Novel Nasal Epithelial Cell Markers of Parkinson’s Disease Identified Using Cells Treated with α-Synuclein Preformed Fibrils
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Nasal Fluid Sampling
2.3. Total RNA Extraction and PCR
2.4. Cell Culture and PFF Treatment
2.5. Western Blotting
2.6. Raw Data Preparation
2.7. Statistical Analyses
3. Results
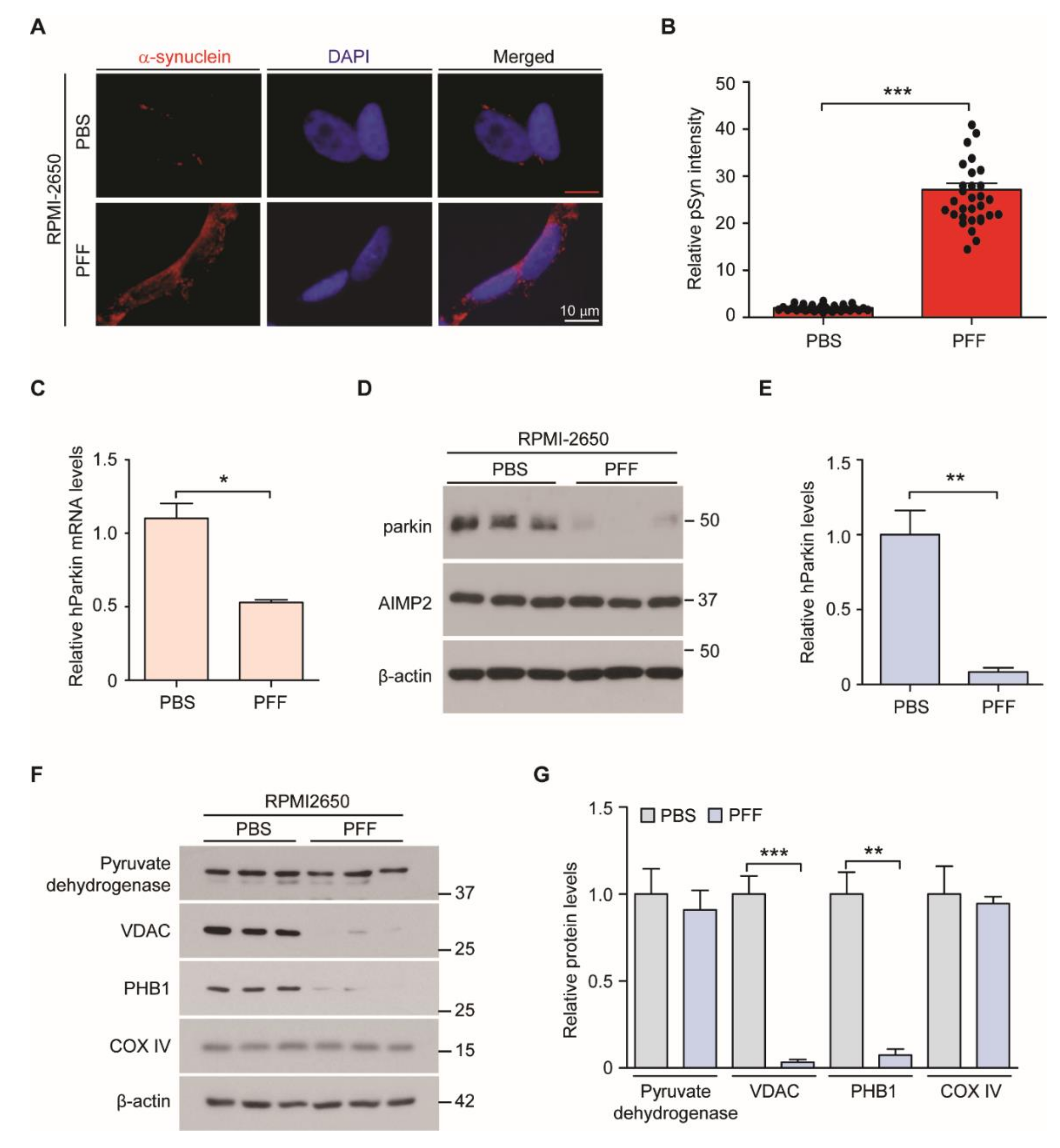
3.1. Generation of PFF-Induced Nasal Cell Model of PD
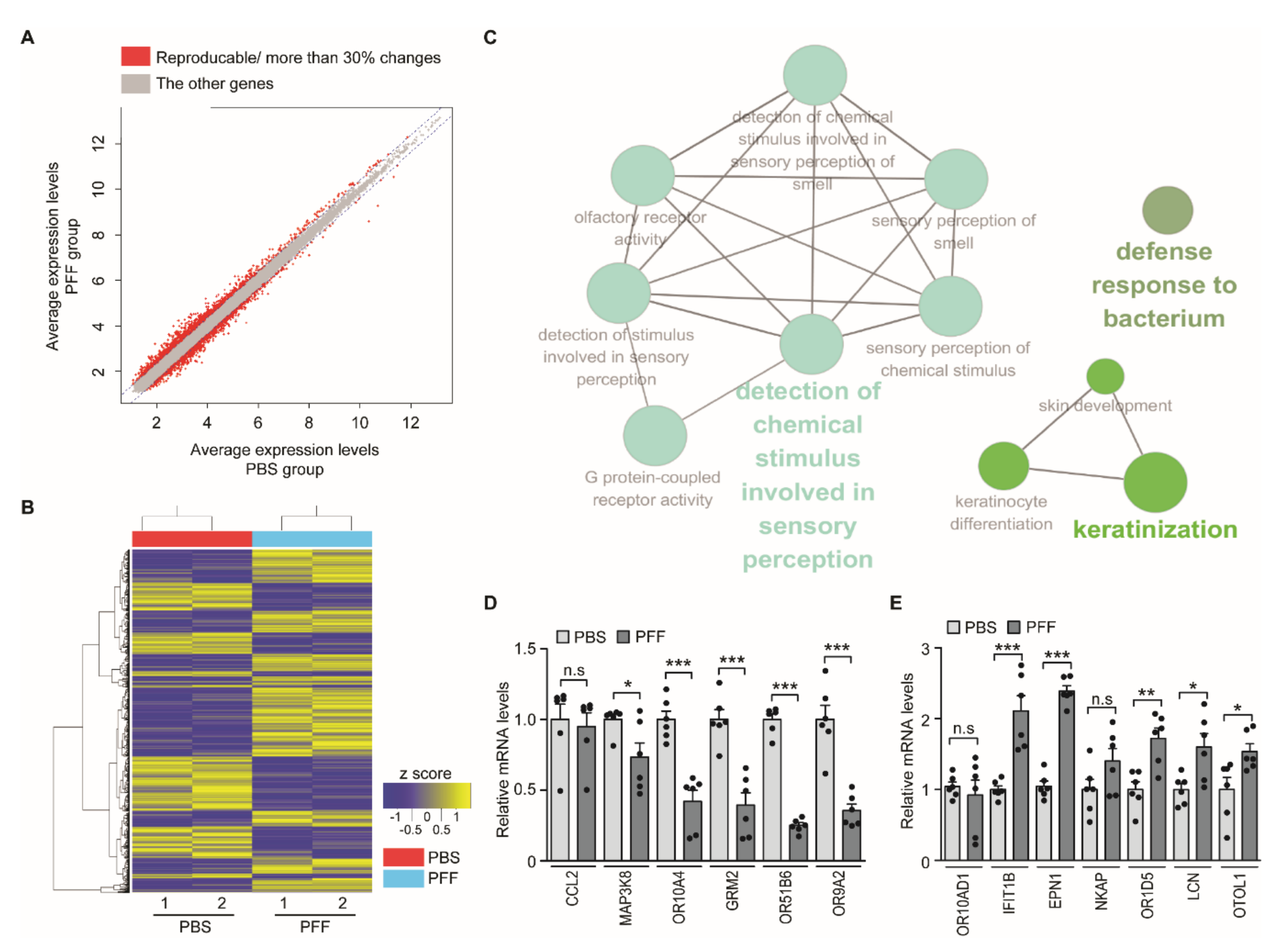
3.2. Screening of PD Biomarkers in the PFF-Exposed Nasal Cell Model
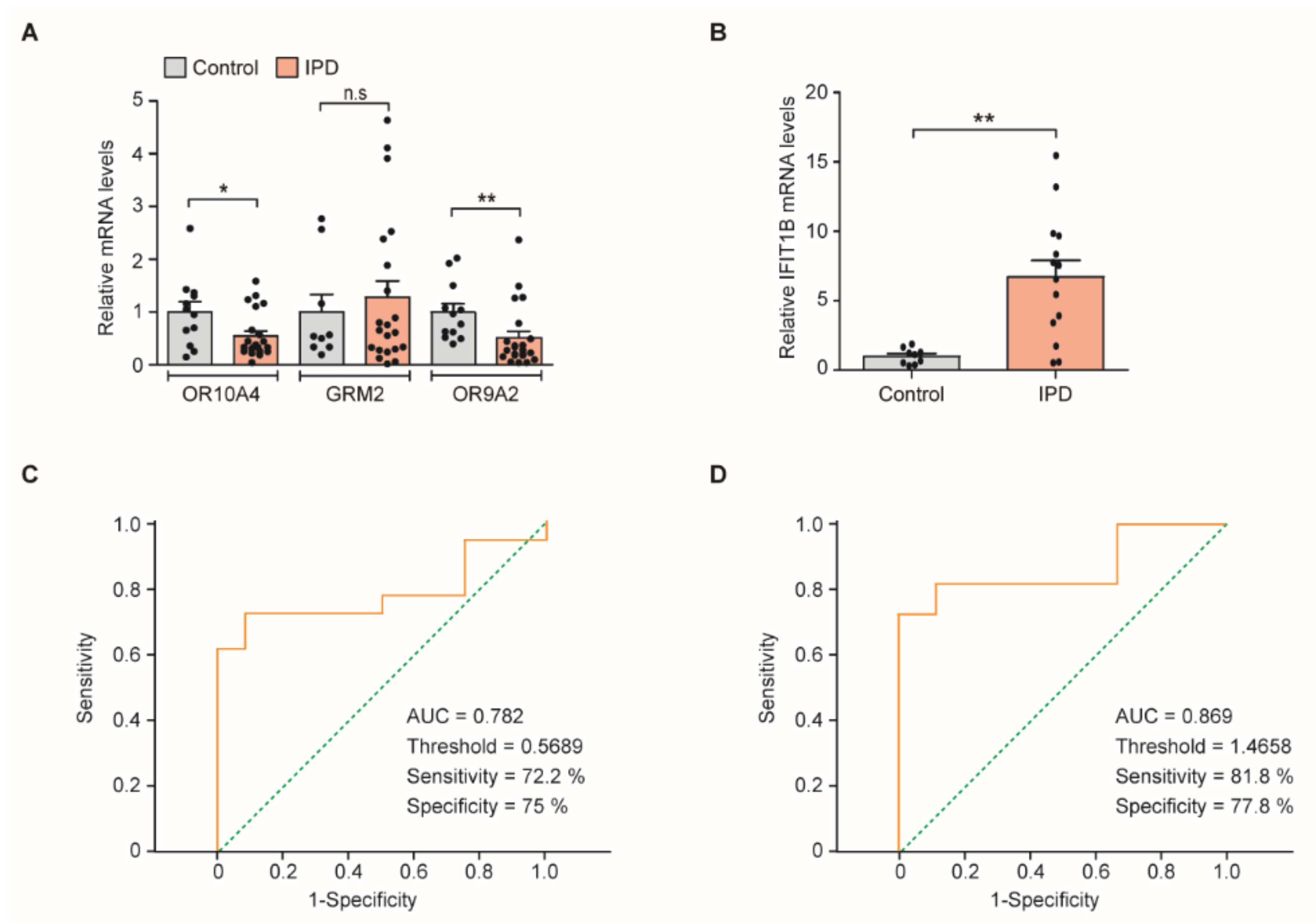
3.3. Quantitative Assessment of PD Marker Gene Transcripts in Cells Obtained from Clinical Nasal Fluid Samples
3.4. Receiver Operating Characteristic (ROC) Curve Threshold Analysis of OR9A2 and IFIT1B mRNA Levels for PD Diagnosis
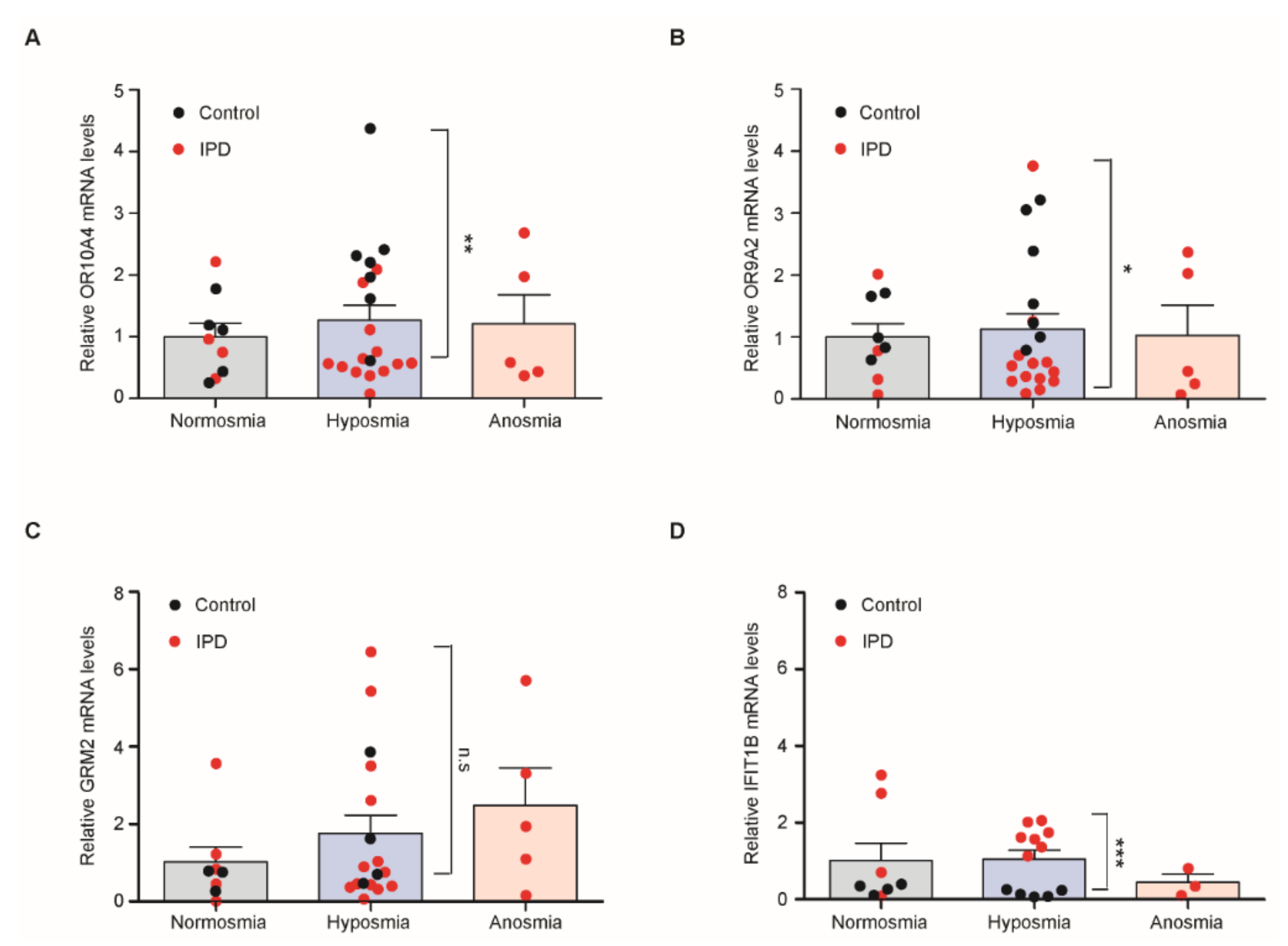
3.5. Quantitative Assessment of Transcripts of PD Marker Genes in Hyposmia Cases
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lang, A.E.; Lozano, A.M. Parkinson’s disease. First of two parts. N. Engl. J. Med. 1998, 339, 1044–1053. [Google Scholar] [CrossRef]
- Moore, D.J.; West, A.B.; Dawson, V.L.; Dawson, T.M. Molecular pathophysiology of Parkinson’s disease. Annu. Rev. Neurosci. 2005, 28, 57–87. [Google Scholar] [CrossRef] [Green Version]
- Visanji, N.P.; Brooks, P.L.; Hazrati, L.N.; Lang, A.E. The prion hypothesis in Parkinson’s disease: Braak to the future. Acta Neuropathol. Commun. 2013, 1, 2. [Google Scholar] [CrossRef] [Green Version]
- Beach, T.G.; White, C.L., 3rd; Hladik, C.L.; Sabbagh, M.N.; Connor, D.J.; Shill, H.A.; Sue, L.I.; Sasse, J.; Bachalakuri, J.; Henry-Watson, J.; et al. Olfactory bulb alpha-synucleinopathy has high specificity and sensitivity for lewy body disorders. Acta Neuropathol. 