CD36 and GPR120 Methylation Associates with Orosensory Detection Thresholds for Fat and Bitter in Algerian Young Obese Children
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants and Study Design
2.2. Preferred Eating Pattern
2.3. Orosensory Detection of Oleic Acid (OA) Thresholds
2.4. Bitter Taste Perception Sensitivity
2.5. DNA Extraction and Conversion
2.6. Methylation-Specific PCR (MS-PCR)
2.7. Statistical Analysis
3. Results
3.1. Clinical and Biochemical Characteristics of the Participants
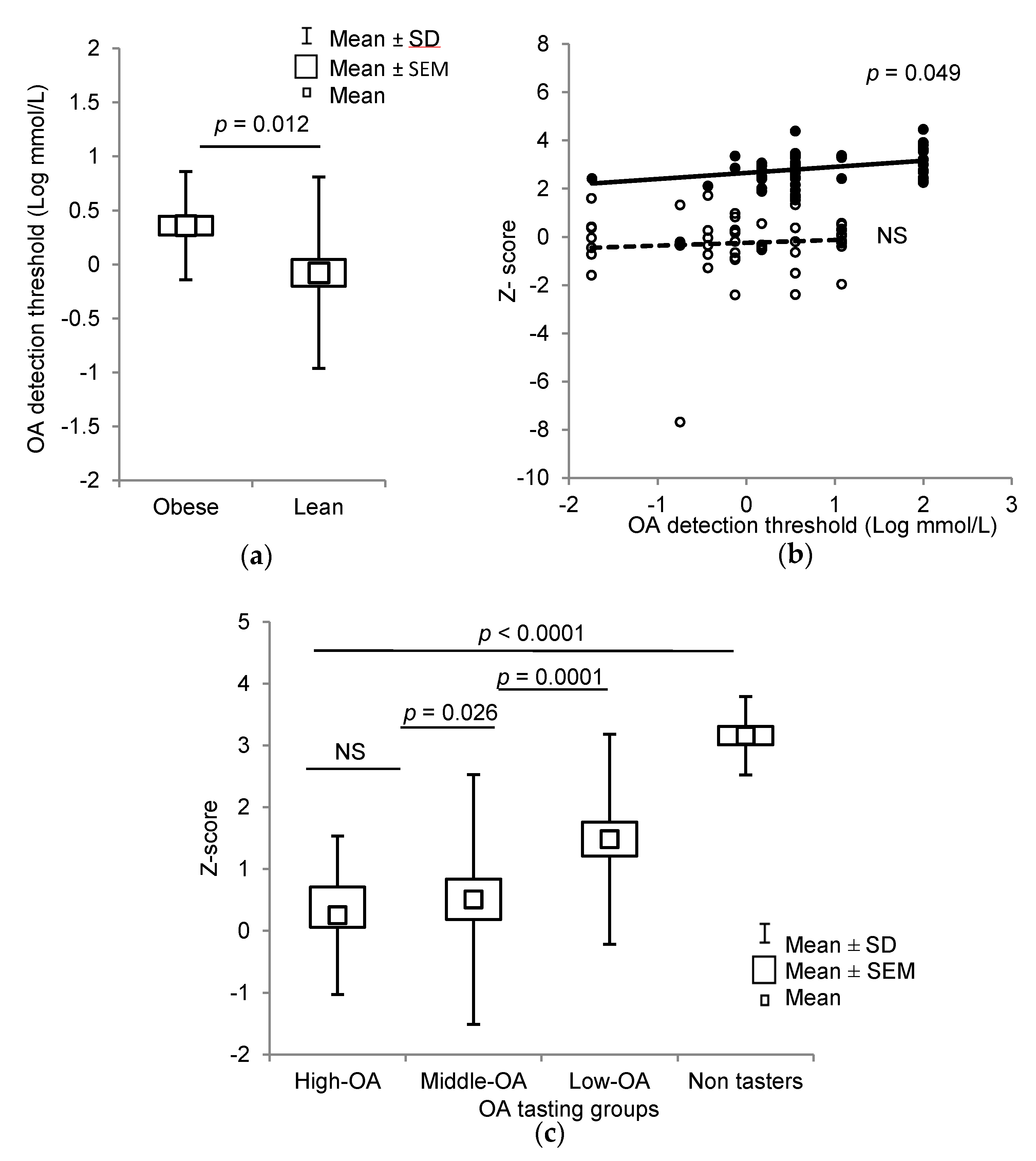
3.2. Orosensory Perceptions of Oleic Acid and PROP
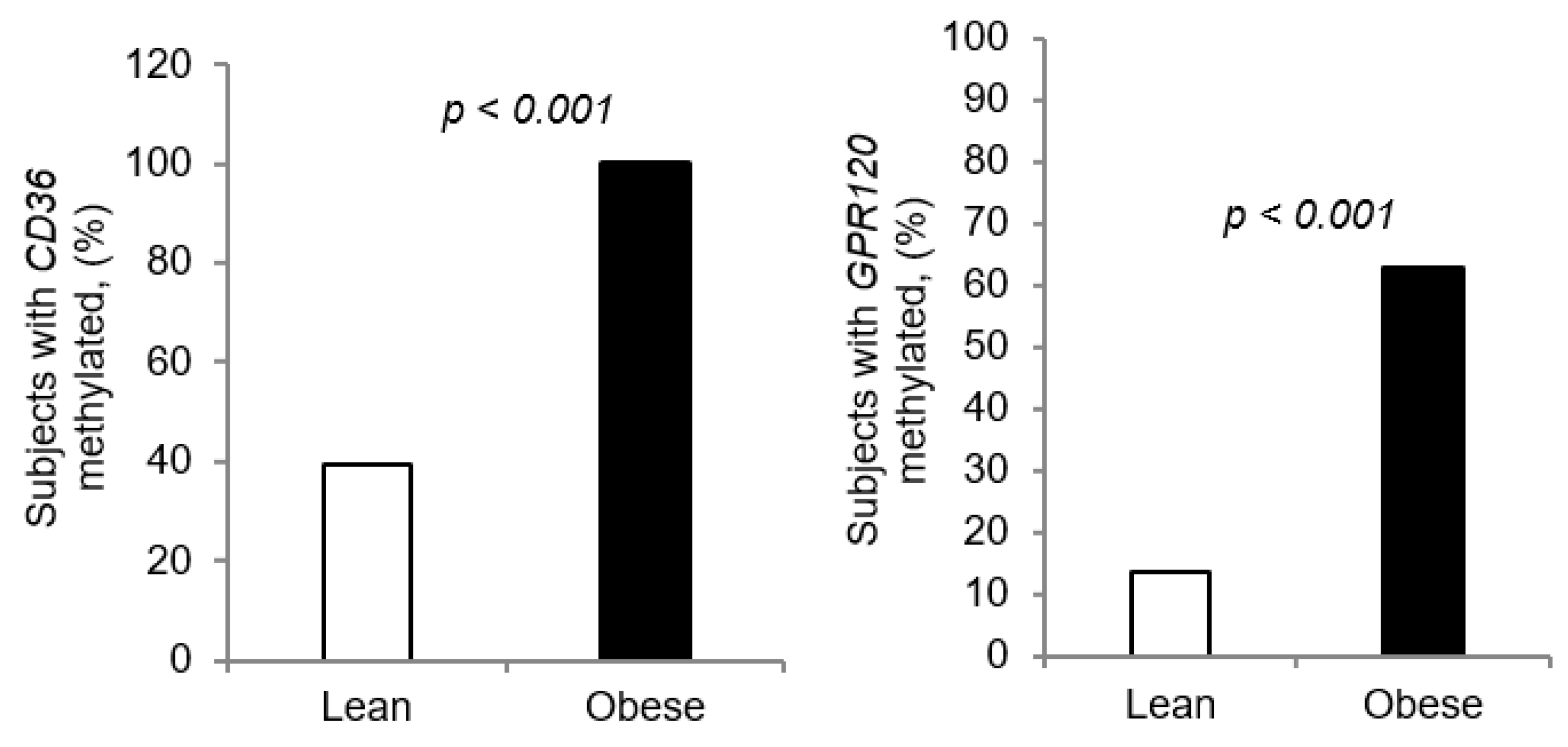
3.3. Methylation of CpG Islands of CD36 and GPR120 Promoters
3.4. Preferred Eating Pattern in Obese Children
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Parameters | Lean Children 1 (n = 51) | Obese Children 1 (n = 51) | p Values 2 |
| Age (years) | 9.31 ± 0.86 | 8.87 ± 1.39 | 0.05 |
| z-score Glycaemia (mg/dL) | −0.25 ± 1.42 0.77 ± 0.1 | 2.89 ± 0.61 0.77 ± 0.10 | <0.001 0.83 |
| Cholesterol (g/L) | 0.56 ± 0.18 | 0.83 ± 0.34 | <0.001 |
| Insulin (pmol/L) | 3.77 ± 0.01 | 3.80 ± 0.04 | <0.001 |
| Triglycerides (g/L) LDL-C (mmol/L) HDL-C (mmol/L) | 1.39 ± 0.22 2.53 ± 0.61 1.10 ± 0.32 | 1.38 ± 0.26 2.60 ± 0.50 1.24 ± 0.42 | 0.85 0.80 0.84 |
| Genes Promoter Regions | Serum Glucose (g/L) | Serum Triglycerides (g/L) | Serum Cholesterol (g/L) | Serum Insulin | z-Score | OA Perception Threshold | PROP Perception Threshold | Calories Intake/Day |
|---|---|---|---|---|---|---|---|---|
| Methylated CD36 | 0.145 p = 0.146 | −0.039 p = 0.516 | 0.252 p = 0.007 | 0.260 p = 0.015 | 0.610 p < 0.001 | 0.407 p < 0.001 | 0.132 p = 0.185 | 0.628 p < 0.01 |
| Methylated GPR120 | −0.067 p = 0.402 | −0.135 p = 0.980 | 0.285 p = 0.004 | 0.211 p = 0.50 | 0.442 p < 0.001 | 0.265 p = 0.007 | 0.163 p = 0.103 | 0.520 p < 0.01 |
References
- Afshin, A. Health Effects of Overweight and Obesity in 195 Countries over 25 Years. New Engl. J. Med. 2017, 377, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Lew, E.A.; Garfinkel, L. Variations in mortality by weight among 750,000 men and women. J. Chronic Dis. 1979, 32, 563–576. [Google Scholar] [CrossRef]
- Rivera-Dommarco, J.; Barquera, S.; Campirano, F.; Campos-Nonato, I.; Safdie, M.; Tovar, V. Epidemiological and nutritional transition in Mexico: Rapid increase of non-communicable chronic diseases and obesity. Public Heal. Nutr. 2002, 5, 113–122. [Google Scholar] [CrossRef]
