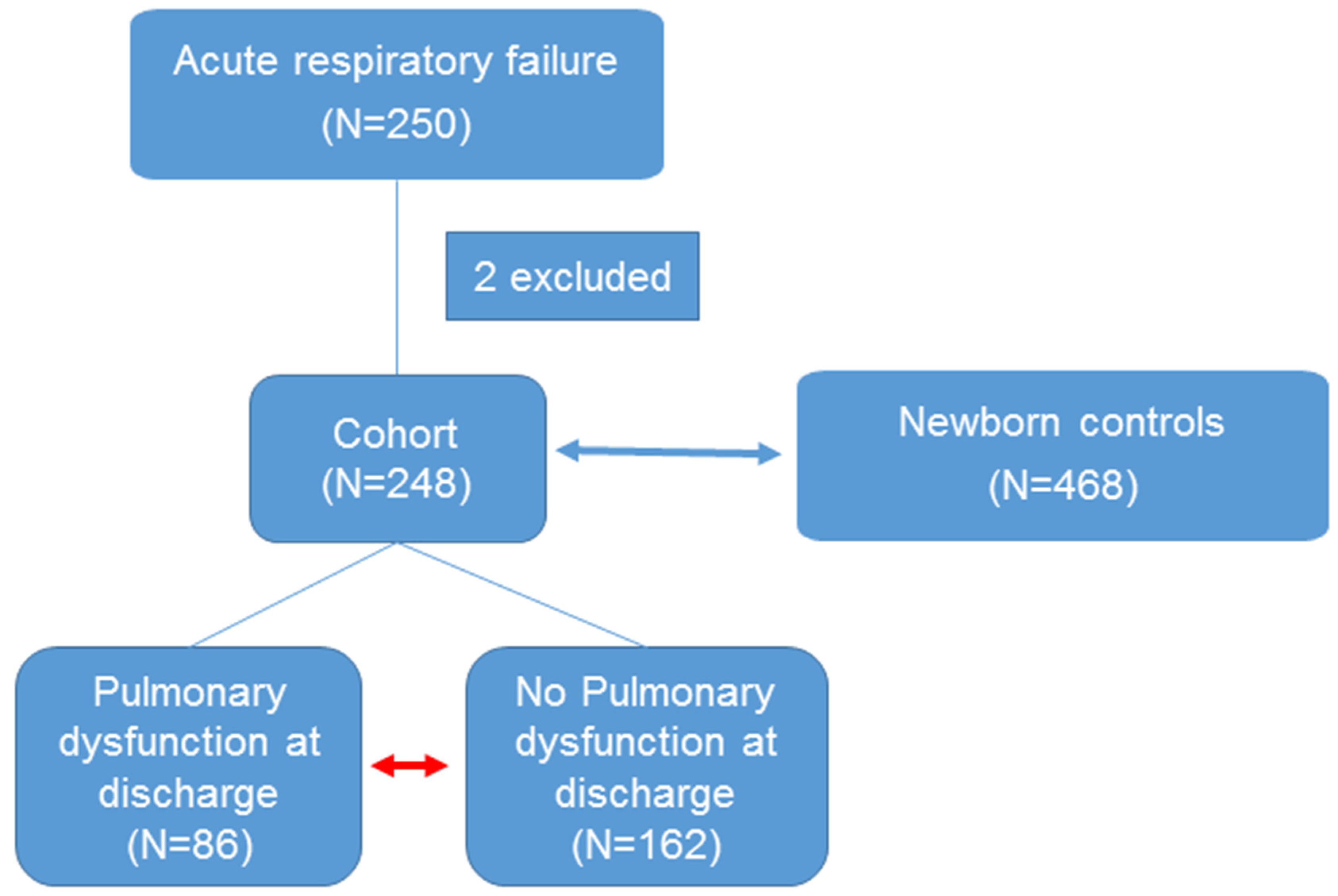
2.1. Study Population
2.2. DNA Isolation
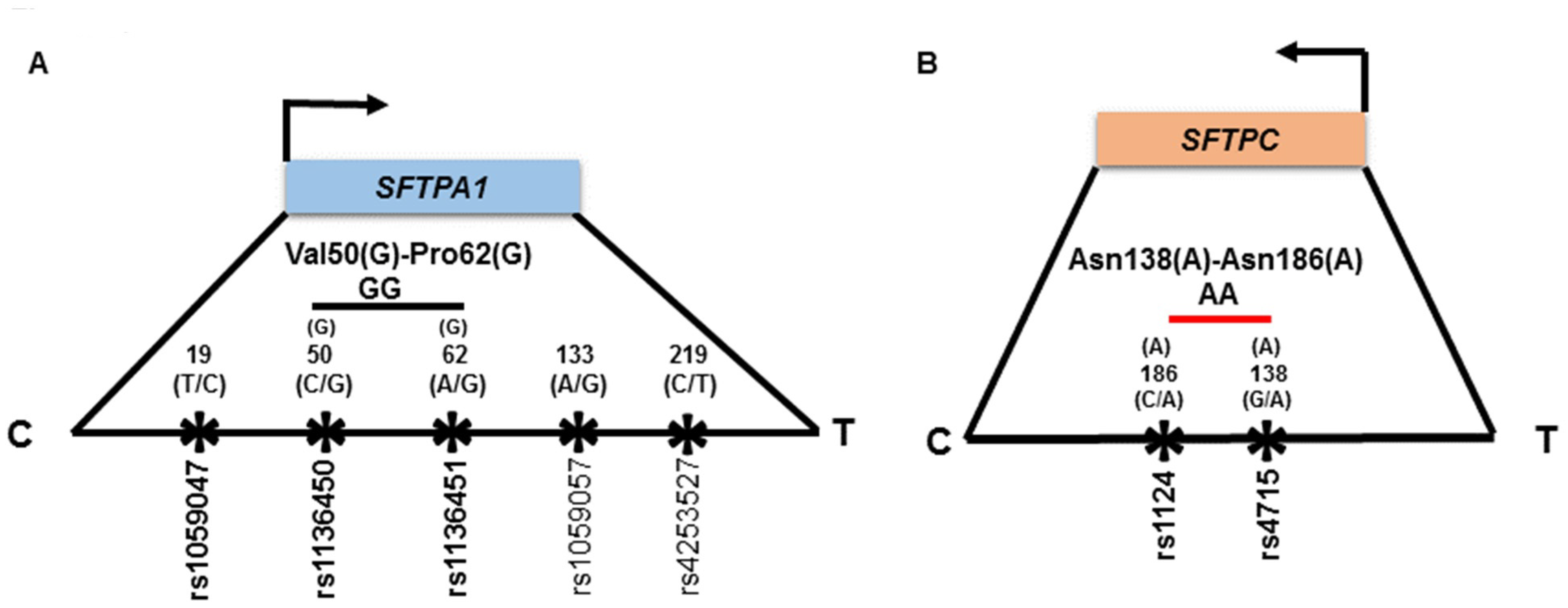
2.3. Genetic Variations Selection for this Study
2.4. Genotype Analysis
2.5. Statistical Analysis
3. Results
3.1. Clinical Characteristics of ARF Study Group
3.2. SNP–SNP Interaction
3.3. Association of SP Genes SNPs with ARF Compared to Newborn Controls after Adjusting for Sex and Ethnicity
3.4. Haplotype Association with ARF Risk
3.5. Association of SP Genes with the PDAD Subgroup Compared to no PDAD after Individual Adjustments of Variables
3.5.1. Age
3.5.2. Positive Bacterial Culture
3.5.3. Ventilator Days
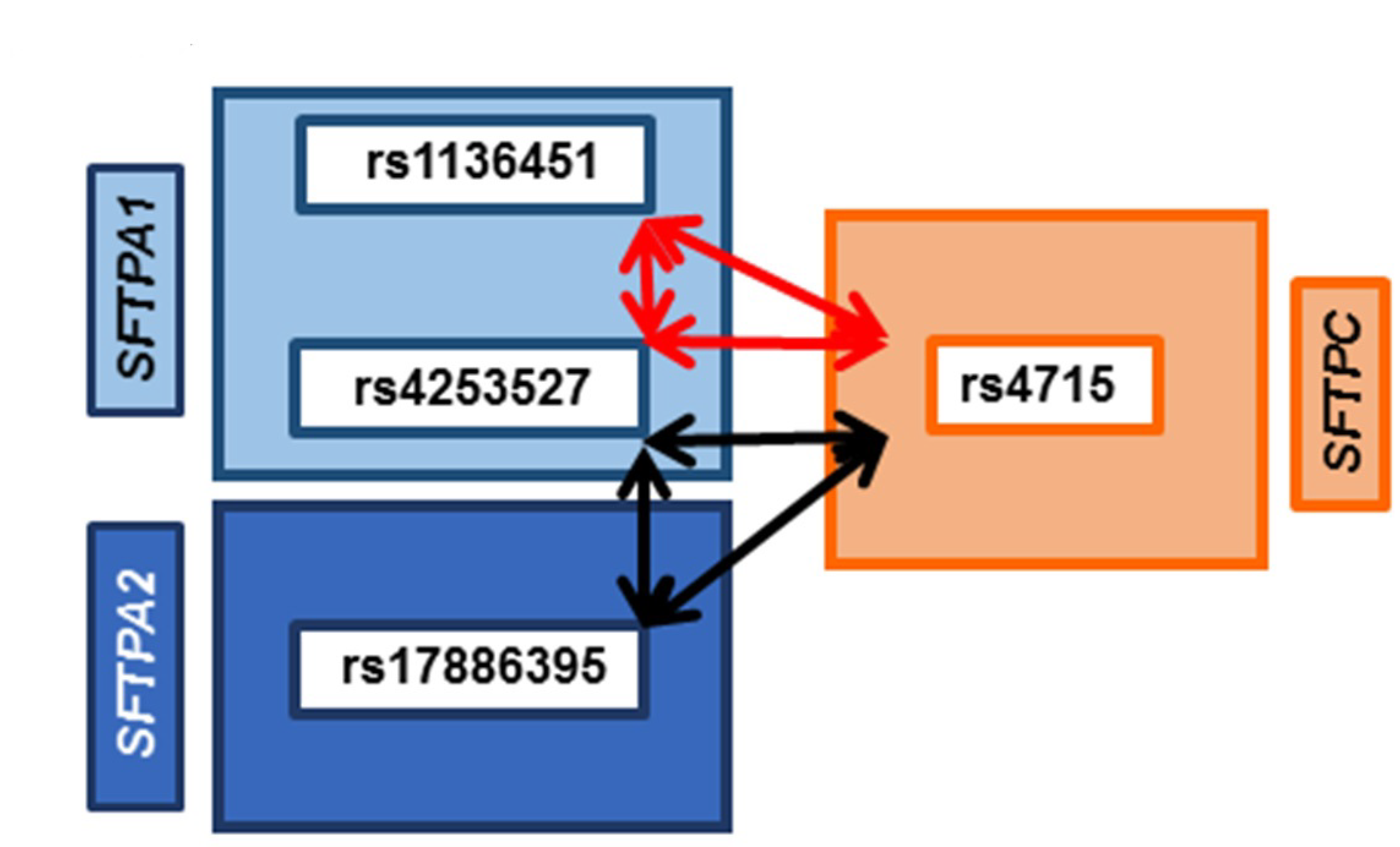
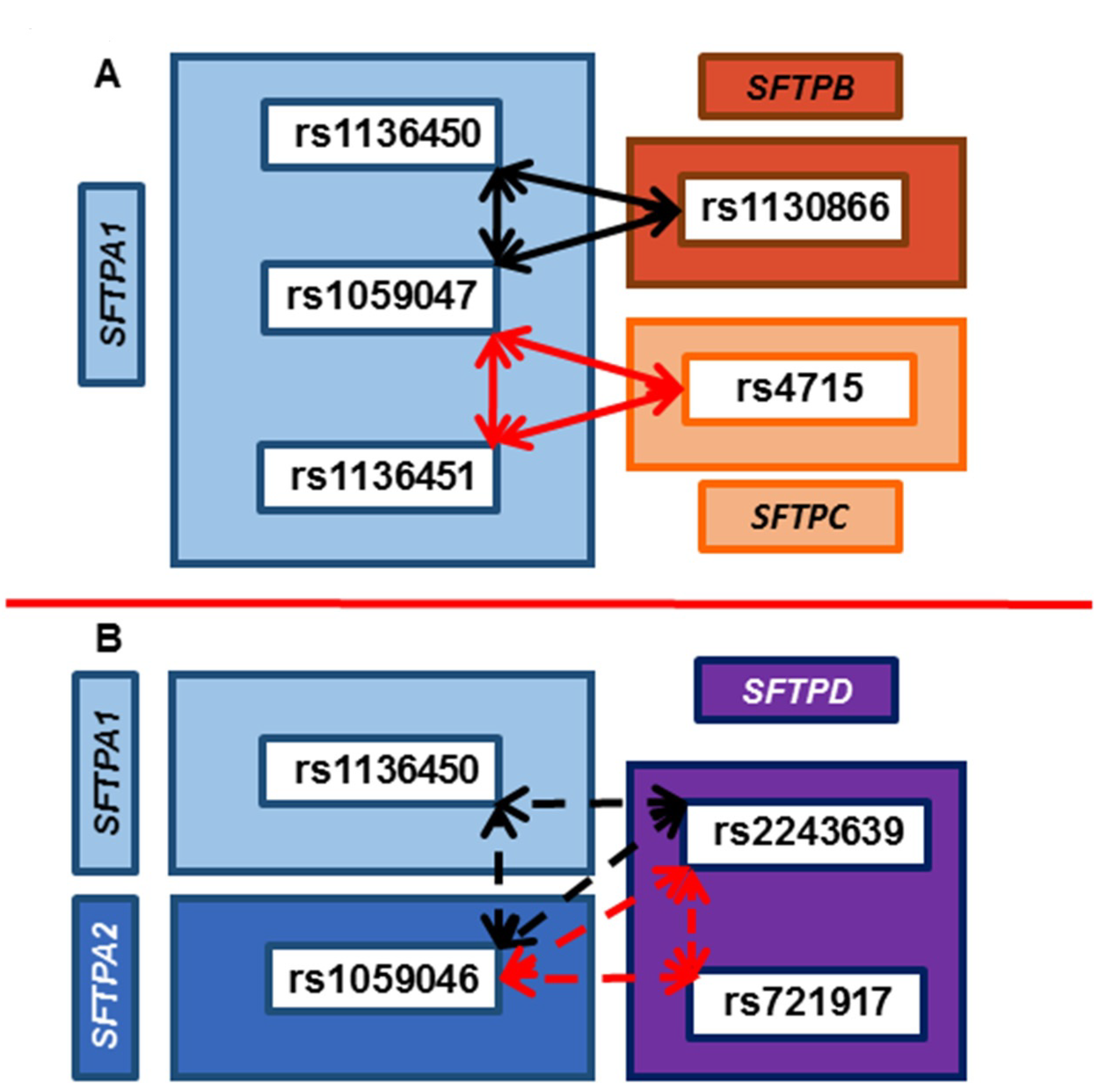
3.6. Common SNP–SNP Interactions
3.6.1. Association with ARF Compared to Controls and PDAD Compared to No PDAD
3.6.2. Associations with PDAD Compared to No PDAD after Adjusting for Age, Positive Bacterial Culture, and Ventilator Days
4. Discussion
4.1. Association of SFTPC with ARF in a Single-SNP Model
4.2. Association of Interactions of SNPs of SP Genes with ARF and PDAD
4.3. Association of SNPs of SFTPA2 with ARF
4.4. Association of SNPs of SFTPA1 with PDAD
4.5. Association of SFTPD with Decreased Risk of PDAD
4.6. Interactions of Hydrophobic SPs (SFTPB and SFTPC) with ARF and PDAD
4.7. Haplotype Association with ARF Risk
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vo, P.; Kharasch, V.S. Respiratory Failure. Pediatr. Rev. 2014, 35, 476–486. [Google Scholar] [CrossRef][Green Version]
- Hammer, J. Acute respiratory failure in children. Paediatr. Respir. Rev. 2013, 14, 64–69. [Google Scholar] [CrossRef]
- Hammer, J.; Eber, E. The peculiarities of infant respiratory physiology. In Paediatric Pulmonary Function Testing; Karger Publishers: Basel, Switzerland, 2005; Volume 33, pp. 2–7. [Google Scholar]
- Mathers, C.D.; Boerma, T.; Ma Fat, D. Global and regional causes of death. Br. Med. Bull. 2009, 92, 7–32. [Google Scholar] [CrossRef]
- Pollack, M.M.; Holubkov, R.; Glass, P.; Dean, J.M.; Meert, K.L.; Zimmerman, J.; Anand, K.J.S.; Carcillo, J.; Newth, C.J.L.; Harrison, R.; et al. Functional Status Scale: New pediatric outcome measure. Pediatrics 2009, 124, e18–e28. [Google Scholar] [CrossRef]
- Yagiela, L.M.; Barbaro, R.P.; Quasney, M.W.; Pfarr, M.A.; Ursu, D.C.; Prosser, L.A.; Odetola, F.O. Outcomes and Patterns of Healthcare Utilization After Hospitalization for Pediatric Critical Illness Due to Respiratory Failure. Pediatr. Crit. Care Med. 2019, 20, 120–127. [Google Scholar] [CrossRef]
- Serrano, A.G.; Perez-Gil, J. Protein-lipid interactions and surface activity in the pulmonary surfactant system. Chem. Phys. Lipids 2006, 141, 105–118. [Google Scholar] [CrossRef]