2009, 117, 169–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doty, R.L. Olfactory dysfunction in parkinson disease. Nat. Rev. Neurol. 2012, 8, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gu, C.Z.; Su, J.B.; Zhu, L.H.; Zhou, Y.; Huang, H.Y.; Liu, C.F. Changes in olfactory bulb volume in Parkinson’s disease: A systematic review and meta-analysis. PLoS ONE 2016, 11, e0149286. [Google Scholar] [CrossRef]
- Rey, N.L.; Steiner, J.A.; Maroof, N.; Luk, K.C.; Madaj, Z.; Trojanowski, J.Q.; Lee, V.M.; Brundin, P. Widespread transneuronal propagation of alpha-synucleinopathy triggered in olfactory bulb mimics prodromal Parkinson’s disease. J. Exp. Med. 2016, 213, 1759–1778. [Google Scholar] [CrossRef]
- Badorrek, P.; Muller, M.; Koch, W.; Hohlfeld, J.M.; Krug, N. Specificity and reproducibility of nasal biomarkers in patients with allergic rhinitis after allergen challenge chamber exposure. Ann. Allergy Asthma Immunol. 2017, 118, 290–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawkes, C.H.; Shephard, B.C.; Daniel, S.E. Olfactory dysfunction in Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry 1997, 62, 436–446. [Google Scholar] [CrossRef] [Green Version]
- Irander, K.; Palm, J.P.; Borres, M.P.; Ghafouri, B. Clara cell protein in nasal lavage fluid and nasal nitric oxide—Biomarkers with anti-inflammatory properties in allergic rhinitis. Clin. Mol. Allergy 2012, 10, 4. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Kang, S.J.; Jo, Y.M.; Kim, M.S.; Lee, Y.; Cho, S.H.; Kim, H.T. Quantitative analysis of nasal transcripts reveals potential biomarkers for Parkinson’s disease. Sci. Rep. 2019, 9, 11111. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Dawson, V.L.; Dawson, T.M. Animal models of Parkinson’s disease: Vertebrate genetics. Cold Spring Harb. Perspect. Med. 2012, 2, a009324. [Google Scholar] [CrossRef] [PubMed]
