Machine Learning Enables Prediction of Cardiac Amyloidosis by Routine Laboratory Parameters: A Proof-of-Concept Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Setting and Study Design
2.2. Diagnostic Procedures
2.2.1. Cardiac Amyloidosis
2.2.2. Amyloidosis-Unrelated Heart Failure
2.3. Statistical Procedures
3. Results
3.1. Patient Baseline Characteristics
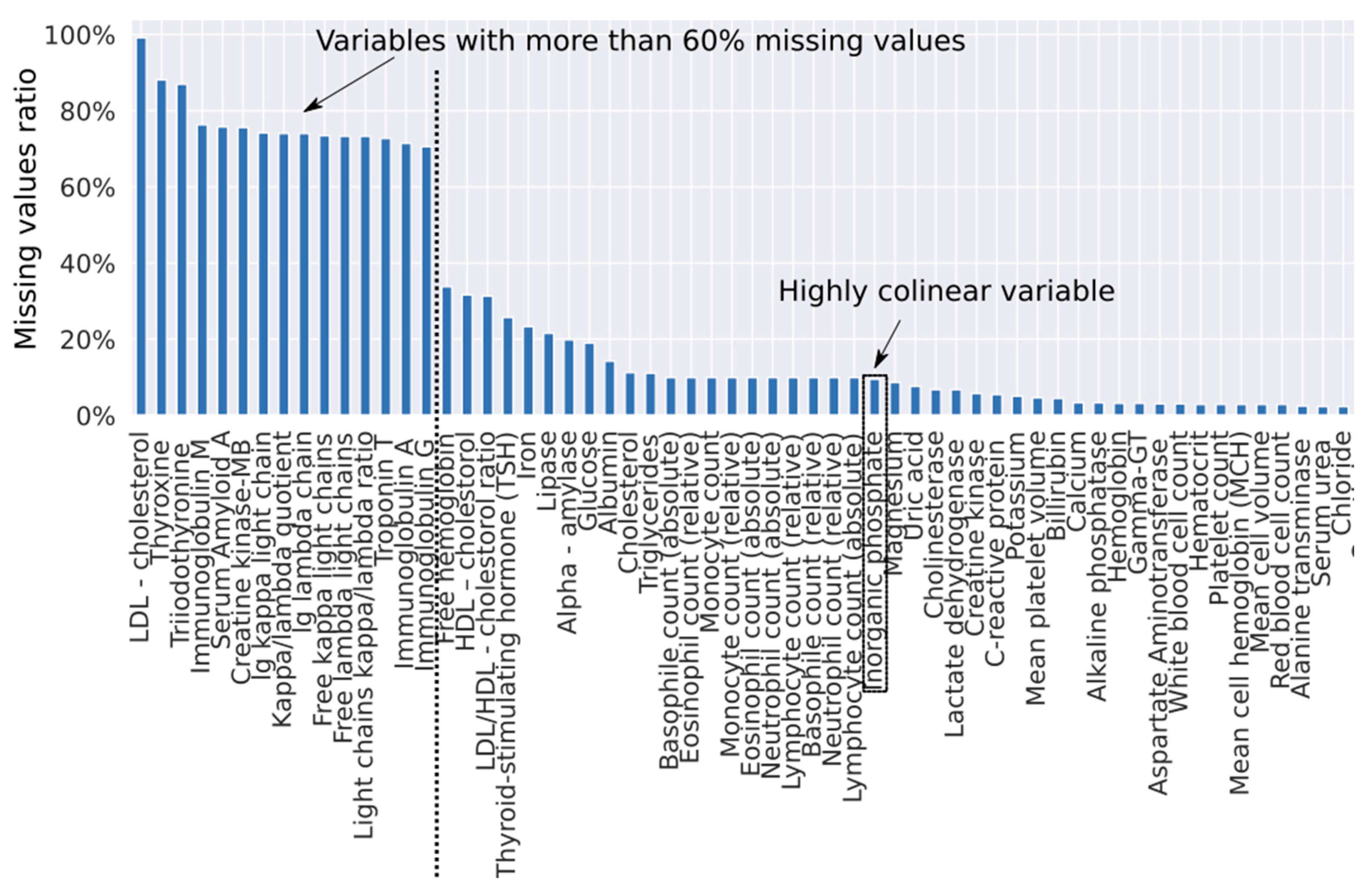
3.2. Development of the Diagnostic Algorithm with Imputation of Missing Values
3.3. Improving the Diagnostic Algorithm with Machine Learning
3.4. Fully Automated Machine Learning Diagnostic Algorithm
4. Discussion
Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ponikowski, P.; Voors, A.A.; Anker, S.D.; Bueno, H.; Cleland, J.G.F.; Coats, A.J.; Falk, V.; González-Juanatey, J.R.; Harjola, V.P.; Jankowska, E.A.; et al. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: The Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC)Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur. Heart J. 2016, 37, 2129–2200. [Google Scholar] [CrossRef] [PubMed]
- Falk, R.H.; Alexander, K.M.; Liao, R.; Dorbala, S. AL (Light-Chain) Cardiac Amyloidosis: A Review of Diagnosis and Therapy. J. Am. Coll. Cardiol. 2016, 68, 1323–1341. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.; Koike, H.; Slama, M.; Coelho, T. Hereditary transthyretin amyloidosis: a model of medical progress for a fatal disease. Nat. Rev. Neurol. 2019, 15, 387–404. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.; Gonzalez-Duarte, A.; O’Riordan, W.D.; Yang, C.-C.; Ueda, M.; Kristen, A.V.; Tournev, I.; Schmidt, H.H.; Coelho, T.; Berk, J.L.; et al. Patisiran, an rnai therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 2018, 379, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Benson, M.D.; Waddington-Cruz, M.; Berk, J.L.; Polydefkis, M.; Dyck, P.J.; Wang, A.K.; Planté-Bordeneuve, V.; Barroso, F.A.; Merlini, G.; Obici, L.; et al. Inotersen Treatment for Patients with Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018, 379, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Maurer, M.S.; Schwartz, J.H.; Gundapaneni, B.; Elliott, P.M.; Merlini, G.; Waddington-Cruz, M.; Kristen, A.V.; Grogan, M.; Witteles, R.; Damy, T.; et al. Tafamidis Treatment for Patients with Transthyretin Amyloid Cardiomyopathy. N. Engl. J. Med. 2018, 379, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- Gillmore, J.D.; Damy, T.; Fontana, M.; Hutchinson, M.; Lachmann, H.J.; Martinez-Naharro, A.; Quarta, C.C.; Rezk, T.; Whelan, C.J.; Gonzalez-Lopez, E.; et al. A new staging system for cardiac transthyretin amyloidosis. Eur. Heart J. 2018, 39, 2799–2806. [Google Scholar] [CrossRef] [PubMed]