- Wright, J.R. Immunoregulatory functions of surfactant proteins. Nat. Rev. Immunol. 2005, 5, 58–68. [Google Scholar] [CrossRef]
- Kishore, U.; Greenhough, T.J.; Waters, P.; Shrive, A.K.; Ghai, R.; Kamran, M.F.; Bernal, A.L.; Reid, K.B.; Madan, T.; Chakraborty, T. Surfactant proteins SP-A and SP-D: Structure, function and receptors. Mol. Immunol. 2006, 43, 1293–1315. [Google Scholar] [CrossRef]
- Thorenoor, N.; Kawasawa, Y.I.; Gandhi, C.K.; Zhang, X.; Floros, J. Differential Impact of Co-expressed SP-A1/SP-A2 Protein on AM miRNome; Sex Differences. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Lopez-Rodriguez, E.; Pascual, A.; Arroyo, R.; Floros, J.; Perez-Gil, J. Human Pulmonary Surfactant Protein SP-A1 Provides Maximal Efficiency of Lung Interfacial Films. Biophys. J. 2016, 111, 524–536. [Google Scholar] [CrossRef]
- DiAngelo, S.; Lin, Z.; Wang, G.; Phillips, S.; Ramet, M.; Luo, J.; Floros, J. Novel, non-radioactive, simple and multiplex PCR-cRFLP methods for genotyping human SP-A and SP-D marker alleles. Dis. Markers 1999, 15, 269–281. [Google Scholar] [CrossRef]
- Wert, S.E.; Whitsett, J.A.; Nogee, L.M. Genetic disorders of surfactant dysfunction. Pediatr. Dev. Pathol. 2009, 12, 253–274. [Google Scholar] [CrossRef]
- Silveyra, P.; Floros, J. Genetic variant associations of human SP-A and SP-D with acute and chronic lung injury. Front. Biosci. (Landmark Ed.) 2012, 17, 407–429. [Google Scholar] [CrossRef]
- Nogee, L.M.; Wert, S.E.; Proffit, S.A.; Hull, W.M.; Whitsett, J.A. Allelic heterogeneity in hereditary surfactant protein B (SP-B) deficiency. Am. J. Respir. Crit. Care Med. 2000, 161, 973–981. [Google Scholar] [CrossRef]
- Ramet, M.; Haataja, R.; Marttila, R.; Floros, J.; Hallman, M. Association between the surfactant protein A (SP-A) gene locus and respiratory-distress syndrome in the Finnish population. Am. J. Hum. Genet. 2000, 66, 1569–1579. [Google Scholar] [CrossRef]
- Kala, P.; Ten Have, T.; Nielsen, H.; Dunn, M.; Floros, J. Association of pulmonary surfactant protein A (SP-A) gene and respiratory distress syndrome: Interaction with SP-B. Pediatr. Res. 1998, 43, 169–177. [Google Scholar] [CrossRef][Green Version]
- Floros, J.; Fan, R.; Diangelo, S.; Guo, X.; Wert, J.; Luo, J. Surfactant protein (SP) B associations and interactions with SP-A in white and black subjects with respiratory distress syndrome. Pediatr. Int. Off. J. Jpn. Pediatr. Soc. 2001, 43, 567–576. [Google Scholar] [CrossRef]
- Lin, Z.; Thorenoor, N.; Wu, R.; DiAngelo, S.L.; Ye, M.; Thomas, N.J.; Liao, X.; Lin, T.R.; Warren, S.; Floros, J. Genetic Association of Pulmonary Surfactant Protein Genes, SFTPA1, SFTPA2, SFTPB, SFTPC, and SFTPD With Cystic Fibrosis. Front. Immunol. 2018, 9, 2256. [Google Scholar] [CrossRef]