- Hisahara, S.; Shimohama, S. Toxin-induced and genetic animal models of Parkinson’s disease. Parkinsons Dis. 2010, 2011, 951709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Kwon, S.H.; Kam, T.I.; Panicker, N.; Karuppagounder, S.S.; Lee, S.; Lee, J.H.; Kim, W.R.; Kook, M.; Foss, C.A.; et al. Transneuronal propagation of pathologic alpha-synuclein from the gut to the brain models Parkinson’s disease. Neuron 2019, 103, 627–641.e7. [Google Scholar] [CrossRef]
- Volpicelli-Daley, L.A.; Luk, K.C.; Lee, V.M. Addition of exogenous alpha-synuclein preformed fibrils to primary neuronal cultures to seed recruitment of endogenous alpha-synuclein to lewy body and lewy neurite-like aggregates. Nat. Protoc. 2014, 9, 2135–2146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perlmutter, J.S. Assessment of Parkinson disease manifestations. Curr. Protoc. Neurosci. 2009, 49, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, A.J.; Daniel, S.E.; Kilford, L.; Lees, A.J. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: A clinico-pathological study of 100 cases. J. Neurol. Neurosurg. Psychiatry 1992, 55, 181–184. [Google Scholar] [CrossRef] [Green Version]
- Chung, J.H.; Lee, Y.J.; Kang, T.W.; Kim, K.R.; Jang, D.P.; Kim, I.Y.; Cho, S.H. Altered quality of life and psychological health (scl-90-r) in patients with chronic rhinosinusitis with nasal polyps. Ann. Otol. Rhinol. Laryngol. 2015, 124, 663–670. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Luk, K.C.; Kehm, V.; Carroll, J.; Zhang, B.; O’Brien, P.; Trojanowski, J.Q.; Lee, V.M. Pathological alpha-synuclein transmission initiates parkinson-like neurodegeneration in nontransgenic mice. Science 2012, 338, 949–953. [Google Scholar] [CrossRef] [Green Version]