- Abraham, R.S.; Geyer, S.M.; Price-Troska, T.L.; Allmer, C.; Kyle, R.A.; Gertz, M.A.; Fonseca, R. Immunoglobulin light chain variable (V) region genes influence clinical presentation and outcome in light chain-associated amyloidosis (AL). Blood 2003, 101, 3801–3808. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.H. Greedy function approximation: a gradient boosting machine. Ann. Statist. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Tuv, E.; Borisov, A.; Runger, G.; Torkkola, K. Feature selection with ensembles, artificial variables, and redundancy elimination. J. Mach. Learn. Res. 2009, 10, 1341–1366. [Google Scholar]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining—KDD ’16, New York, NY, USA, 13–17 August 2016; pp. 785–794. [Google Scholar] [CrossRef] [Green Version]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Statist. Soc. Ser. B (Methodol.) 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Ruopp, M.D.; Perkins, N.J.; Whitcomb, B.W.; Schisterman, E.F. Youden Index and optimal cut-point estimated from observations affected by a lower limit of detection. Biom. J. 2008, 50, 419–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cariou, E.; Bennani Smires, Y.; Victor, G.; Robin, G.; Ribes, D.; Pascal, P.; Petermann, A.; Fournier, P.; Faguer, S.; Roncalli, J.; et al. Diagnostic score for the detection of cardiac amyloidosis in patients with left ventricular hypertrophy and impact on prognosis. Amyloid 2017, 24, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Nicol, M.; Baudet, M.; Brun, S.; Harel, S.; Royer, B.; Vignon, M.; Lairez, O.; Lavergne, D.; Jaccard, A.; Attias, D.; et al. Diagnostic score of cardiac involvement in AL amyloidosis. Eur. Heart J. Cardiovasc. Imaging 2020, 21, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Samsky, M.D.; Patel, C.B.; DeWald, T.A.; Smith, A.D.; Felker, G.M.; Rogers, J.G.; Hernandez, A.F. Cardiohepatic interactions in heart failure: an overview and clinical implications. J. Am. Coll. Cardiol. 2013, 61, 2397–2405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruberg, F.L.; Grogan, M.; Hanna, M.; Kelly, J.W.; Maurer, M.S. Transthyretin Amyloid Cardiomyopathy: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2019, 73, 2872–2891. [Google Scholar] [CrossRef] [PubMed]
