Abstract
This study aimed to explore the associations between the TP53 rs1042522 (TP53 Arg72Pro), MDM2 rs2279744 (MDM2 309T>G), rs3730485 (MDM2 del1518), MDM4 rs4245739 (MDM4 34091 C>A) variants and odds of developing acute myeloid leukemia (AML) in a cohort of 809 adult subjects, consisting of 406 healthy controls and 403 AML patients. Model-based multifactor dimensionality reduction (MB-MDR) framework was used to identify the interactions of the mentioned variants and their association with AML risk. Associations of the mentioned variants with clinical features of AML, somatic mutations, and response to treatment were also evaluated. Significant associations between TP53 rs1042522 and MDM4 rs4245739 variants and AML susceptibility were noticed. MB-MDR and logistic regression analysis revealed an interaction between MDM2 rs2279744 and TP53 rs1042522, between MDM4 rs4245739 and MDM2 rs3730485, as well as significant associations with AML susceptibility. Several associations between the mentioned variants and clinical features of AML and somatic mutations were also noticed. Individually, the variant genotypes of TP53 rs1042522 and MDM4 rs4245739 were associated with AML susceptibility, but their interaction with MDM2 rs2279744 and rs3730485 modulated the risk for AML. The variant genotypes of TP53 rs1042522 were associated with adverse molecular and cytogenetic risk and also with NPM1 mutations.
1. Introduction
Acute myeloid leukemia (AML) patients may have one or several genetic abnormalities. More than 90% of the AML patients had genetic abnormalities. Frequently, the genetic alteration involved Tumor protein p53 (TP53) gene. This gene has an important role in apoptosis and DNA-damage response [1]. TP53 gene encodes the p53 protein. TP53 gene expression is inhibited by Mouse double minute 2 homolog (MDM2) gene and by MDM4 gene (homolog of MDM2) [2,3], MDM2, and MDM4 genes being negative regulators of p53 [4].
Previously it was reported that approximately 30% of acute lymphoid leukemia (ALL) patients and 47% of AML patients had over-expressed the MDM2 protein compared to the control group (at least 10-fold). Over-expression of MDM2 protein being correlated for AML patients with a worse prognostic [1,5] MDM4 gene is the homolog of the MDM2 gene and their codified proteins showed very high structural and functional similarities, both inhibiting p53 protein by complex mechanisms described by Qin L et al. [6]. Therefore, the inhibition of p53 by over-expression of MDM2 and MDM4 thereby can contribute to leukemogenesis.
In malignancies, including leukemia, TP53 gene expression was reported to be affected by a missense variant, rs1042522 (TP53 Arg72Pro) [7,8]. Moreover, the variant allele of rs1042522 was reported to decrease apoptotic activity, increase the risk of cancer (including leukemia), and to be associated with poor overall survival (OS) in patients with AML and higher rate of refractory disease [2,7,9,10,11].
However, the role of rs1042522 in AML susceptibility and OS is not fully clarified due to different results reported in the literature. Six studies performed on AML patients were included in a recent meta-analysis [7] including 200 Japanese patients [12], 171 patients from UK and USA [13], 411 Chinese patients from two studies (231+180) [1,14], and 272 Indian patients from two studies (131+141) [15,16]. While no association was found between AML risk and variant allele of rs1042522, different associations with OS and response to treatment were noticed. On the other hand, one recent study performed on 189 Brazilian AML patients described an association between rs1042522 and AML risk [11]. One cause of contradictory results may be the small cohorts of the mentioned studies and different ethnicity of the subjects [2,11].
Similarly, it was reported that expression of MDM2 and MDM4 genes are affected by MDM2 rs2279744 (MDM2 309T>G) and rs3730485 (MDM2 del1518) respectively by MDM4 rs4245739 (MDM4 34091 C>A) variants, the mentioned variants being associated with increased risk for different types of cancer and higher incidence of advanced tumor stages [17,18,19,20]. In contradiction with the abovementioned results, Gansmo LB et al. [21] in their study among endometrial cancer patients found that rs3730485 was in strong linkage disequilibrium (LD) with rs2279744 and the variant allele of rs3730485 was associated with reduced cancer risk among patients with the wildtype genotype for rs2279744 [21]. Lian T et al. [22] in their meta-analysis suggested that variant allele of rs4245739 was associated with reduced cancer risk, especially in Asian populations [22]. Another meta-analysis performed by Hua W et al. [23] on Asian and Caucasian populations reported no associations between rs3730485 variant and cancer risk. However, none of the mentioned meta-analyses included AML patients.
Regarding the risk for AML, Soleymannejad M et al. [24] noticed an association between MDM2 rs2279744 and AML susceptibility. Also, Phillips CL et al. [25] reported the same association on childhood AML patients and no association with treatment response [25]. Moreover, Falk IJ et al. [26] showed association with lower OS in adult AML patients. On the other hand, Abdel TM et al. [27] did not find an association between rs2279744 and lower OS. In other hematological malignancies, such as chronic lymphoid leukemia, the variant genotype of rs2279744 was reported to be associated with a nine-fold increase in the risk of death [28]. Moreover, the simultaneous presence of wildtype allele of TP53 rs1042522 and the variant allele of MDM2 rs2279744 may influence the risk of therapy-related myeloid neoplasms [29]. MDM2 rs3730485 and MDM4 rs4245739 variants were studied in different types of cancer, together or in combination with one or with the other two mentioned variants, but from our knowledge, AML patients were not investigated. None of the published studies investigated the TP53 rs1042522, MDM2 rs2279744, rs3730485, and MDM4 rs4245739 variants simultaneously. Moreover, none of the published studies focused on identifying the interactions of the mentioned variants and their association with AML (or other cancers) risk.
First of all, our study aimed to explore the associations between the TP53 rs1042522 (TP53 Arg72Pro), MDM2 rs2279744 (MDM2 309T>G), rs3730485 (MDM2 del1518), and MDM4 rs4245739 (MDM4 34091 C>A) variants and odds of developing AML in a cohort of 809 adult subjects, consisting of 406 healthy controls and 403 AML patients. In addition, we analyzed the pairwise and higher-order interactions of investigated TP53, MDM2, and MDM4 variants and their association with odds of AML. Moreover, we evaluated the associations of the mentioned variants with the clinical features of AML, somatic mutations, and response to treatment.
2. Materials and Methods
2.1. Patients and Controls
The Board of the Ethical Committee of the Clinical and Emergency Hospital of Targu Mures, Romania, approved this case-control study (10665/2019). The subjects included signed a written informed consent form. The study was performed in accordance with the fundamental principles of the Declaration of Helsinki. A total number of 809 adult subjects were included, 406 healthy controls and 403 AML patients. AML group consisted of 215 males and 188 females while the control group was comprised of 181 males and 225 females. We included only subjects who signed the written informed consent form, with complete laboratory and clinical records, in which the genotyping investigation was successfully performed. AML diagnosis was based on clinical examination and laboratory investigation (including complete blood count, blood smear, bone marrow and/or blood microscopic examination, flow cytometry, cytogenetics, fusion gene investigation as reported previously [30], DNA copy number variations analysis as reported previously [31], fragment analysis for FLT3-ITD and NPM1 mutations as reported previously [32,33,34], and target next-generation sequencing as reported previously [34]).