- Yun, S.P.; Kam, T.I.; Panicker, N.; Kim, S.; Oh, Y.; Park, J.S.; Kwon, S.H.; Park, Y.J.; Karuppagounder, S.S.; Park, H.; et al. Block of a1 astrocyte conversion by microglia is neuroprotective in models of Parkinson’s disease. Nat. Med. 2018, 24, 931–938. [Google Scholar] [CrossRef]
- Kim, C.; Ho, D.H.; Suk, J.E.; You, S.; Michael, S.; Kang, J.; Joong Lee, S.; Masliah, E.; Hwang, D.; Lee, H.J.; et al. Neuron-released oligomeric alpha-synuclein is an endogenous agonist of tlr2 for paracrine activation of microglia. Nat. Commun. 2013, 4, 1562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bieri, G.; Gitler, A.D.; Brahic, M. Internalization, axonal transport and release of fibrillar forms of alpha-synuclein. Neurobiol. Dis. 2018, 109, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Ou, M.T.; Karuppagounder, S.S.; Kam, T.I.; Yin, X.; Xiong, Y.; Ge, P.; Umanah, G.E.; Brahmachari, S.; Shin, J.H.; et al. Pathological alpha-synuclein transmission initiated by binding lymphocyte-activation gene 3. Science 2016, 353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benson, M.; Jansson, L.; Adner, M.; Luts, A.; Uddman, R.; Cardell, L.O. Gene profiling reveals decreased expression of uteroglobin and other anti-inflammatory genes in nasal fluid cells from patients with intermittent allergic rhinitis. Clin. Exp. Allergy 2005, 35, 473–478. [Google Scholar] [CrossRef]
- Bergoin, C.; Gosset, P.; Lamblin, C.; Bolard, F.; Turck, D.; Tonnel, A.B.; Wallaert, B. Cell and cytokine profile in nasal secretions in cystic fibrosis. J. Cyst. Fibros. 2002, 1, 110–115. [Google Scholar] [CrossRef]
- Zilocchi, M.; Finzi, G.; Lualdi, M.; Sessa, F.; Fasano, M.; Alberio, T. Mitochondrial alterations in Parkinson’s disease human samples and cellular models. Neurochem. Int. 2018, 118, 61–72. [Google Scholar] [CrossRef]