- Perfetto, F.; Bergesio, F.; Emdin, M.; Cappelli, F. Troponins in cardiac amyloidosis: multipurpose markers. Nat. Rev. Cardiol. 2014, 11, 179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Attia, Z.I.; Kapa, S.; Lopez-Jimenez, F.; McKie, P.M.; Ladewig, D.J.; Satam, G.; Pellikka, P.A.; Enriquez-Sarano, M.; Noseworthy, P.A.; Munger, T.M.; et al. Screening for cardiac contractile dysfunction using an artificial intelligence-enabled electrocardiogram. Nat. Med. 2019, 25, 70–74. [Google Scholar] [CrossRef] [PubMed]


| Amyloidosis-Unrelated HF (n = 415) | Amyloidosis-Related HF (n = 121) | p-Value | |
|---|---|---|---|
| Variable, median (Q1–Q3)] | |||
| Age, years | 69.0 (58.0, 75.0) | 73.0 (62.0, 78.0) | 0.004 |
| NT-proBNP, pg/mL | 680.0 (226.1, 1621.0) | 3132.5 (1343.8, 6994.2) | < 0.001 |
| Variable, mean (SD) | |||
| Body mass index, kg/m2 | 29.8 (6.6) | 26.0 (4.1) | < 0.001 |
| Variable, n (%) | |||
| Gender, males | 167 (40.2) | 87 (71.9) | < 0.001 |
| Coronary artery disease | 112 (27.7) | 23 (19.5) | 0.114 |
| Atrial fibrillation | 173 (42.6) | 47 (39.5) | 0.679 |
| Arterial hypertension | 346 (83.8) | 67 (57.3) | < 0.001 |
| Diabetes mellitus | 118 (28.6) | 11 (9.2) | < 0.001 |
| Hyperlipidemia | 220 (54.3) | 26 (22.4) | < 0.001 |
| MRA | 104 (26.3) | 51 (45.1) | 0.001 |
| Calcium channel blocker | 93 (23.1) | 9 (7.7) | 0.001 |
| Beta-blocker | 278 (68.6) | 58 (52.7) | 0.004 |
| Diuretics | 213 (53.8) | 80 (70.2) | 0.004 |
| ACEI/ARB | 253 (62.5) | 49 (43.8) | 0.001 |
| Oral anticoagulant | 184 (45.7) | 54 (47.0) | 0.888 |
| Statin | 181 (44.6) | 29 (25.0) | 0.001 |
| Amyloidosis-Unrelated HF (n = 124) | Amyloidosis-Related HF (n = 36) | p-Value | |
|---|---|---|---|
| Variable, median (Q1-Q3) | |||
| Age, years | 75.0 (69.0,78.0) | 78.0 (71.8,83.0) | 0.066 |
| NT-proBNP, pg/mL | 851.0 (372.1,1939.0) | 2568.5 (1543.8,4334.0) | <.001 |
| Variable, mean (SD) | |||
| Body mass index, kg/m2 | 29.5 (5.5) | 26.0 (4.5) | 0.003 |
| Variable, n (%) | |||
| Gender, males | 36 (29.5) | 28 (77.8) | <0.001 |
| Coronary artery disease | 33 (26.8) | 16 (44.4) | 0.222 |