- Seifart, C.; Plagens, A.; Brodje, D.; Muller, B.; von Wichert, P.; Floros, J. Surfactant protein B intron 4 variation in German patients with COPD and acute respiratory failure. Dis. Markers 2002, 18, 129–136. [Google Scholar] [CrossRef]
- Lin, Z.; Pearson, C.; Chinchilli, V.; Pietschmann, S.M.; Luo, J.; Pison, U.; Floros, J. Polymorphisms of human SP-A, SP-B, and SP-D genes: Association of SP-B Thr131Ile with ARDS. Clin. Genet. 2000, 58, 181–191. [Google Scholar] [CrossRef]
- Floros, J.; Lin, H.M.; Garcia, A.; Salazar, M.A.; Guo, X.; DiAngelo, S.; Montano, M.; Luo, J.; Pardo, A.; Selman, M. Surfactant protein genetic marker alleles identify a subgroup of tuberculosis in a Mexican population. J. Infect. Dis. 2000, 182, 1473–1478. [Google Scholar] [CrossRef] [PubMed]
- Thomas, N.J.; DiAngelo, S.; Hess, J.C.; Fan, R.; Ball, M.W.; Geskey, J.M.; Willson, D.F.; Floros, J. Transmission of surfactant protein variants and haplotypes in children hospitalized with respiratory syncytial virus. Pediatr. Res. 2009, 66, 70–73. [Google Scholar] [CrossRef] [PubMed]
- Randolph, A.G.; Meert, K.L.; O’neil, M.E.; Hanson, J.H.; Luckett, P.M.; Arnold, J.H.; Gedeit, R.G.; Cox, P.N.; Roberts, J.S.; Venkataraman, S.T. The feasibility of conducting clinical trials in infants and children with acute respiratory failure. Am. J. Respir. Crit. Care Med. 2003, 167, 1334–1340. [Google Scholar] [CrossRef] [PubMed]
- Royall, J.A.; Levin, D.L. Adult respiratory distress syndrome in pediatric patients. I. Clinical aspects, pathophysiology, pathology, and mechanisms of lung injury. J. Pediatr. 1988, 112, 169–180. [Google Scholar] [CrossRef]
- Skelton, R.; Holland, P.; Darowski, M.; Chetcuti, P.; Morgan, L.; Harwood, J. Abnormal surfactant composition and activity in severe bronchiolitis. Acta Paediatr. 1999, 88, 942–946. [Google Scholar] [CrossRef] [PubMed]
- Gregory, T.J.; Longmore, W.J.; Moxley, M.A.; Whitsett, J.A.; Reed, C.R.; Fowler, A.A., 3rd; Hudson, L.D.; Maunder, R.J.; Crim, C.; Hyers, T.M. Surfactant chemical composition and biophysical activity in acute respiratory distress syndrome. J. Clin. Investig. 1991, 88, 1976–1981. [Google Scholar] [CrossRef]
- Cheverud, J.M.; Routman, E.J. Epistasis and its contribution to genetic variance components. Genetics 1995, 139, 1455–1461. [Google Scholar]
- Garcia-Closas, M.; Lubin, J.H. Power and sample size calculations in case-control studies of gene-environment interactions: Comments on different approaches. Am. J. Epidemiol. 1999, 149, 689–692. [Google Scholar] [CrossRef]
- Moore, J.H. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum. Hered. 2003, 56, 73–82. [Google Scholar] [CrossRef]
- Moore, J.H.; Williams, S.M. New strategies for identifying gene-gene interactions in hypertension. Ann. Med. 2002, 34, 88–95. [Google Scholar] [CrossRef]