2.2. Genotyping Investigation
For DNA isolation, we used the manufacturer’s protocol of PureLink Genomic DNA kit (Thermo Fisher Scientific, Carlsbad, CA, USA) or Quick-DNA Miniprep Plus Kit (ZymoResearch, Irvine, CA, USA). Total DNA was purified from nucleated blood cells. Genotyping protocols used for TP53 rs1042522, MDM2 rs3730485, and MDM4 rs4245739 variants were previously published [21,35,36,37,38]. The protocols were in-silico checked using the following free tools: Primer-BLAST (National Center for Biotechnology Information, NCBI), in-silico PCR program (University of California, Santa Cruz, UCSC In-Silico PCR—UCSC Genome Browser), NEBcutter V2.0 for RFLP-PCR technique (New England BioLabs, Ipswich, MA, USA) and SNPCheck 3 program (National Genetics Reference Laboratories, NGRL Manchester). For MDM2 rs2279744 genotyping we used TaqMan assay (C__15968533_20, Thermo Fisher Scientific, Carlsbad, CA, USA) and 7500 Fast Dx Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). The genotypes of 10% of the subjects were confirmed by capillary sequencing (3500 Genetic Analyzer, Applied Biosystems, Foster City, CA, USA).
NCBI’s (NCBI Homo sapiens Annotation Release 109), Ensembl’s (Ensembl Release 98) and European Variation Archive’s genome browser were used for alleles annotation of the investigated variants.
2.3. Statistical Analysis
2.3.1. Descriptive Analysis
The demographic, clinical, and genetic data were presented via descriptive measures as mean ± standard deviation or percentages and absolute frequencies.
2.3.2. Inferential Analysis
The Hardy–Weinberg equilibrium (HWE) and pairwise linkage disequilibrium (LD), using the standardized D, were performed both in AML and control group.
The statistical associations of TP53 rs1042522, MDM2 rs2279744, rs3730485, and MDM4 rs4245739 variants with demographic, clinical features, and somatic mutations were assessed using Chi-square/Fisher’s Exact test. Post-hoc pairwise Chi-square or Fisher tests were performed after significant association have been identified and adjusted p-values computed via Benjamini–Hochberg procedure were reported.
The associations between selected genetic variants and odds of AML were estimated in different genetic models: Codominant, dominant, recessive, and overdominant using generalized linear models (GLMs) with a binomial distribution (logit as link function). In addition, the univariate effect of selected variants on odds of AML was adjusted for age group (≥60 years) and gender. The Benjamini–Hochberg procedure based on false discovery rate (FDR) criterion was performed in order to adjust for multiple genetic comparisons.
The MB-MDR method was performed to identify the potential higher-order gene–gene interactions among the selected genetic variants [39]. The mbmdr package (version 2.6) for R (version 3.6.1) obtained from the CRAN repository was used to apply the MB-MDR method [40]. For all studied genetic variants, codominant genetic model was assumed and the second, third, and four-order variants combinations were investigated.
We used the default settings of mbmdr R package, and the genotypes combinations having the beta coefficients > 0 and p-value smaller than 0.10 were assigned to the “high-risk” group while the genotype combinations with the beta coefficients < 0 and p-value smaller than 0.10 were assigned to the “low-risk” group. In the case when p-value ≥ 0.10, genotype combinations were assigned to the “no-evidence” group. In the next step of MB-MDR analysis, the multi-locus genotypes with the same risk category were merged and a new variable with three categories (H = high risk, L = low risk, 0 = no-evidence) was created. The H and L risk groups were tested for association with odds of AML versus the remaining two groups by logistic regression with adjustment to main effects of single nucleotide polymorphisms (SNPs), age, and gender. The results were expressed by the beta regression coefficients for each risk category: βH for high-risk group and βL for high-risk group. Significance of the regression coefficients was evaluated by Wald statistics (WH and WL) adjusted for the number of genotype combinations included in each risk category. Based on WH and WL, p-values were calculated (pH and pL) and the minimum of these two values was considered as the result of statistics test for the studied interaction effect. In order to adjust for multiple testing, the statistics (defined as the maximum between WH and WL) was compared with the permutational distribution of the Wald statistic, using 1000 permutations, and the p-values obtained by permutation test were considered as corrected p-values.
All statistical tests were two-sided and significant at an estimated significance level p <0.05. All statistical analysis of data was performed in R software, version 3.6.1.
3. Results
3.1. Description of AML and Control Groups
The mean ± standard deviation of age at diagnosis in AML cases was 56.51 ± 16.10 years and 56.85 ± 15.60 for control group. There was no statistically significant difference in overall mean age between the two groups (p = 0.760). In the present study, the frequency distribution of age-group categories of AML cases was found to be 48.64% (196 cases) in the ≥60 years age group while for the control group, 54.43% (221 controls) were classified in the ≥60 years age group (p = 0.106).
Regarding gender distribution, there was a significant difference in sex distribution between these two groups (p = 0.013), male individuals being more frequent in the AML group than in the control group (215, 53.34% versus 181, 44.58%) with a male-to-female ratio of 1.14 for AML group.
Regarding AML types, 316 (78.41%) of our patients were with de novo AML, 82 (20.35%) with secondary AML (sAML) and 5 (1.24%) developed therapy related AML. According to cytogenetic risk and European Leukemia Net (ELN) 2017 risk stratification scores, 81 (20.1%) and 105 (26.05%) AML patients were low risk, 223 (55.33%) and 183 (45.41%) were intermediate risk, and 89 (22.08%) and 78 (19.35%) were high risk, respectively.
3.2. TP53 rs1042522, MDM2 rs2279744, rs3730485, and MDM4 rs4245739 Variants and Odds of AML
Distribution of TP53 rs1042522, MDM2 rs2279744, rs3730485, and MDM4 rs4245739 variants were tested for Hardy–Weinberg equilibrium (HWE) and no significant differences in the genotype frequencies were found except for TP53 rs1042522 variant (p < 0.001) in the AML group and the MDM4 rs4245739 variant in the two groups (p = 0.0004 for AML cases and p < 0.001 for controls).
The linkage disequilibrium (LD) analysis was performed both in AML cases and controls and there was a strong LD in controls (D’ = 0.89, p < 0.001) and AML group (D’ = 0.80, p < 0.001).
Distribution of the investigated variant genotypes is illustrated in Table 1. Four genetic models (codominant, dominant, recessive, and overdominant) were used to compare the distribution of genotypes between groups. The variant genotypes of TP53 rs1042522 and MDM4 rs4245739 were associated with odds of AML and they also remained significant predictors of AML risk after adjusting for gender and age-group.