| Atrial fibrillation | 69 (58.0) | 18 (50.0) | 0.638 |
| Arterial hypertension | 110 (93.2) | 25 (69.4) | 0.003 |
| Diabetes mellitus | 36 (30.8) | 8 (22.2) | 0.638 |
| Hyperlipidemia | 63 (52.9) | 16 (44.4) | 0.638 |
| MRA | 60 (50.4) | 19 (52.8) | 0.954 |
| Calcium channel blocker | 26 (21.7) | 4 (11.1) | 0.534 |
| Beta-blocker | 85 (70.8) | 18 (50.0) | 0.127 |
| Diuretics | 80 (67.8) | 28 (77.8) | 0.634 |
| ACEI/ARB | 88 (73.3) | 23 (63.9) | 0.634 |
| Oral anticoagulant | 75 (61.0) | 19 (52.8) | 0.638 |
| Statin | 59 (50.0) | 16 (44.4) | 0.804 |
| Model | ROC AUC | Sensitivity % | Specificity % | PPV % | NPV (FOR) % |
|---|---|---|---|---|---|
| LR-L1 | 0.58 | 67.6 | 53.2 | 30.0 | 84.6 (15.4) |
| LR-XGBoost | 0.75 | 84.6 | 71.7 | 33.3 | 96.6 (3.4) |
| XGBoost-62 | 0.86 | 89.2 | 78.2 | 55.0 | 96.1 (3.9) |
| XGBoost-46 | 0.66 | 67.6 | 70.2 | 40.3 | 87.9 (12.1) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agibetov, A.; Seirer, B.; Dachs, T.-M.; Koschutnik, M.; Dalos, D.; Rettl, R.; Duca, F.; Schrutka, L.; Agis, H.; Kain, R.; et al. Machine Learning Enables Prediction of Cardiac Amyloidosis by Routine Laboratory Parameters: A Proof-of-Concept Study. J. Clin. Med. 2020, 9, 1334. https://doi.org/10.3390/jcm9051334
Agibetov A, Seirer B, Dachs T-M, Koschutnik M, Dalos D, Rettl R, Duca F, Schrutka L, Agis H, Kain R, et al. Machine Learning Enables Prediction of Cardiac Amyloidosis by Routine Laboratory Parameters: A Proof-of-Concept Study. Journal of Clinical Medicine. 2020; 9(5):1334. https://doi.org/10.3390/jcm9051334
Chicago/Turabian StyleAgibetov, Asan, Benjamin Seirer, Theresa-Marie Dachs, Matthias Koschutnik, Daniel Dalos, René Rettl, Franz Duca, Lore Schrutka, Hermine Agis, Renate Kain, and et al. 2020. "Machine Learning Enables Prediction of Cardiac Amyloidosis by Routine Laboratory Parameters: A Proof-of-Concept Study" Journal of Clinical Medicine 9, no. 5: 1334. https://doi.org/10.3390/jcm9051334
APA StyleAgibetov, A., Seirer, B., Dachs, T.-M., Koschutnik, M., Dalos, D., Rettl, R., Duca, F., Schrutka, L., Agis, H., Kain, R., Auer-Grumbach, M., Binder, C., Mascherbauer, J., Hengstenberg, C., Samwald, M., Dorffner, G., & Bonderman, D. (2020). Machine Learning Enables Prediction of Cardiac Amyloidosis by Routine Laboratory Parameters: A Proof-of-Concept Study. Journal of Clinical Medicine, 9(5), 1334. https://doi.org/10.3390/jcm9051334