- Lin, Z.; deMello, D.E.; Batanian, J.R.; Khammash, H.M.; DiAngelo, S.; Luo, J.; Floros, J. Aberrant SP-B mRNA in lung tissue of patients with congenital alveolar proteinosis (CAP). Clin. Genet. 2000, 57, 359–369. [Google Scholar] [CrossRef]
- Selman, M.; Lin, H.M.; Montano, M.; Jenkins, A.L.; Estrada, A.; Lin, Z.; Wang, G.; DiAngelo, S.L.; Guo, X.; Umstead, T.M.; et al. Surfactant protein A and B genetic variants predispose to idiopathic pulmonary fibrosis. Hum. Genet. 2003, 113, 542–550. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, T.; Lin, Z.; Hegarty, J.; Koltun, W.A.; Wu, R. A general model for multilocus epistatic interactions in case-control studies. PLoS ONE 2010, 5, e11384. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Thalamuthu, A.; Liu, J.J.; Chen, C.; Wang, Z.; Wu, R. Asymptotic distribution for epistatic tests in case-control studies. Genomics 2011, 98, 145–151. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hochberg, Y.B.A.Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol. ) 1995, 57, 289–300. [Google Scholar]
- Hope, A.C.A. A Simplified Monte Carlo Significance Test Procedure. J. R. Stat. Soc. Ser. B 1968, 30, 582–598. [Google Scholar] [CrossRef]
- Day, N.E.; Byar, D.P. Testing Hypotheses in Case-Control Studies-Equivalence of Mantel-Haenszel Statistics and Logit Score Tests. Biometrics 1979, 35, 623–630. [Google Scholar] [CrossRef]
- Lin, M.; Aquilante, C.; Johnson, J.A.; Wu, R. Sequencing drug response with HapMap. Pharm. J. 2005, 5, 149–156. [Google Scholar] [CrossRef]
- Lahti, M.; Marttila, R.; Hallman, M. Surfactant protein C gene variation in the Finnish population–association with perinatal respiratory disease. Eur. J. Hum. Genet. 2004, 12, 312–320. [Google Scholar] [CrossRef]
- Glasser, S.W.; Burhans, M.S.; Korfhagen, T.R.; Na, C.-L.; Sly, P.D.; Ross, G.F.; Ikegami, M.; Whitsett, J.A. Altered stability of pulmonary surfactant in SP-C-deficient mice. Proc. Natl. Acad. Sci. USA 2001, 98, 6366–6371. [Google Scholar] [CrossRef]
- Lofgren, J.; Ramet, M.; Renko, M.; Marttila, R.; Hallman, M. Association between surfactant protein A gene locus and severe respiratory syncytial virus infection in infants. J. Infect. Dis. 2002, 185, 283–289. [Google Scholar] [CrossRef] [PubMed]
- El Saleeby, C.M.; Li, R.; Somes, G.W.; Dahmer, M.K.; Quasney, M.W.; DeVincenzo, J.P. Surfactant protein A2 polymorphisms and disease severity in a respiratory syncytial virus-infected population. J. Pediatr. 2010, 156, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Umstead, T.M.; Phelps, D.S.; Al-Mondhiry, H.; Floros, J. The effect of ozone exposure on the ability of human surfactant protein a variants to stimulate cytokine production. Environ. Health Perspect. 