Table 1.
TP53 rs1042522, MDM2 rs2279744, rs3730485, and MDM4 rs4245739 genotypes distribution.
3.3. TP53 rs1042522, MDM2 rs2279744, rs3730485, and MDM4 rs4245739 Interactions and Odds of AML
The results of epistatic pairwise interactions between studied variants are illustrated in Table 2. In recessive model, there was a significant effect of interaction between MDM2 rs2279744 and TP53 rs1042522 variants (p = 0.044 < 0.05) and between MDM4 rs4245739 and MDM2 rs3730485 variants (p = 0.035 < 0.05). The mentioned interactions were also confirmed by logistic regression, which showed that the effect of the TP53 rs1042522 variant was modified by MDM2 rs2279744. Table 3 illustrates the results of logistic regression. In subjects with a combination of the homozygous genotypes with the variant allele of MDM2 rs2279744 and TP53 rs1042522 (GG + Pro/Pro) the odds of AML was significantly increased (OR = 5.64) compared to those with wildtype and heterozygous genotype of MDM2 rs2279744 and homozygous genotype with the variant allele of TP53 rs1042522 (OR = 1.67). Regarding MDM4 rs4245739 and MDM2 rs3730485 interaction, logistic regression showed that in subjects with combined homozygous genotypes with the variant allele of MDM4 rs4245739 and MDM2 rs3730485 (AA + DD), the odds of AML was significantly decreased compared to those with wildtype and heterozygous genotype of MDM4 rs4245739 and homozygous genotype with the variant allele of MDM2 rs3730485.

Table 2.
Epistatic pairwise interactions between single nucleotide polymorphisms (SNPs) in odds of acute myeloid leukemia (AML).

Table 3.
Logistic regression results with the main effects and gene–gene interaction terms according to the recessive genetic model.
In Table 4, we illustrate the results of model-based multifactor dimensionality reduction (MB-MDR) analysis used to identify the higher-order interactions between the mentioned genetic variants and their association with odds of AML.

Table 4.
Results of the model-based multifactor dimensionality reduction method in the second-step analysis.
The synergistic effect and antagonism effect of on odds of AML are illustrated in Table 4.
The MB-MDR analysis suggested that that the four-locus model involving MDM4 rs4245739, TP53 rs1042522, MDM2 rs2279744, and MDM2 rs3730485 was significantly associated with increased odds of AML (p = 0.004) while the three-locus models involving MDM4 rs4245739, TP53 rs1042522, and MDM2 rs2279744 were significantly associated with a decreased odds of AML (p = 0.001). The results of the permutation test established the sensitivity of the findings obtained by MB-MDR analysis.
3.4. TP53 rs1042522, MDM2 rs2279744, rs3730485, and MDM4 rs4245739 Variants and Clinical Features of AML Patients
The associations between TP53 rs1042522, MDM2 rs2279744, rs3730485, and MDM4 rs4245739 variants and the clinical features of AML patients are illustrated as Supplementary Materials (Supplementary Materials Tables S1–S4. Table S1: Associations between demographic and clinical features and TP53 rs1042522 variant in codominant, dominant, and recessive models, Table S2: Associations between demographic and clinical features and MDM2 rs2279744 variant in codominant, dominant, and recessive models, Table S3: Associations between demographic and clinical features and MDM2 rs3730485 variant in codominant, dominant, and recessive models, Table S4: Associations between demographic and clinical features and MDM4 rs4245739 variant in codominant, dominant, and recessive models).
Significant associations were found between TP53 rs1042522 and ELN risk, cytogenetic risk, NPM1 mutation, and platelet (PLT) count. The post-hoc pairwise analysis performed after a significant association was found, revealed that high ELN risk, high cytogenetic risk, and homozygous variant genotype of TP53 rs1042522 (adjusted pFDR = 0.0345, respectively pFDR = 0.0016). Also, the same analysis showed an association between low PLT count (<50,000 cells/mm3) and heterozygous genotype of TP53 rs1042522 (adjusted pFDR = 0.0248). MDM2 rs2279744 variant was associated with AML type and PLT count. Post-hoc analysis also revealed that sAML was associated with wildtype and heterozygous (TT and TG) genotypes of MDM2 rs2279744 (adjusted pFDR = 0.0107) and PLT count (≥50,000 cells/mm3) was associated with homozygous genotype of the variant allele (GG) of MDM2 rs2279744 (adjusted pFDR = 0.00407). There was no statistical evidence for an association between TP53 rs1042522 and MDM2 rs2279744 variants and changes in white blood cells (WBC) count, Hemoglobin level, Hematocrit level, blasts percentage, lactate dehydrogenase (LDH) level, Eastern Cooperative Oncology Group (ECOG) performance status, treatment response, or treatment toxicity (p > 0.05). In the dominant model, the variant genotypes (II + DD) of MDM2 rs3730485 were significantly associated with treatment toxicity (p = 0.019). Other significant associations for the MDM2 rs3730485 variant were not noticed. Regarding the MDM4 rs4245739 variant, there were significant associations with age at the time of diagnosis, Hemoglobin, and Hematocrit level (p < 0.05). The post-hoc pairwise analysis revealed that wildtype (AA) and heterozygous (AC) genotypes of MDM4 rs4245739 were associated with older age at diagnosis (≥60 years) of AML patients (adjusted pFDR = 0.0780) and the homozygous genotype with the variant allele (AA) was associated with Hemoglobin level ≥ 10 (adjusted pFDR = 0.0303). Other significant associations between the MDM4 rs4245739 variant and the abovementioned clinical features were not observed.
4. Discussion
TP53 gene had an essential role in carcinogenesis and tumor progression being mutated in approximately half of human tumors. Several genetic abnormalities directly can influence the TP53 activity and p53 functions. Alternatively, TP53 activity and p53 functions may be suppressed by negative regulators such as MDM2 and MDM4 proteins (codified by MDM2, respectively MDM4 gene) [6].
TP53 rs1042522 (TP53 Arg72Pro) was reported to affect the gene and protein function [7,8] due to the fact that the Pro amino acid of p53 protein is weaker for apoptosis induction and also for suppressing cellular transformation compared to Arg amino acid [1]. MDM2 rs2279744 (MDM2 309T>G) and rs3730485 (MDM2 del1518) and MDM4 rs4245739 (MDM4 34091 C>A) variants were reported to influence the genes activity being associated with carcinogenesis and tumor progression [17,18,19,20]. Moreover, the interaction between TP53 rs1042522 and MDM2 rs2279744 [11,13,29] and between MDM2 rs2279744 and rs3730485 [21] was reported. Therefore, our study evaluated the associations between the mentioned variants and odds of developing AML and the interactions of investigated TP53, MDM2, and MDM4 variants and their association with odds of AML.