- Popkin, B.M.; Adair, L.S.; Ng, S.W. Global nutrition transition and the pandemic of obesity in developing countries. Nutr. Rev. 2012, 70, 3–21. [Google Scholar] [CrossRef] [Green Version]
- Flegal, K.M.; Carroll, M.; Ogden, C.L.; Curtin, L.R. Prevalence and Trends in Obesity Among US Adults, 1999–2008. JAMA 2010, 303, 235. [Google Scholar] [CrossRef] [Green Version]
- Reilly, J.J.; Kelly, J. Long-term impact of overweight and obesity in childhood and adolescence on morbidity and premature mortality in adulthood: Systematic review. Int. J. Obes. 2010, 35, 891–898. [Google Scholar] [CrossRef] [Green Version]
- Daniels, S.R.; Arnett, D.K.; Eckel, R.H.; Gidding, S.S.; Hayman, L.L.; Kumanyika, S.; Robinson, T.N.; Scott, B.J.; Jeor, S.S.; Williams, C.L. Overweight in Children and Adolescents. Circulation 2005, 111, 1999–2012. [Google Scholar] [CrossRef] [Green Version]
- Chaudhari, N.; Roper, S. The cell biology of taste. J. Cell Boil. 2010, 190, 285–296. [Google Scholar] [CrossRef] [Green Version]
- Gilbertson, T.; Khan, N.A. Cell signaling mechanisms of oro-gustatory detection of dietary fat: Advances and challenges. Prog. Lipid Res. 2014, 53, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Ozdener, M.H.; Subramaniam, S.; Sundaresan, S.; Šerý, O.; Hashimoto, T.; Asakawa, Y.; Besnard, P.; Abumrad, N.A.; Khan, N.A. CD36- and GPR120-mediated Ca2+ signaling in human taste bud cells mediates differential responses to fatty acids and is altered in obese mice. Gastroenterology 2014, 146, 995–1005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Besnard, P.; Passilly-Degrace, P.; Khan, N.A. Taste of Fat: A Sixth Taste Modality? Physiol. Rev. 2016, 96, 151–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, A.S.; Keast, R.; Khan, N.A. Preference for dietary fat: From detection to disease. Prog. Lipid Res. 2020, 78, 101032. [Google Scholar] [CrossRef] [PubMed]
- Sayed, A.; Šerý, O.; Plesník, J.; Daoudi, H.; Rouabah, A.; Rouabah, L.; Khan, N.A.; Hadjer, D. CD36 AA genotype is associated with decreased lipid taste perception in young obese, but not lean, children. Int. J. Obes. 2015, 39, 920–924. [Google Scholar] [CrossRef] [PubMed]
- Daoudi, H.; Plesník, J.; Sayed, A.; Šerý, O.; Rouabah, A.; Rouabah, L.; Khan, N.A. Oral Fat Sensing and CD36 Gene Polymorphism in Algerian Lean and Obese Teenagers. Nutrients 2015, 7, 9096–9104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mrizak, I.; Šerý, O.; Plesník, J.; Arfa, A.; Fekih, M.; Bouslema, A.; Zaouali, M.; Tabka, Z.; Khan, N.A. The A allele of cluster of differentiation 36 (CD36) SNP 1761667 associates with decreased lipid taste perception in obese Tunisian women. Br. J. Nutr. 2015, 113, 1330–1337. [Google Scholar] [CrossRef] [Green Version]