- Dutta, D.; Ali, N.; Banerjee, E.; Singh, R.; Naskar, A.; Paidi, R.K.; Mohanakumar, K.P. Low levels of prohibitin in substantia nigra makes dopaminergic neurons vulnerable in Parkinson’s disease. Mol. Neurobiol. 2018, 55, 804–821. [Google Scholar] [CrossRef]
- Bouman, L.; Schlierf, A.; Lutz, A.K.; Shan, J.; Deinlein, A.; Kast, J.; Galehdar, Z.; Palmisano, V.; Patenge, N.; Berg, D.; et al. Parkin is transcriptionally regulated by atf4: Evidence for an interconnection between mitochondrial stress and er stress. Cell Death Differ. 2011, 18, 769–782. [Google Scholar] [CrossRef] [Green Version]
- Ham, S.; Lee, Y.I.; Jo, M.; Kim, H.; Kang, H.; Jo, A.; Lee, G.H.; Mo, Y.J.; Park, S.C.; Lee, Y.S.; et al. Hydrocortisone-induced parkin prevents dopaminergic cell death via creb pathway in Parkinson’s disease model. Sci. Rep. 2017, 7, 525. [Google Scholar] [CrossRef] [Green Version]
- Cai, M.; Tian, J.; Zhao, G.H.; Luo, W.; Zhang, B.R. Study of methylation levels of parkin gene promoter in Parkinson’s disease patients. Int. J. Neurosci. 2011, 121, 497–502. [Google Scholar] [CrossRef]
- Olzscha, H.; Schermann, S.M.; Woerner, A.C.; Pinkert, S.; Hecht, M.H.; Tartaglia, G.G.; Vendruscolo, M.; Hayer-Hartl, M.; Hartl, F.U.; Vabulas, R.M. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 2011, 144, 67–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grison, A.; Zucchelli, S.; Urzi, A.; Zamparo, I.; Lazarevic, D.; Pascarella, G.; Roncaglia, P.; Giorgetti, A.; Garcia-Esparcia, P.; Vlachouli, C.; et al. Mesencephalic dopaminergic neurons express a repertoire of olfactory receptors and respond to odorant-like molecules. BMC Genomics 2014, 15, 729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, N.; Koo, J. Olfactory receptors in non-chemosensory tissues. BMB Rep. 2012, 45, 612–622. [Google Scholar] [CrossRef] [Green Version]