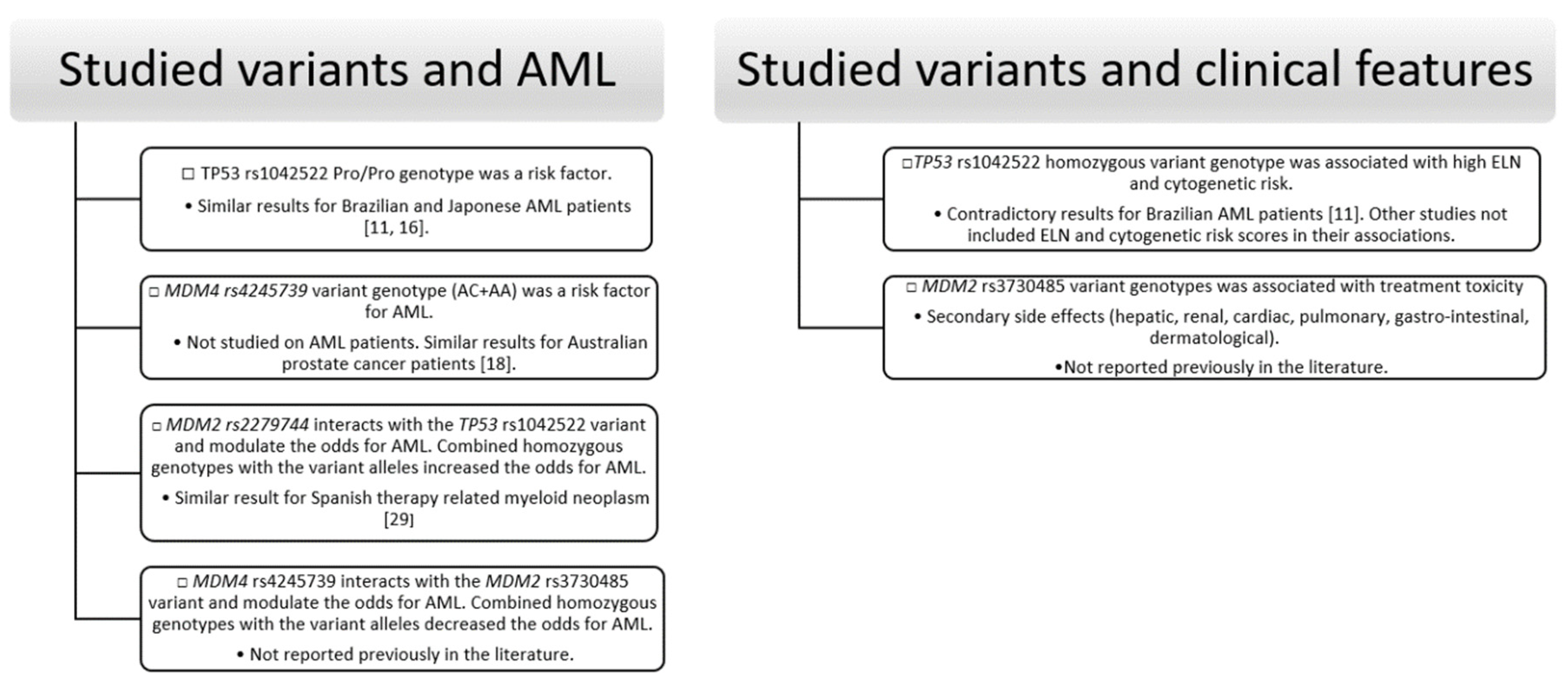
Our results showed that the variant genotypes of TP53 rs1042522 are associated with higher odds of AML. Our results regarding AML susceptibility are similar to those reported recently by Bezerra MF et al. and Dunna NR et al. [11,16]. Studies included in a meta-analysis by Tian X et al. [7], investigating the TP53 rs1042522 on AML patients [1,12,13,14,15], did not find associations between the variant genotypes of TP53 rs1042522 and AML susceptibility, except the study performed by Dunna NR et al. [16] on 141 Japanese AML patients. The contradictory results may be explained by the different ethnicity of the investigated patients (Asian, Caucasian, Brazilian) or by the small number of AML patients included. However, to the best of our knowledge, our study reports the results of the largest cohort of AML patients. Moreover, our study focused on clinical features as well as somatic mutations; and associations between the homozygous variant genotype of TP53 rs1042522 and high cytogenetic and ELN risk were also noticed. In turn, cytogenetic and ELN risk stratification scores were correlated with the outcome of AML patients [41]. An association between NPM1 mutation (which is included in ELN 2017 risk stratification) and TP53 rs1042522 was also noticed. Other associations between the investigated variants and genes mutations were not found. The study performed by Bezerra MF et al. [11] on 189 Brazilian AML patients investigated the molecular and cytogenetic risk scores according to TP53 rs1042522, but they did not find any associations. The role of TP53 rs1042522 in cancer susceptibility and progression may be explained by the fact that the p53 protein codified by the TP53 gene with wild type genotype of rs1042522, interacts more efficiently with the MDM2 protein and in consequence the apoptosis is more efficient [42]. As a consequence, (theoretically) the variant genotypes of TP53 rs1042522 might be risk factors for cancer development and/or progression. Part of published studies, such as the study performed by Furuya T et al. [42] found associations with cancer risk and progression of cancer, while others such as our study just for susceptibility.
MDM4 rs4245739 was intensively studied in different types of cancer [18,22,35,36,43,44,45]. Part of them [35,36] found no association of this variant with cancer susceptibility, while others [22,43,44,45] found an association with reduced risk of cancer, mainly of breast cancer but not only. The published results are contradictory, depending on the cancer type. For example, the study performed by Stegeman S. et al. [18] reported an association of the MDM4 rs4245739 A allele with an increased risk for prostate cancer [18]. On the other hand, Gonsmo L et al. [46] performed a study on ovarian and endometrial cancers, showing that the MDM4 rs4245739 C allele represents a risk factor only for ovarian cancer [46]. According to our knowledge, none of the published studies included leukemic patients. Our study used the latest version for allele description based on NCBI’s and Ensemble’s genome browser and we found a significant association of the variant genotypes (AC and AA) with odds of AML, the results being similar to those reported for prostate cancer by Stegeman S et al. [18]. In previously published studies, the alleles were opposite annotated (A > C). Currently, even if A allele of MDM4 rs4245739 is the ancestral allele, the C allele is considered as reference (C > A). Thus, we are unable to consider our results contradictory to those who reported associations of the variant genotypes of MDM4 rs4245739 with a reduced risk of cancer.
In our study, MDM2 rs2279744 and rs3730485 variants were not associated with AML susceptibility. Significant associations were noticed between MDM2 rs2279744 variant and sAML, high PLT count, as well as between MDM2 rs3730485 variant and treatment toxicity.