- Pepino, M.Y.; Love-Gregory, L.; Klein, S.; Abumrad, N.A. The fatty acid translocase gene CD36 and lingual lipase influence oral sensitivity to fat in obese subjects. J. Lipid Res. 2011, 53, 561–566. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.S.; Murtaza, B.; Hichami, A.; Khan, N.A. A crosstalk between fat and bitter taste modalities. Biochimie 2019, 159, 3–8. [Google Scholar] [CrossRef]
- Khan, A.S.; Hichami, A.; Khan, N.A. Taste perception and its effects on oral nutritional supplements in younger life phases. Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 411–415. [Google Scholar] [CrossRef]
- Melis, M.; Sollai, G.; Muroni, P.; Crnjar, R.; Barbarossa, I.T. Associations between Orosensory Perception of Oleic Acid, the Common Single Nucleotide Polymorphisms (rs1761667 and rs1527483) in the CD36 Gene, and 6-n-Propylthiouracil (PROP) Tasting. Nutrients 2015, 7, 2068–2084. [Google Scholar] [CrossRef] [Green Version]
- Karmous, I.; Plesník, J.; Khan, A.S.; Šerý, O.; Abid, A.; Mankai, A.; Aouidet, A.; Khan, N.A. Orosensory detection of bitter in fat-taster healthy and obese participants: Genetic polymorphism of CD36 and TAS2R38. Clin. Nutr. 2018, 37, 313–320. [Google Scholar] [CrossRef]
- Bird, A. Perceptions of epigenetics. Nature 2007, 447, 396–398. [Google Scholar] [CrossRef]
- Feinberg, A.P.; Tycko, B. The history of cancer epigenetics. Nat. Rev. Cancer 2004, 4, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, M. DNA methylation in cancer: Too much, but also too little. Oncogene 2002, 21, 5400–5413. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; Lian, T.; Gu, C.; Yu, K.; Gao, Y.Q.; Su, X.-D. The effects of cytosine methylation on general transcription factors. Sci. Rep. 2016, 6, 29119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caiafa, P.; Zampieri, M. DNA methylation and chromatin structure: The puzzling CpG islands. J. Cell. Biochem. 2005, 94, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Sawan, C.; Vaissière, T.; Murr, R.; Herceg, Z. Epigenetic drivers and genetic passengers on the road to cancer. Mutat. Res. Mol. Mech. Mutagen. 2008, 642, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.-L.; Yu, J.; Zhang, H.-Y.; Sun, M.-H.; Gu, J.; Du, X.; Shi, D.-R.; Wang, P.; Yang, Z.-H.; Zhu, J.-D. Methylation profile of the promoter CpG islands of 31 genes that may contribute to colorectal carcinogenesis. World J. Gastroenterol. 2004, 10, 3441–3454. [Google Scholar] [CrossRef]