2002, 110, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Mikerov, A.N.; Wang, G.; Umstead, T.M.; Zacharatos, M.; Thomas, N.J.; Phelps, D.S.; Floros, J. Surfactant protein A2 (SP-A2) variants expressed in CHO cells stimulate phagocytosis of Pseudomonas aeruginosa more than do SP-A1 variants. Infect. Immun. 2007, 75, 1403–1412. [Google Scholar] [CrossRef][Green Version]
- Mikerov, A.N.; Umstead, T.M.; Gan, X.; Huang, W.; Guo, X.; Wang, G.; Phelps, D.S.; Floros, J. Impact of ozone exposure on the phagocytic activity of human surfactant protein A (SP-A) and SP-A variants. Am. J. Physiol. Lung Cell. Mol. Physiol. 2008, 294, L121–L130. [Google Scholar] [CrossRef]
- Wang, G.; Myers, C.; Mikerov, A.; Floros, J. Effect of cysteine 85 on biochemical properties and biological function of human surfactant protein A variants. Biochemistry 2007, 46, 8425–8435. [Google Scholar] [CrossRef]
- Palaniyar, N.; Ikegami, M.; Korfhagen, T.; Whitsett, J.; McCormack, F.X. Domains of surfactant protein A that affect protein oligomerization, lipid structure and surface tension. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2001, 129, 109–127. [Google Scholar] [CrossRef]
- Crouch, E.C. Collectins and pulmonary host defense. Am. J. Respir. Cell Mol. Biol. 1998, 19, 177–201. [Google Scholar] [CrossRef] [PubMed]
- Hickling, T.P.; Malhotra, R.; Sim, R.B. Human lung surfactant protein A exists in several different oligomeric states: Oligomer size distribution varies between patient groups. Mol. Med. 1998, 4, 266–275. [Google Scholar] [CrossRef]
- Sanchez-Barbero, F.; Rivas, G.; Steinhilber, W.; Casals, C. Structural and functional differences among human surfactant proteins SP-A1, SP-A2 and co-expressed SP-A1/SP-A2: Role of supratrimeric oligomerization. Biochem. J. 2007, 406, 479–489. [Google Scholar] [CrossRef]
- Thorenoor, N.; Zhang, X.; Umstead, T.M.; Scott Halstead, E.; Phelps, D.S.; Floros, J. Differential effects of innate immune variants of surfactant protein-A1 (SFTPA1) and SP-A2 (SFTPA2) in airway function after Klebsiella pneumoniae infection and sex differences. Respir. Res. 2018, 19, 23. [Google Scholar] [CrossRef]
- Guo, X.; Lin, H.M.; Lin, Z.; Montano, M.; Sansores, R.; Wang, G.; DiAngelo, S.; Pardo, A.; Selman, M.; Floros, J. Surfactant protein gene A, B, and D marker alleles in chronic obstructive pulmonary disease of a Mexican population. Eur. Respir. J. 2001, 18, 482–490. [Google Scholar] [CrossRef]