Contrary to our results regarding the AML susceptibility, the homozygous genotype with the variant allele of MDM2 rs2279744 was reported as risk factor for AML in a cohort of 231 Chinese patients [1]. In a study performed on 575 pediatric AML patients of different races, significant association between the homozygous genotype with the variant allele of MDM2 rs2279744 and AML susceptibility was noticed for Black and Hispanic races [25]. Similar results were obtained in the meta-analysis performed by He X et al. [47] where leukemia patients were included (including AML patients). Similar to our results regarding AML susceptibility, Falk I et al. [26] reported no association between MDM2 rs2279744 variant and AML risk on a cohort of 189 Swedish patients, but association between the variant genotypes and low OS was observed. They suggested that MDM2 rs2279744 variant and TP53 mutational status might be used for prognostication, risk stratification, and treatment choice [26]. The associations between the investigated MDM2 variants and cancer susceptibility and progression have a scientific substrate, being well known that these variants increase the MDM2 expression and attenuate the TP53 suppressor pathway. Recently, in other types of cancer, the variant genotypes of MDM2 rs2279744 and/or rs3730485 were reported as risk factors for breast cancer but with a trend towards a good prognosis [48], for laryngeal [49], gynecological cancers [50], and in haplotype analysis for papillary thyroid carcinoma [17].
As we mentioned, the investigated variants influence the gene activity and the investigated genes may interact. In consequence, we performed an interaction analysis for the investigated variants. Our study demonstrated that MDM2 rs2279744 interacts with the TP53 rs1042522 variant and MDM4 rs4245739 with the MDM2 rs3730485 variant. Moreover, the mentioned variant interactions were associated with odds of AML. The homozygous genotype with the variant allele of TP53 rs1042522 was associated with AML susceptibility, but combined with the homozygous genotype with the variant allele of MDM2 rs2279744 increased the odds for AML 5.64 times. The variant allele of MDM2 rs2279744 enhances the risk effect of the variant allele of TP53 rs1042522. MDM2 rs2279744 and TP53 rs1042522 interaction and their association with the risk for cancer were recently reported by Cabezas M et al. [29] in therapy-related myeloid neoplasms. Our results are explained by the fact that TP53 rs1042522 variant allele was reported to modify the p53 function and interaction with MDM2 protein [42], thus being reported by several studies as a risk factor for cancer. While, separately MDM2 rs2279744 variant allele was reported to modify the MDM2 protein function [51] also being reported by several studies as a risk factor for cancer. Our study demonstrated their combined effect to the odds for AML.
In our study, the variant genotypes of MDM4 rs4245739 were also found to be associated with AML susceptibility. Moreover, the combination of homozygous genotypes with the variant alleles of MDM4 rs4245739 and MDM2 rs3730485 decreased the odds of AML, while the combination of wildtype or heterozygous genotype of MDM4 rs4245739 with homozygous genotype with the variant allele of MDM2 rs3730485 increased the odds of AML. Our results suggesting that in subjects with homozygous genotype with the variant allele of MDM2 rs3730485 the risk effect of the variant genotypes of MDM4 rs4245739 is inverted (reversed effect).
Regarding OS, the investigated TP53 rs1042522, MDM2 rs2279744, rs3730485, and MDM4 rs4245739 variants were not significantly associated with OS (p > 0.05) in either of the genetic models. Clinical characteristics such as treatment (low dose treatment), patients’ outcome, high LDH level, high PLT count, high WBC count, high cytogenetic and ELN risk, high age (>60 years) at diagnosis, and FLT3 ITD mutation were associated with lower OS (p < 0.01), as expected. Part of the mentioned OS associations have been previously reported in the literature [2,41,52,53] and our group also reported part of these clinical associations on smaller cohorts of AML patients [31,32,53].
Briefly, our study showed association between TP53 rs1042522 and MDM4 rs4245739 variants and AML susceptibility, between TP53 rs1042522 and PLT count, NPM1 mutations (type A-D insertion), ELN, and cytogenetic risk. MDM4 rs4245739 variant was also associated with age at diagnosis, and changes in Hemoglobin and Hematocrit level. MDM2 rs2279744 variant was associated with secondary AML type and changes in PLT count and MDM2 rs3730485 with secondary (hepatic, renal, cardiac, pulmonary, gastro-intestinal, dermatological) events as a result of treatment toxicity. MB-MDR framework and logistic regression demonstrated the interaction between MDM2 rs2279744 and TP53 rs1042522 variants and between MDM4 rs4245739 and MDM2 rs3730485 variants and also their association with AML susceptibility (Figure 1).
Figure 1.
Summary diagram of significant results of the association analysis.
The novelty of our study consists of the simultaneous analysis of the four variants on a large cohort of adult AML patients. Also, according to our best knowledge, the present study is the first one to report the association between high ELN and cytogenetic risk scores and the TP53 rs1042522 variant. Moreover, even if MDM2 rs3730485 and MDM4 rs4245739 were studied in several types of cancer, none of them included AML patients. Our study also focused on identifying the interactions of the mentioned variants and their association with odds of AML. However, our study has several limitations as well. One limitation is the lack of gene expression and protein level analysis. In addition, although the statistical models containing higher-order interactions between studied variant were internally validated using permutation samples, more studies with larger number of subjects would be needed to validate our associations. It is important to notice that MDM4 rs4245739 variant was not in HWE in controls and AML group and once again this may be a demographic characteristic (considering that some unraveled modifying factors, at gene and environmental level, may be responsible). However, our frequencies of all investigates variants were similar to the allele frequencies reported by Ensembl Genome browser. Regarding other Romanian studies, similar frequencies of TP53 rs1042522 alleles were found in a case-control study where Romanian colorectal cancer patients were included [54]. Another case-control study, where Romanian and German cholangiocarcinoma patients were included, investigated TP53 rs1042522 (TP53 Arg72Pro) and MDM2 rs2279744 (MDM2 309T>G) and similar genotypes frequencies were reported [55].
5. Conclusions
Our findings provide evidence regarding the association between TP53 rs1042522, MDM4 rs4245739 variants, and AML susceptibility. A significant effect of interaction was found between MDM2 rs2279744 and TP53 rs1042522 variants and between MDM4 rs4245739 and MDM2 rs3730485 variants. The results of pairwise interactions showed that the effect of the TP53 rs1042522 variant was modified by MDM2 rs2279744, and patients with combined variant homozygous genotypes for MDM2 rs2279744 and TP53 rs1042522 have increased odds of AML. The results of MB-MDR analysis revealed significant higher-order interactions between the TP53 rs1042522, MDM2 rs2279744, rs3730485, and MDM4 rs4245739 variants. The variant genotypes of TP53 rs1042522 were significantly associated with adverse molecular and cytogenetic risk scores and also with NPM1 mutation in AML patients.