- García-Cardona, M.C.; Huang, F.; García-Vivas, J.M.; López-Camarillo, C.; Navarro, B.E.D.R.; Olivos, E.N.; Hong-Chong, E.; Bolaños-Jiménez, F.; A Marchat, L. DNA methylation of leptin and adiponectin promoters in children is reduced by the combined presence of obesity and insulin resistance. Int. J. Obes. 2014, 38, 1457–1465. [Google Scholar] [CrossRef]
- Keller, M.; Hopp, L.; Liu, X.; Wohland, T.; Rohde, K.; Cancello, R.; Klös, M.; Bacos, K.; Kern, M.; Eichelmann, F.; et al. Genome-wide DNA promoter methylation and transcriptome analysis in human adipose tissue unravels novel candidate genes for obesity. Mol. Metab. 2016, 6, 86–100. [Google Scholar] [CrossRef]
- Díaz, M.; García, C.; Sebastiani, G.; López-Bermejo, A.; Ibáñez, L.; de Zegher, F. Placental and Cord Blood Methylation of Genes Involved in Energy Homeostasis: Association with Fetal Growth and Neonatal Body Composition. Diabetes 2016, 66, 779–784. [Google Scholar] [CrossRef] [Green Version]
- Mennella, J.A.; Pepino, M.Y.; Reed, D.R. Genetic and environmental determinants of bitter perception and sweet preferences. Pediatrics 2005, 115, e216–e222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liman, E.R. Salty Taste: From Transduction to Transmitter Release, Hold the Calcium. Neuron 2020, 106, 709–711. [Google Scholar] [CrossRef] [PubMed]
- Anliker, J.A.; Bartoshuk, L.; Ferris, A.M.; Hooks, L.D. Children’s food preferences and genetic sensitivity to the bitter taste of 6-n-propylthiouracil (PROP). Am. J. Clin. Nutr. 1991, 5, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Bolhuis, D.; Costanzo, A.; Newman, L.P.; Keast, R. Salt Promotes Passive Overconsumption of Dietary Fat in Humans. J. Nutr. 2015, 146, 838–845. [Google Scholar] [CrossRef] [Green Version]
- Li, L.-C.; Dahiya, R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics 2002, 18, 1427–1431. [Google Scholar] [CrossRef] [Green Version]
- Vanderwall, C.; Eickhoff, J.C.; Clark, R.R.; Carrel, A.L. BMI z-score in obese children is a poor predictor of adiposity changes over time. BMC Pediatr. 2018, 18, 187. [Google Scholar] [CrossRef] [Green Version]
- Haryono, R.Y.; Sprajcer, M.A.; Keast, R. Measuring oral fatty acid thresholds, fat perception, fatty food liking, and papillae density in humans. J. Vis. Exp. 2014, e51236. [Google Scholar] [CrossRef] [Green Version]