- Hilgendorff, A.; Heidinger, K.; Bohnert, A.; Kleinsteiber, A.; König, I.R.; Ziegler, A.; Lindner, U.; Frey, G.; Merz, C.; Lettgen, B.; et al. Association of polymorphisms in the human surfactant protein-D (SFTPD) gene and postnatal pulmonary adaptation in the preterm infant. Acta Paediatr. 2009, 98, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.J.; Akiyama, J.; Allen, L.; Hawgood, S. Localization and developmental expression of surfactant proteins D and A in the respiratory tract of the mouse. Pediatr. Res. 1996, 39, 930–937. [Google Scholar] [CrossRef] [PubMed]
- Cordell, H.J. Detecting gene-gene interactions that underlie human diseases. Nat. Rev. Genet. 2009, 10, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Haataja, R.; Rämet, M.; Marttila, R.; Hallman, M. Surfactant proteins A and B as interactive genetic determinants of neonatal respiratory distress syndrome. Hum. Mol. Genet. 2000, 9, 2751–2760. [Google Scholar] [CrossRef]
- Stram, D.O. Multi-SNP Haplotype Analysis Methods for Association Analysis. In Statistical Human Genetics: Methods and Protocols; Elston, R.C., Ed.; Springer: New York, NY, USA, 2017; pp. 485–504. [Google Scholar] [CrossRef]
- Puthothu, B.; Krueger, M.; Heinze, J.; Forster, J.; Heinzmann, A. Haplotypes of surfactant protein C are associated with common paediatric lung diseases. Pediatr. Allergy Immunol. 2006, 17, 572–577. [Google Scholar] [CrossRef]
- Willson, D.F.; Jiao, J.H.; Bauman, L.A.; Zaritsky, A.; Craft, H.; Dockery, K.; Conrad, D.; Dalton, H. Calf’s lung surfactant extract in acute hypoxemic respiratory failure in children. Crit. Care Med. 1996, 24, 1316–1322. [Google Scholar] [CrossRef]
- Willson, D.F.; Thomas, N.J.; Markovitz, B.P.; Bauman, L.A.; DiCarlo, J.V.; Pon, S.; Jacobs, B.R.; Jefferson, L.S.; Conaway, M.R.; Egan, E.A.; et al. Effect of Exogenous Surfactant (Calfactant) in Pediatric Acute Lung InjuryA Randomized Controlled Trial. JAMA 2005, 293, 470–476. [Google Scholar] [CrossRef]
| Variable | No PDAD (n = 162) | PDAD (n = 86) | p value |
|---|---|---|---|
| Demographics Age (months) Female/male (%/%) | 2.8 ± 3.5 63/99 (39/61) | 5.4 ± 5.6 36/50 (42/58) | <0.001 0.89 |
| Race White Black Mixed/Other | 129 (80) 23 (14) 10 (6) | 66 (77) 15 (17) 5 (6) | 0.74 |
| Ethnicity Hispanic Non-Hispanic | 31 (19) 131 (81) | 21 (24) 65 (76) | 0.61 |
| Admission diagnosis (%) RSV bronchiolitis Other bronchiolitis Other pneumonia Other respiratory failure Non-pulmonary | 89 (55) 39 (24) 13 (8) 17 (10) 4 (2) | 38 (44) 17 (20) 17 (20) 12 (14) 2 (2) | 0.08 |
| PRISM III score | 4.6 ± 3.6 | 4.8 ± 4.0 | 0.69 |
| Specific virus (%) (n = 187) RSV Influenza Parainfluenza Adenovirus | 102 (79) 3 (2) 1 (1) 1 (1) | 38 (64) 1 (1) 2 (2) 2 (3) | 0.03 1 0.33 0.24 |
| Positive bacterial culture (%)(n = 195) | 81 (63) | 54 (82) | 0.009 |
| Duration of support Ventilator days Oxygen days PICU days | 6.5 ± 4.0 10.0 ± 5.1 8.3 ± 4.6 | 9.5 ± 12.1 12.7 ± 14.5 12.1 ± 13.5 | 0.004 0.03 0.001 |