Supplementary Materials
The following are available online at https://www.mdpi.com/2077-0383/9/6/1672/s1, Table S1: Associations between demographic and clinical features and TP53 rs1042522 variant in codominant, dominant and recessive models, Table S2: Associations between demographic and clinical features and MDM2 rs2279744 variant in codominant, dominant and recessive models, Table S3: Associations between demographic and clinical features and MDM2 rs3730485 variant in codominant, dominant and recessive models, Table S4: Associations between demographic and clinical features and MDM4 rs4245739 variant in codominant, dominant and recessive models. The data used in the current study may be available upon reasonable request.
Author Contributions
Conceptualization, F.T.; data curation, F.T., A.T., G.A.C., A.B., B.B., A.C., D.D., M.C., E.L., and L.J.; formal analysis, F.T. and C.B.; funding acquisition, F.T. and C.B.; investigation, F.T., A.T., G.A.C., A.B., B.B., A.C., L.J., and C.B.; methodology, F.T., M.I., A.T., and C.B.; project administration, F.T., A.T., and C.B.; resources, A.T., D.D., M.C., E.L., and C.B.; software, M.I.; supervision, E.L. and C.B.; validation, M.I., A.T., and C.B.; visualization, F.T.; writing—original draft, F.T.; writing—review and editing, M.I., A.T., and C.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by an internal grant of the George Emil Palade University of Medicine, Pharmacy, Science, and Technology of Targu Mures, Romania, grant number 615/1/17.01.2019 and by a grant of the Romanian National Authority for Scientific Research and Innovation, CNCS/CCCDI—UEFISCDI, project no. PN—III—P2—2.1—PED—2016—1076 within PNCDI III, contract no. 147 PED/2017.
Acknowledgments
Part of this work was performed using the infrastructure of Center for Advanced Medical and Pharmaceutical Research of the “George Emil Palade” University of Medicine, Pharmacy, Science, and Technology of Targu Mures, Romania.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Xiong, X.; Wang, M.; Wang, L.; Liu, J.; Zhao, X.; Tian, Z.; Wang, J. Risk of MDM2 SNP309 alone or in combination with the p53 codon 72 polymorphism in acute myeloid leukemia. Leuk. Res. 2009, 33, 1454–1458. [Google Scholar] [CrossRef]
- Megías-Vericat, J.E.; Fernández, P.M.; Herrero, M.J.; Boso, V.; Martínez-Cuadrón, D.; Poveda, J.L.; Sanz, M.A.; Aliño, S.F. Pharmacogenomics and the treatment of acute myeloid leukemia. Pharmacogenomics 2016, 17, 1245–1272. [Google Scholar] [CrossRef]
- Tan, B.X.; Khoo, K.H.; Lim, T.M.; Lane, D.P. High Mdm4 levels suppress p53 activity and enhance its half-life in acute myeloid leukaemia. Oncotarget 2014, 5, 933–943. [Google Scholar] [CrossRef]
- Li, L.; Tan, Y.; Chen, X.; Xu, Z.; Yang, S.; Ren, F.; Guo, H.; Wang, X.; Chen, Y.; Li, G.; et al. MDM4 Overexpressed in Acute Myeloid Leukemia Patients with Complex Karyotype and Wild-Type TP53. PLoS ONE 2014, 9, e113088. [Google Scholar] [CrossRef] [PubMed]
- Rayburn, E.; Zhang, R.; He, J.; Wang, H. MDM2 and human malignancies: Expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr. Cancer Drug Targets 2005, 5, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lozano, G. Molecular pathways: Targeting Mdm2 and Mdm4 in cancer therapy. Clin. Cancer Res. 2013, 19, 34–41. [Google Scholar] [CrossRef]
- Tian, X.; Dai, S.; Sun, J.; Jiang, S.; Jiang, Y. Association between TP53 Arg72Pro polymorphism and leukemia risk: A meta-analysis of 14 case-control studies. Sci. Rep. 2016, 6, 24097. [Google Scholar] [CrossRef]
- Pim, D.; Banks, L. p53 polymorphic variants at codon 72 exert different effects on cell cycle progression. Int. J. Cancer 2004, 108, 196–199. [Google Scholar] [CrossRef]
- Megías-Vericat, J.E.; Martínez-Cuadrón, D.; Sanz, M.Á.; Poveda, J.L.; Montesinos, P. Daunorubicin and cytarabine for certain types of poor-prognosis acute myeloid leukemia: A systematic literature review. Expert Rev. Clin. Pharmacol. 2019, 12, 197–218. [Google Scholar] [CrossRef]
- Dumont, P.; Leu, J.I.-J.; Della Pietra, A.C., 3rd; George, D.L.; Murphy, M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat. Genet. 2003, 33, 357–365. [Google Scholar] [CrossRef]
- Bezerra, M.F.; Coelho-Silva, J.L.; Nascimento, J.C.; Benicio, M.T.; Rocha, C.R.; Machado, C.G.; Rego, E.M.; Bezerra, M.A.; Lucena-Araujo, A.R.; Beltrão, E.I. Association between the TP53 Arg72Pro polymorphism and clinical outcomes in acute myeloid leukemia. Haematologica 2017, 102, e43–e46. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Naoe, T.; Kiyoi, H.; Kunishima, S.; Minami, S.; Miyawaki, S.; Asou, N.; Kuriyama, K.; Saito, H.; Ohno, R. Poor clinical significance of p53 gene polymorphism in acute myeloid leukemia. Leuk. Res. 2000, 24, 349–352. [Google Scholar] [CrossRef]
- Ellis, N.A.; Huo, D.; Yildiz, O.; Worrillow, L.J.; Banerjee, M.; Le Beau, M.M.; Larson, R.A.; Allan, J.M.; Onel, K. MDM2 SNP309 and TP53 Arg72Pro interact to alter therapy-related acute myeloid leukemia susceptibility. Blood 2008, 112, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.-Y.; Ren, Z.-H.; Jiao, B.; Xiao, R.; Yun, H.-Y.; Chen, B.; Zhao, W.-L.; Zhu, Q.; Chen, Z.; Chen, S. Genetic variations of DNA repair genes and their prognostic significance in patients with acute myeloid leukemia. Int. J. Cancer 2011, 128, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, P.S.; Ihsan, R.; Mishra, A.K.; Yadav, D.S.; Saluja, S.; Mittal, V.; Saxena, S.; Kapur, S. High order interactions of xenobiotic metabolizing genes and P53 codon 72 polymorphisms in acute leukemia. Environ. Mol. Mutagen. 2012, 53, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Dunna, N.R.; Vure, S.; Sailaja, K.; Surekha, D.; Raghunadharao, D.; Rajappa, S.; Satti, V. TP53 codon 72 polymorphism and risk of acute leukemia. Asian Pac. J. Cancer Prev. 2012, 13, 347–350. [Google Scholar] [CrossRef]
- Maruei-Milan, R.; Heidari, Z.; Salimi, S. Role of MDM2 309T>G (rs2279744) and I/D (rs3730485) polymorphisms and haplotypes in risk of papillary thyroid carcinoma, tumor stage, tumor size, and early onset of tumor: A case control study. J. Cell. Physiol. 2019, 234, 12934–12940. [Google Scholar] [CrossRef]