- Chevrot, M.; Passilly-Degrace, P.; Ancel, D.; Bernard, A.; Enderli, G.; Gomes, M.; Robin, I.; Issanchou, S.; Vergès, B.; Nicklaus, S.; et al. Obesity interferes with the orosensory detection of long-chain fatty acids in humans. Am. J. Clin. Nutr. 2014, 99, 975–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tepper, B.J.; Koelliker, Y.; Zhao, L.; Ullrich, N.V.; Lanzara, C.; D’Adamo, P.; Ferrara, A.; Ulivi, S.; Esposito, L.; Gasparini, P.; et al. Variation in the Bitter-taste Receptor GeneTAS2R38, and Adiposity in a Genetically Isolated Population in Southern Italy. Obesity 2008, 16, 2289–2295. [Google Scholar] [CrossRef] [PubMed]
- Pittman, D.; Labban, C.E.; Anderson, A.A.; O’Connor, H.E. Linoleic and Oleic Acids Alter the Licking Responses to Sweet, Salt, Sour, and Bitter Tastants in Rats. Chem. Senses 2006, 31, 835–843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattes, R.D. Effects of linoleic acid on sweet, sour, salty, and bitter taste thresholds and intensity ratings of adults. Am. J. Physiol. Liver Physiol. 2007, 292, G1243–G1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, A.; Owyang, C. Sugars, Sweet Taste Receptors, and Brain Responses. Nutrients 2017, 9, 653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dotson, C.D.; Zhang, L.; Xu, H.; Shin, Y.-K.; Vigues, S.; Ott, S.H.; Elson, A.; Choi, H.J.; Shaw, H.; Egan, J.M.; et al. Bitter Taste Receptors Influence Glucose Homeostasis. PLoS ONE 2008, 3, e3974. [Google Scholar] [CrossRef] [Green Version]
- Hirasawa, A.; Tsumaya, K.; Awaji, T.; Katsuma, S.; Adachi, T.; Yamada, M.; Sugimoto, Y.; Miyazaki, S.; Tsujimoto, G. Free fatty acids regulate gut incretin glucagon-like peptide-1 secretion through GPR120. Nat. Med. 2004, 11, 90–94. [Google Scholar] [CrossRef]
- Chen, M.C.; Wu, S.V.; Reeve, J.R.; Rozengurt, E. Bitter stimuli induce Ca2+ signaling and CCK release in enteroendocrine STC-1 cells: Role of L-type voltage-sensitive Ca2+ channels. Am. J. Physiol. Physiol. 2006, 291, C726–C739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichimura, A.; Hirasawa, A.; Poulain-Godefroy, O.; Bonnefond, A.; Hara, T.; Yengo, L.; Kimura, I.; Leloire, A.; Liu, N.; Iida, K.; et al. Dysfunction of lipid sensor GPR120 leads to obesity in both mouse and human. Nature 2012, 483, 350–354. [Google Scholar] [CrossRef] [Green Version]
- Waguri, T.; Goda, T.; Kasezawa, N.; Yamakawa-Kobayashi, K. The combined effects of genetic variations in the GPR120 gene and dietary fat intake on obesity risk. Biomed. Res. 2013, 34, 69–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newell-Price, J.; Clark, A.J.; King, P. DNA Methylation and Silencing of Gene Expression. Trends Endocrinol. Metab. 2000, 11, 142–148. [Google Scholar] [CrossRef]