| Gene | SNP #1 ID | Gene | SNP #2 ID | Gene | SNP #3 ID | Interaction | p value | FDR | OR (95% CI) | |
|---|---|---|---|---|---|---|---|---|---|---|
| One dominant effect | ||||||||||
| 1 * | SFTPA2 | rs1059046 | SFTPA2 | rs1965707 | SFTPA1 | rs1059047 | a1 × d2 × a3 | 0.001285994 | 0.001 | 2.2 (1.3–3.6) |
| 2 * | SFTPA2 | rs1059046 | SFTPA2 | rs1965707 | SFTPA1 | rs1136451 | 0.001052925 | 0.00105 | 2.2 (1.3–3.7) | |
| 3 * | SFTPA2 | rs1059046 | SFTPA2 | rs1965707 | SFTPA1 | rs4253527 | 0.000833915 | 0.00083 | 2.3 (1.4–4.0) | |
| 4 * | SFTPA2 | rs1059046 | SFTPA2 | rs1965708 | SFTPA1 | rs1136451 | 0.000662744 | 0.00066 | 2.6 (1.4–4.7) | |
| 5 #a | SFTPA2 | rs1059046 | SFTPA2 | rs17886395 | SFTPA2 | rs1965708 | a1 × a2 × d3 | 0.000229167 | 0.00020 | 0.4 (0.2–0.7) |
| Two dominant effect | ||||||||||
| 1 *a | SFTPA2 | rs1059046 | SFTPA2 | rs17886395 | SFTPA2 | rs1965707 | a1 × d2 × d3 | 0.00008108 | 0.00008 | 2.1 (1.4–3.2) |
| 2 *a | SFTPA2 | rs1059046 | SFTPA2 | rs17886395 | SFTPA2 | rs1965708 | 0.001549286 | 0.00155 | 1.8 (1.2–2.7) | |
| 3 * | SFTPA2 | rs1059046 | SFTPA2 | rs1965707 | SFTPA1 | rs1059047 | 0.00341059 | 0.00341 | 1.8 (1.2–2.7) | |
| 4 * | SFTPA2 | rs1059046 | SFTPA2 | rs1965707 | SFTPA1 | rs1136451 | 0.002675982 | 0.00268 | 1.8 (1.2–2.7) | |
| 5 * | SFTPA2 | rs1059046 | SFTPA2 | rs1965708 | SFTPA1 | rs1059047 | 0.000228776 | 0.00023 | 2.2 (1.4–3.5) | |
| 6 * | SFTPA2 | rs1059046 | SFTPA1 | rs1059047 | SFTPA1 | rs1136450 | 0.001111869 | 0.00111 | 2.1 (1.3–3.3) | |
| 7 # | SFTPA2 | rs1965707 | SFTPA2 | rs1965708 | SFTPC | rs4715 | 0.00053651 | 0.00050 | 0.3 (0.1–0.6) | |
| 8 #a | SFTPA1 | rs1059047 | SFTPA1 | rs1136451 | SFTPA1 | rs4253527 | 0.004439716 | 0.00400 | 0.3 (0.1–0.8) | |
| 9 *a | SFTPA2 | rs1059046 | SFTPA2 | rs17886395 | SFTPA2 | rs1965707 | d1 × a2 × d3 | 0.000424175 | 0.00042 | 1.8 (1.3–2.5) |
| 10 *a | SFTPA2 | rs1059046 | SFTPA2 | rs17886395 | SFTPA2 | rs1965708 | 0.001836416 | 0.00184 | 1.7 (1.2–2.3) | |
| 11 * | SFTPA2 | rs17886395 | SFTPA1 | rs1136450 | SFTPC | rs4715 | 0.000648071 | 0.00049 | 4.0 (1.7–10.1) | |
| 12 # | SFTPA2 | rs17886395 | SFTPA1 | rs1059047 | SFTPA1 | rs1136450 | 0.000490381 | 0.00060 | 0.6 (0.5–0.8) | |
| 13 * | SFTPA2 | rs17886395 | SFTPA2 | rs1965707 | SFTPA1 | rs1136450 | d1 × d2 × a3 | 0.000191969 | 0.00019 | 2.2 (1.4–3.4) |
| 14 # | SFTPA2 | rs17886395 | SFTPA1 | rs1136450 | SFTPC | rs4715 | 0.000158449 | 0.00010 | 0.2 (0.1–0.5) | |
| Three dominant effect | ||||||||||
| 1 * | SFTPA2 | rs17886395 | SFTPA1 | rs1136451 | SFTPA1 | rs4253527 | d1 × d2 × d3 | 0.000333494 | 0.00005 | 1.6 (1.3–2.1) |
| 2 #a | SFTPA2 | rs1059046 | SFTPA2 | rs17886395 | SFTPA2 | rs1965708 | 0.005639344 | 0.00030 | 0.6 (0.5–0.8) | |
| 3 # | SFTPA2 | rs1059046 | SFTPA2 | rs1965707 | SFTPC | rs4715 | 0.004814545 | 0.00600 | 0.6 (0.4–0.9) | |