- Stegeman, S.; Moya, L.; Selth, L.A.; Spurdle, A.B.; Clements, J.A.; Batra, J. A genetic variant of MDM4 influences regulation by multiple microRNAs in prostate cancer. Endocrine-Related Cancer 2015, 22, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Moazeni-Roodi, A.; Ghavami, S.; Hashemi, M. The 40bp indel polymorphism of MDM2 increase the risk of cancer: An updated meta-analysis. Mol. Biol. Res. Commun 2019, 8, 1–8. [Google Scholar] [PubMed]
- Gansmo, L.B.; Vatten, L.; Romundstad, P.; Hveem, K.; Ryan, B.M.; Harris, C.C.; Knappskog, S.; Lønning, P.E. Associations between the MDM2 promoter P1 polymorphism del1518 (rs3730485) and incidence of cancer of the breast, lung, colon and prostate. Oncotarget 2016, 7, 28637–28646. [Google Scholar] [CrossRef] [PubMed]
- Gansmo, L.B.; Bjørnslett, M.; Halle, M.K.; Salvesen, H.B.; Romundstad, P.; Hveem, K.; Vatten, L.; Dørum, A.; Lønning, P.E.; Knappskog, S. MDM2 promoter polymorphism del1518 (rs3730485) and its impact on endometrial and ovarian cancer risk. BMC Cancer 2017, 17, 97. [Google Scholar] [CrossRef] [PubMed]
- Lian, T.; Zhu, J.; He, J.; Li, C.; Tang, R.; Jiang, L.; Qi, T.; Ke, L.; Liu, R.; Wu, B. The associations between MDM4 rs4245739 A>C polymorphism and cancer risk: A meta-analysis. Int. J. Clin. Exp. Med. 2019, 12, 10411–10421. [Google Scholar]
- Hua, W.; Zhang, A.; Duan, P.; Zhu, J.; Zhao, Y.; He, J.; Zhang, Z. MDM2 promoter del1518 polymorphism and cancer risk: Evidence from 22,931 subjects. OncoTargets Ther. 2017, 10, 3773–3780. [Google Scholar] [CrossRef]
- Soleymannejad, M.; Sheikhha, M.H.; Neamatzadeh, H. Association of Mouse Double Minute 2 -309T>G Polymorphism with Acute Myeloid Leukemia in an Iranian Population: A Case- Control Study. Asian Pac. J. Cancer Prev. 2019, 20, 3037–3041. [Google Scholar] [CrossRef] [PubMed]
- Phillips, C.L.; Gerbing, R.; Alonzo, T.; Perentesis, J.P.; Harley, I.T.; Meshinchi, S.; Bhatla, D.; Radloff, G.; Davies, S.M. MDM2 polymorphism increases susceptibility to childhood acute myeloid leukemia: A report from the Children’s Oncology Group. Pediatr. Blood Cancer 2010, 55, 248–253. [Google Scholar] [CrossRef][Green Version]
- Falk, I.J.; Willander, K.; Chaireti, R.; Lund, J.; Nahi, H.H.; Hermanson, M.; Gréen, H.; Lotfi, K.; Söderkvist, P. TP53 mutations and MDM2 SNP309 identify subgroups of AML patients with impaired outcome. Eur. J. Haematol. 2014, 94, 355–362. [Google Scholar] [CrossRef]
- Hamid, T.M.A.; El Gammal, M.M.; Eibead, G.T.; Saber, M.M.; Elazm, O.M.A.; Ibead, G.T.; Azm, O.M.A. Clinical impact of SNP of P53 genes pathway on the adult AML patients. Hematology 2015, 20, 328–335. [Google Scholar] [CrossRef]
- Gryshchenko, I.; Hofbauer, S.; Stoecher, M.; Daniel, P.T.; Steurer, M.; Gaiger, A.; Eigenberger, K.; Greil, R.; Tinhofer, I. MDM2 SNP309 Is Associated with Poor Outcome in B-Cell Chronic Lymphocytic Leukemia. J. Clin. Oncol. 2008, 26, 2252–2257. [Google Scholar] [CrossRef]
- Cabezas, M.; García-Quevedo, L.; Alonso, C.; Manubens, M.; Álvarez, Y.; Barquinero, J.-F.; Cajal, S.R.Y.; Ortega, M.; Blanco, A.; Caballín, M.R.; et al. Polymorphisms in MDM2 and TP53 Genes and Risk of Developing Therapy-Related Myeloid Neoplasms. Sci. Rep. 2019, 9, 150. [Google Scholar] [CrossRef]
- Ruminy, P.; Marchand, V.; Buchbinder, N.; Larson, T.; Joly, B.; Penther, D.; Lemasle, E.; Lepretre, S.; Angot, E.; Mareschal, S.; et al. Multiplexed targeted sequencing of recurrent fusion genes in acute leukaemia. Leukemia 2016, 30, 757–760. [Google Scholar] [CrossRef]
- Bănescu, C.; Tripon, F.; Trifa, A.P.; Crauciuc, A.G.; Bogliș, A.; Lazar, E.; Dima, D.; Macarie, I.; Duicu, C.; Iancu, M. Presence of copy number aberration and clinical prognostic factors in patients with acute myeloid leukemia: An analysis of effect modification. Pol. Arch. Intern. Med. 2019, 129, 898–906. [Google Scholar] [CrossRef] [PubMed]
- Tripon, F.; Iancu, M.; Trifa, A.; Crauciuc, G.A.; Boglis, A.; Dima, D.; Lazar, E.; Banescu, C. Modelling the Effects of MCM7 Variants, Somatic Mutations, and Clinical Features on Acute Myeloid Leukemia Susceptibility and Prognosis. J. Clin. Med. 2020, 9, 158. [Google Scholar] [CrossRef] [PubMed]
- Banescu, C.; Tripon, F.; Trifa, A.P.; Crauciuc, A.G.; Moldovan, V.G.; Bogliş, A.; Benedek, I.; Dima, D.; Cândea, M.; Duicu, C.; et al. Cytokine rs361525, rs1800750, rs1800629, rs1800896, rs1800872, rs1800795, rs1800470, and rs2430561 SNPs in relation with prognostic factors in acute myeloid leukemia. Cancer Med. 2019, 8, 5492–5506. [Google Scholar] [CrossRef]
- Tripon, F.; Crauciuc, G.A.; Moldovan, V.G.; Bogliș, A.; Benedek, I.J.; Lázár, E.; Banescu, C. Simultaneous FLT3, NPM1 and DNMT3A mutations in adult patients with acute myeloid leukemia—Case study. Rev. Romana Med. Lab. 2019, 27, 245–254. [Google Scholar] [CrossRef]
- Pedram, N.; Pouladi, N.; Feizi, M.A.; Montazeri, V.; Sakhinia, E.; Estiar, M.A. Analysis of the Association between MDM4 rs4245739 Single Nucleotide Polymorphism and Breast Cancer Susceptibility. Clin. Lab. 2016, 62, 1303–1308. [Google Scholar] [CrossRef]
- Khanlou, Z.M.; Pouladi, N.; Feizi, M.H.; Pedram, N. Lack of Associations of the MDM4 rs4245739 Polymorphism with Risk of Thyroid Cancer among Iranian-Azeri Patients: A Case-Control Study. Asian Pac. J. Cancer Prev. 2017, 18, 1133–1138. [Google Scholar]
- Omrani-Nava, V.; Hedayatizadeh-Omran, A.; Alizadeh-Navaei, R.; Mokhberi, V.; Jalalian, R.; Janbabaei, G.; Amjadi, O.; Rahmatpour, G.; Mozaffari, A. TP53 single nucleotide polymorphism (rs1042522) in Iranian patients with coronary artery disease. Biomed. Rep. 2018, 9, 259–265. [Google Scholar] [CrossRef]
- Dong, N.; Gao, X.; Zhu, Z.; Yu, Q.; Bian, S.; Gao, Y. A 40-bp insertion/deletion polymorphism in the constitutive promoter of MDM2 confers risk for hepatocellular carcinoma in a Chinese population. Gene 2012, 497, 66–70. [Google Scholar] [CrossRef]
- Moore, J.H.; Gilbert, J.C.; Tsai, C.-T.; Chiang, F.-T.; Holden, T.; Barney, N.; White, B. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J. Theor. Biol. 2006, 241, 252–261. [Google Scholar] [CrossRef]
- Urrea, V.; Calle, M.; Van Steen, K.; Malats, N. mbmdr: Model Based Multifactor Dimensionality Reduction. R Package Version 2.6. Available online: https://CRAN.R-project.org/package=mbmdr (accessed on 13 March 2020).