- Fensterl, V.; Sen, G.C. Interferon-induced ifit proteins: Their role in viral pathogenesis. J. Virol. 2015, 89, 2462–2468. [Google Scholar] [CrossRef] [Green Version]
- Baker, C.A.; Lu, Z.Y.; Manuelidis, L. Early induction of interferon-responsive mrnas in creutzfeldt-jakob disease. J. Neurovirol. 2004, 10, 29–40. [Google Scholar] [CrossRef] [Green Version]
- Ishibashi, D.; Homma, T.; Nakagaki, T.; Fuse, T.; Sano, K.; Satoh, K.; Mori, T.; Atarashi, R.; Nishida, N. Type i interferon protects neurons from prions in in vivo models. Brain 2019, 142, 1035–1050. [Google Scholar] [CrossRef] [Green Version]
- Doty, R.L. Olfactory dysfunction in neurodegenerative diseases: Is there a common pathological substrate? Lancet Neurol. 2017, 16, 478–488. [Google Scholar] [CrossRef]
- Moon, J.; Lee, S.T.; Kong, I.G.; Byun, J.I.; Sunwoo, J.S.; Shin, J.W.; Shim, J.Y.; Park, J.H.; Jeon, D.; Jung, K.H.; et al. Early diagnosis of alzheimer’s disease from elevated olfactory mucosal mir-206 level. Sci. Rep. 2016, 6, 20364. [Google Scholar] [CrossRef]
- Menendez-Gonzalez, M.; Alvarez-Avellon, T.; Salas-Pacheco, J.M.; de Celis-Alonso, B.; Wyman-Chick, K.A.; Arias-Carrion, O. Frontotemporal lobe degeneration as origin of scans without evidence of dopaminergic deficit. Front. Neurol. 2018, 9, 335. [Google Scholar] [CrossRef]
| Control | PD | SWEDD | p-Value | |
|---|---|---|---|---|
| N | 13 | 23 | 8 | |
| Male: female | 6:7 | 9:14 | 2:6 | 0.6251 |
| Age (years) | 63 ± 2.62 | 67 ± 2.66 | 80 ± 1.88 | 0.3356 |
| H&Y scale | 2.30 ± 0.25 | |||
| UPDRS | 26.4 ± 3.75 | |||
| Olfaction (TDI score) | 29.5 ± 2.41 | 23.4 ± 2.07 | 21.3 ± 2.48 | 0.0887 |
| Target Gene | Sequence (5′→3′) |
|---|---|
| GAPDH | Forward: AAACCCATCACCATCTTCCAG |
| Reverse: AGGGGCCATCCACAGTCTTCT | |
| β-ACTIN | Forward: AGAGCTACGAGCTGCCTGAC |
| Reverse: AGCACTGTGTTGGCGTACAG | |
| MAP3K8 | Forward: CTCCCCAAAATGGACGTTACC |