| 4 # | SFTPA2 | rs1059046 | SFTPA1 | rs1059047 | SFTPA1 | rs1136450 | 0.00000000002 | 0.00500 | 0.7 (0.6–0.9) | |
| 5 # | SFTPA2 | rs1059046 | SFTPA1 | rs1136450 | SFTPC | rs4715 | 0.00336572 | 0.00000 | 0.3 (0.2–0.4) | |
| 6 # | SFTPA2 | rs1059046 | SFTPA1 | rs1136451 | SFTPC | rs4715 | 0.000000227 | 0.00300 | 0.5 (0.4–0.8) | |
| 7 #a | SFTPA2 | rs17886395 | SFTPA2 | rs1965707 | SFTPA2 | rs1965708 | 0.000004297 | 0.00000 | 0.5 (0.4–0.7) | |
| 8 # | SFTPA2 | rs17886395 | SFTPA1 | rs1059047 | SFTPA1 | rs1136450 | 0.000809901 | 0.00000 | 0.6 (0.5–0.7) | |
| 9 # | SFTPA2 | rs17886395 | SFTPA1 | rs1059047 | SFTPA1 | rs1136451 | 0.000054837 | 0.00080 | 0.7 (0.5–0.9) | |
| 10 # | SFTPA2 | rs17886395 | SFTPA1 | rs1136451 | SFTPC | rs4715 | 0.000094992 | 0.00009 | 0.4 (0.3–0.7) | |
| 11 # | SFTPA2 | rs17886395 | SFTPA1 | rs4253527 | SFTPC | rs4715 | 0.00329539 | 0.00300 | 0.5 (0.3–0.8) | |
| 12 # | SFTPA2 | rs1965707 | SFTPA2 | rs1965708 | SFTPA1 | rs1136450 | 0.000141004 | 0.00010 | 0.7 (0.5–0.8) | |
| 13 # | SFTPA2 | rs1965707 | SFTPA1 | rs1136450 | SFTPC | rs4715 | 0.003330483 | 0.00300 | 0.6 (0.4–0.8) | |
| 14 # | SFTPA1 | rs1059047 | SFTPA1 | rs1136450 | SFTPA1 | rs1136451 | 0.005263705 | 0.00500 | 0.7 (0.6–0.9) | |
| 15 # | SFTPA1 | rs1136451 | SFTPA1 | rs4253527 | SFTPC | rs4715 | 0.00126707 | 0.00100 | 0.5 (0.3–0.8) | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gandhi, C.K.; Chen, C.; Wu, R.; Yang, L.; Thorenoor, N.; Thomas, N.J.; DiAngelo, S.L.; Spear, D.; Keim, G.; Yehya, N.; et al. Association of SNP–SNP Interactions of Surfactant Protein Genes with Pediatric Acute Respiratory Failure. J. Clin. Med. 2020, 9, 1183. https://doi.org/10.3390/jcm9041183
Gandhi CK, Chen C, Wu R, Yang L, Thorenoor N, Thomas NJ, DiAngelo SL, Spear D, Keim G, Yehya N, et al. Association of SNP–SNP Interactions of Surfactant Protein Genes with Pediatric Acute Respiratory Failure. Journal of Clinical Medicine. 2020; 9(4):1183. https://doi.org/10.3390/jcm9041183
Chicago/Turabian StyleGandhi, Chintan K., Chixiang Chen, Rongling Wu, Lili Yang, Nithyananda Thorenoor, Neal J. Thomas, Susan L. DiAngelo, Debbie Spear, Garrett Keim, Nadir Yehya, and et al. 2020. "Association of SNP–SNP Interactions of Surfactant Protein Genes with Pediatric Acute Respiratory Failure" Journal of Clinical Medicine 9, no. 4: 1183. https://doi.org/10.3390/jcm9041183
APA StyleGandhi, C. K., Chen, C., Wu, R., Yang, L., Thorenoor, N., Thomas, N. J., DiAngelo, S. L., Spear, D., Keim, G., Yehya, N., & Floros, J. (2020). Association of SNP–SNP Interactions of Surfactant Protein Genes with Pediatric Acute Respiratory Failure. Journal of Clinical Medicine, 9(4), 1183. https://doi.org/10.3390/jcm9041183