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef]
- Furuya, T.K.; Tomitão, M.T.P.; Camacho, L.C.C.; Ramos, M.F.K.P.; Eluf-Neto, J.; Alves, V.A.F.; Zilberstein, B.; Cecconello, I.; Ribeiro, U., Jr.; Chammas, R. Association between Polymorphisms in Inflammatory Response-Related Genes and the Susceptibility, Progression and Prognosis of the Diffuse Histological Subtype of Gastric Cancer. Genes 2018, 9, 631. [Google Scholar] [CrossRef]
- Gansmo, L.B.; Romundstad, P.R.; Birkeland, E.; Hveem, K.; Vatten, L.; Knappskog, S.; Lønning, P.E. MDM4 SNP34091 (rs4245739) and its effect on breast-, colon-, lung-, and prostate cancer risk. Cancer Med. 2015, 4, 1901–1907. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhu, J.; Fu, W.; Liang, Z.; Song, S.; Zhao, Y.; Lyu, L.; Zhang, A.; He, J.; Duan, P. MDM4 rs4245739 A > C polymorphism correlates with reduced overall cancer risk in a meta-analysis of 69477 subjects. Oncotarget 2016, 7, 71718–71726. [Google Scholar] [CrossRef]
- Zhai, Y.; Dai, Z.; He, H.; Gao, F.; Yang, L.; Dong, Y.; Lu, J. A PRISMA-compliant meta-analysis of MDM4 genetic variants and cancer susceptibility. Oncotarget 2016, 7, 73935–73944. [Google Scholar] [CrossRef]
- Gansmo, L.B.; Bjørnslett, M.; Halle, M.K.; Salvesen, H.B.; Dørum, A.; Birkeland, E.; Hveem, K.; Romundstad, P.; Vatten, L.; Lønning, P.E.; et al. The MDM4 SNP34091 (rs4245739) C-allele is associated with increased risk of ovarian-but not endometrial cancer. Tumour Biol. 2016, 37, 10697–10702. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Chen, P.; Yang, K.; Liu, B.; Zhang, Y.; Wang, F.; Guo, Z.; Liu, X.; Lou, J.; Chen, H. Association of MDM2 Polymorphism with Risk and Prognosis of Leukemia: A Meta-Analysis. Acta Haematol. 2015, 133, 365–371. [Google Scholar] [CrossRef]
- Miedl, H.; Lebhard, J.; Ehart, L.; Schreiber, M. Association of the MDM2 SNP285 and SNP309 Genetic Variants with the Risk, Age at Onset and Prognosis of Breast Cancer in Central European Women: A Hospital-Based Case-Control Study. Int. J. Mol. Sci. 2019, 20, 509. [Google Scholar] [CrossRef]
- Fernández-Mateos, J.; Seijas-Tamayo, R.; Klain, J.C.A.; Borgoñón, M.P.; Pérez-Ruiz, E.; Mesía, R.; Del Barco, E.; Coloma, C.S.; Dominguez, A.R.; Daroqui, J.C.; et al. Genetic Susceptibility in Head and Neck Squamous Cell Carcinoma in a Spanish Population. Cancers 2019, 11, 493. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Y.; Zhang, Z. Association of rs2279744 and rs117039649 promoter polymorphism with the risk of gynecological cancer A meta-analysis of case–control studies. Medicine (Baltimore) 2018, 97, e9554. [Google Scholar] [CrossRef]
- Ma, Y.; Bian, J.; Cao, H. MDM2 SNP309 rs2279744 Polymorphism and Gastric Cancer Risk: A Meta-Analysis. PLoS ONE 2013, 8, e56918. [Google Scholar] [CrossRef]
- Antohe, I.; Dascalescu, A.; Danaïla, C.; Zlei, M.; Ivanov, I.; Sireteanu, A.; Boca, O.; Oana, R.; Cianga, P. FLT-3 ITD Positive Acute Basophilic Leukemia with Rare Complex Karyotype Presenting with Acute Respiratory Failure: Case Report. Rev. Romana de Med. de Lab. 2018, 26, 87–94. [Google Scholar] [CrossRef]
- Bănescu, C.; Skrypnyk, C. The Value of FLT3, NPM1 and DNMT3A Gene Mutation Analysis in Acute Myeloid Leukemia Diagnosis. Rev. Romana de Med. de Lab. 2019, 27, 239–243. [Google Scholar] [CrossRef]
- Murarasu, D.; Puiu, L.; Mihalcea, C.E.; Pitica, A.; Madelina, I.; Mambet, C.; Elena, R.; Matei, L.; Dragu, D.; Simion, L.; et al. Characterization of TP53 polymorphisms in Romanian colorectal cancer patients. Rom. Biotechnol. Lett. 2018, 23, 14124–14134. [Google Scholar] [CrossRef]
- Zimmer, V.; Hoblinger, A.; Mihalache, F.; Assmann, G.; Acalovschi, M.; Lammert, F. Potential genotype-specific single nucleotide polymorphism interaction of common variation in p53 and its negative regulator mdm2 in cholangiocarcinoma susceptibility. Oncol. Lett. 2012, 4, 101–106. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).