| Reverse: GGATTTCCACATCAGATGGCTTA | |
| OR10A4 | Forward: AGCTGCCTCTTGGTTCTCAG |
| Reverse: ACAGACCAGTGCAATAACAGG | |
| GRM2 | Forward: CCGCATTGCACGCATCTTC |
| Reverse: GGCCCGAGATAAGTGCCAG | |
| OR51B6 | Forward: ATGGGGCTCAATAAGTCTGCT |
| Reverse: GAGGTCTGTAGCTGCCAACA | |
| OR9A2 | Forward: ATGTATCGCTCAACTTTTCCTGT |
| Reverse: CCCATGACACTATTACCACCCAA | |
| CCL2 | Forward: CAGCCAGATGCAATCAATGCC |
| Reverse: TGGAATCCTGAACCCACTTCT | |
| OR10AD1 | Forward: CTAAGGAATGGCAGCATAGTGAC |
| Reverse: AGCAGACATCCAGGAGAGACA | |
| IFIT1B | Forward: AATCAAGGAAGCTACAAACTGGC |
| Reverse: TCTTCATGCGTAACCCTTTCTG | |
| EPN1 | Forward: CAAGAACTGGCGTCACGTTTA |
| Reverse: CTGCTTAGCTTTCTCACGCA | |
| NKAP | Forward: CAGTCTATTACGGCAGCTACTCG |
| Reverse: CTCCACTGGTGTATGTTCATCAG | |
| OR1D5 | Forward: TTCACACAGCGTTGATTGCC |
| Reverse: CAAGCGTCCCATAAAAGAGGG | |
| LCN2 | Forward: GAAGTGTGACTACTGGATCAGGA |
| Reverse: ACCACTCGGACGAGGTAACT | |
| OTOL1 | Forward: GTCCTGTCACTGGGAAGTTTAAC |
| Reverse: CCAGACTGATTCGAGCAGGTC |
| Gene Symbol | Gene Description | FC (Microarray) | FC (RT qPCR) |
|---|---|---|---|
| BAGE2 | B melanoma antigen family, member 2 | 0.58 | ND |
| TMC3 | Transmembrane channel like 3 | 0.59 | ND |
| MAP3K8 | Mitogen-activated protein kinase kinase kinase 8 | 0.60 | 0.73 |
| OR10A4 | Olfactory receptor, family 10, subfamily A, member 4 | 0.61 | 0.42 |
| GRM2 | Glutamate receptor, metabotropic 2 | 0.62 | 0.39 |
| KRTAP10-2 | Keratin associated protein 10-2 | 0.63 | ND |
| OR51B6 | Olfactory receptor, family 51, subfamily B, member 6 | 0.63 | 0.25 |
| OR9A2 | Olfactory receptor, family 9, subfamily A, member 2 | 0.64 | 0.35 |
| KLK15 | Kallikrein related peptidase 15 | 0.64 | ND |
| CCL2 | Chemokine (C-C motif) ligand 2 | 0.64 | 0.95 |
| GSTA2 | Glutathione S-transferase alpha 2 | 1.91 | ND |