- Love-Gregory, L.; Kraja, A.T.; Allum, F.; Aslibekyan, S.; Hedman, Å.K.; Duan, Y.; Borecki, I.B.; Arnett, N.K.; McCarthy, M.I.; Deloukas, P.; et al. Higher chylomicron remnants and LDL particle numbers associate withCD36SNPs and DNA methylation sites that reduceCD36. J. Lipid Res. 2016, 57, 2176–2184. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Sears, R.; Xing, X.; Zhang, B.; Li, D.; Rockweiler, N.; Jang, H.S.; Choudhary, M.N.; Lee, H.J.; Lowdon, R.F.; et al. Tissue-specific DNA methylation is conserved across human, mouse, and rat, and driven by primary sequence conservation. BMC Genom. 2017, 18, 724. [Google Scholar] [CrossRef]
- Amaral, C.L.; Milagro, F.I.; Curi, R.; Martínez, J.A. DNA Methylation Pattern in Overweight Women under an Energy-Restricted Diet Supplemented with Fish Oil. BioMed Res. Int. 2014, 2014, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.-L.; Dong, S.; Gao, L.-F.; Li, L.; Xi, Y.-D.; Ma, W.-W.; Yuan, L.-H.; Xiao, R. Global DNA methylation was changed by a maternal high-lipid, high-energy diet during gestation and lactation in male adult mice liver. Br. J. Nutr. 2015, 113, 1032–1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milagro, F.I.; Gómez-Abellán, P.; Campión, J.; Martínez, J.A.; Ordovás, J.M.; Garaulet, M. CLOCK, PER2 and BMAL1 DNA Methylation: Association with Obesity and Metabolic Syndrome Characteristics and Monounsaturated Fat Intake. Chrono Int. 2012, 29, 1180–1194. [Google Scholar] [CrossRef] [PubMed]
- Hermsdorff, H.H.M.; Mansego, M.L.; Campion, J.; Milagro, F.I.; Zulet, M.A.; Martínez, J.A. TNF-alpha promoter methylation in peripheral white blood cells: Relationship with circulating TNFα, truncal fat and n-6 PUFA intake in young women. Cytokine 2013, 64, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.E.; Bartoshuk, L.M.; Kidd, J.R.; Duffy, V.B. Supertasting and PROP Bitterness Depends on More Than the TAS2R38 Gene. Chem. Senses 2008, 33, 255–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aribi, M.; Sari, B.-E.; Saari, B. Effect of Endogamy and Consanguinity on the Development of LabialVenous Malformations in rea of Tlemcen (West Algeria). Open Genom. J. 2008, 1, 1–8. [Google Scholar] [CrossRef]
- Madsen, M.S.; Siersbæk, R.D.; Boergesen, M.; Nielsen, R.; Mandrup, S. Peroxisome Proliferator-Activated Receptor γ and C/EBPα Synergistically Activate Key Metabolic Adipocyte Genes by Assisted Loading. Mol. Cell. Boil. 2013, 34, 939–954. [Google Scholar] [CrossRef] [Green Version]