| OR10AD1 | Olfactory receptor, family 10, subfamily AD, member 1 | 1.71 | 0.88 |
| OR5M1 | Olfactory receptor, family 5, subfamily M, member 1 | 1.61 | ND |
| DAPL1 | Death associated protein like 1 | 1.60 | ND |
| IFIT1B | Interferon-induced protein with tetratricopeptide repeats 1B | 1.60 | 2.11 |
| EPN1 | Epsin 1 | 1.59 | 2.28 |
| OR2T4 | Olfactory receptor, family 2, subfamily T, member 4 | 1.59 | ND |
| TSTD3 | Thiosulfate sulfurtransferase (rhodanese)-like domain containing 3 | 1.58 | ND |
| TAS2R43 | Taste receptor, type 2, member 43 | 1.57 | ND |
| SPANXN5 | SPANX family, member N5 | 1.57 | ND |
| NKAP | NFKB activating protein | 1.44 | 1.40 |
| OR1D5 | olfactory receptor, family 1, subfamily D, member 5 | 1.46 | 1.72 |
| LCN2 | lipocalin 2 | 1.50 | 1.60 |
| OTOL1 | otolin 1 | 1.56 | 1.54 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.; Kang, S.-J.; Jo, Y.M.; Park, S.; Yun, S.P.; Lee, Y.-S.; Kim, H.-T.; Lee, N.-E.; Kim, Y.-S.; Cho, S.H.; et al. Novel Nasal Epithelial Cell Markers of Parkinson’s Disease Identified Using Cells Treated with α-Synuclein Preformed Fibrils. J. Clin. Med. 2020, 9, 2128. https://doi.org/10.3390/jcm9072128
Kim H, Kang S-J, Jo YM, Park S, Yun SP, Lee Y-S, Kim H-T, Lee N-E, Kim Y-S, Cho SH, et al. Novel Nasal Epithelial Cell Markers of Parkinson’s Disease Identified Using Cells Treated with α-Synuclein Preformed Fibrils. Journal of Clinical Medicine. 2020; 9(7):2128. https://doi.org/10.3390/jcm9072128
Chicago/Turabian StyleKim, Hyojung, Seok-Jae Kang, Young Mi Jo, Sanggyu Park, Seung Pil Yun, Yun-Song Lee, Hee-Tae Kim, Nae-Eung Lee, Yong-Sang Kim, Seok Hyun Cho, and et al. 2020. "Novel Nasal Epithelial Cell Markers of Parkinson’s Disease Identified Using Cells Treated with α-Synuclein Preformed Fibrils" Journal of Clinical Medicine 9, no. 7: 2128. https://doi.org/10.3390/jcm9072128