| Groups | Lean Children (n = 51) | Obese Children (n = 51) | ||||
|---|---|---|---|---|---|---|
| Total (n) | CD36 Methy. (n) | GPR120 Methy. (n) | Total (n) | CD36 Methy. (n) | GPR120 Methy. (n) | |
| High-OA | 7 | 1 | 2 | 1 | 1 | 1 |
| Middle-OA | 26 | 9 | 2 | 12* | 12 | 8 |
| Low-OA | 18 | 6 | 3 | 20 | 20* | 11* |
| Non-tasters | 0 | 0 | 0 | 18 | 18 | 12 |
| Total (n) | 51 | 16 (31.3%) | 7 (13.7%) | 51 | 51 * (100%) | 32 * (62.7%) |
| Groups | Lean Children (n = 51) | Obese Children (n = 51) | ||||
|---|---|---|---|---|---|---|
| Total (n) | CD36 Methy. (n) | GPR120 Methy. (n) | Total (n) | CD36 Methy. (n) | GPR120 Methy. (n) | |
| High-PROP | 20 | 6 | 3 | 17 | 17 | 10 * |
| Middle-PROP | 29 | 9 | 3 | 23 ** | 23 * | 13 ** |
| Low-PROP | 2 | 1 | 1 | 11 | 11 | 9 |
| Total (n) | 51 | 16 (31.3%) | 7 (13.7) | 51 | 51 * (100%) | 32 * (62.7%) |
| OA Thresholds (mmol/L) | PROP Detection Threshold (mmol/L) | |||
|---|---|---|---|---|
| 0.032 | 0.32 | 3.2 | Total | |
| 0.018 | 4 | 4 | 0 | 8 |
| 0.18 | 1 | 4 | 0 | 5 |
| 0.37 | 4 | 3 | 0 | 7 |
| 0.75 | 6 | 7 | 0 | 13 |
| 1.5 | 5 | 4 | 4 | 13 |
| 3.6 | 11 | 12 | 2 | 25 |
| 12 | 1 | 10 | 2 | 13 |
| Non taster | 5 | 8 | 5 | 18 |
| Total (n) | 37 | 52 | 13 | 102 |
| Food Contents | Unmethylated CD36 | Methylated CD36 | Unmethylated GPR120 | Methylated GPR120 |
|---|---|---|---|---|
| Total (n = 35) | Total (n = 67) | Total (n = 63) | Total (n = 38) | |
| Meat, chips, cheese, breads and chocolate | 7 | 49 * | 25 | 31 * |
| Home prepared food (parents restrictions) and candy | 28 | 18* | 38 | 8 * |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berrichi, M.; Hichami, A.; Addou-Klouche, L.; Sayed Khan, A.; Khan, N.A. CD36 and GPR120 Methylation Associates with Orosensory Detection Thresholds for Fat and Bitter in Algerian Young Obese Children. J. Clin. Med. 2020, 9, 1956. https://doi.org/10.3390/jcm9061956
Berrichi M, Hichami A, Addou-Klouche L, Sayed Khan A, Khan NA. CD36 and GPR120 Methylation Associates with Orosensory Detection Thresholds for Fat and Bitter in Algerian Young Obese Children. Journal of Clinical Medicine. 2020; 9(6):1956. https://doi.org/10.3390/jcm9061956
Chicago/Turabian StyleBerrichi, Moustafa, Aziz Hichami, Lynda Addou-Klouche, Amira Sayed Khan, and Naim Akhtar Khan. 2020. "CD36 and GPR120 Methylation Associates with Orosensory Detection Thresholds for Fat and Bitter in Algerian Young Obese Children" Journal of Clinical Medicine 9, no. 6: 1956. https://doi.org/10.3390/jcm9061956






