Targeted Urine Metabolomics for Monitoring Renal Allograft Injury and Immunosuppression in Pediatric Patients
Abstract
1. Introduction
2. Experimental Section
2.1. Patients and Samples
2.2. Urine Collection, Initial Processing, Storage, and GC/MS-TOF Analysis
2.3. Raw Data Processing and Statistics
2.4. Data Availability
3. Results
3.1. Metabolites in Urine Are Perturbed in Different Transplant Injuries in Kidney Transplantation
3.2. Metabolite Marker Panel for Alloimmune Injury
3.3. Metabolite Marker Panel for Acute Rejection
3.4. Metabolite Marker Panel for BK Virus Nephritis
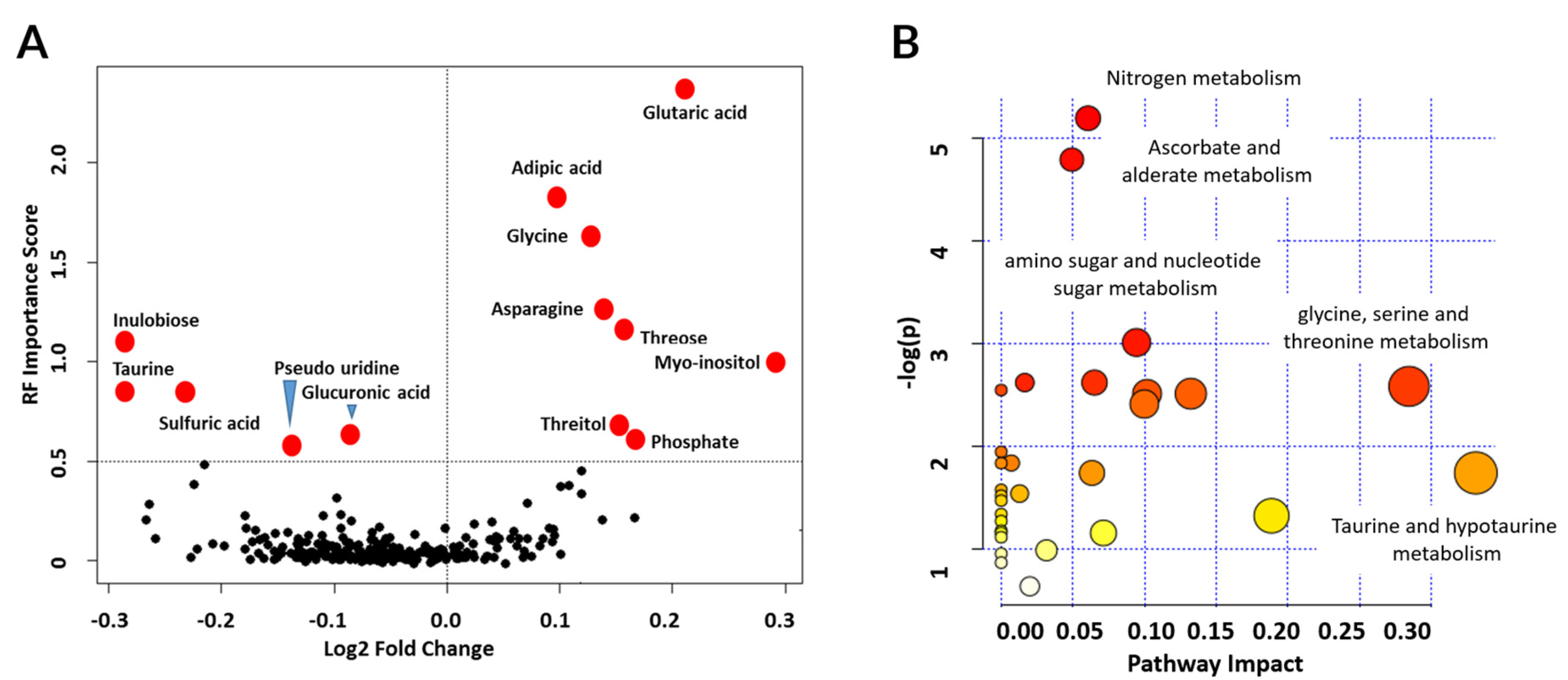
3.5. Metabolic Pathways Associated with Graft Injury
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abecassis, M.; Bartlett, S.T.; Collins, A.J.; Davis, C.L.; Delmonico, F.L.; Friedewald, J.J.; Hays, R.; Howard, A.; Jones, E.; Leichtam, A.B.; et al. Kidney transplantation as primary therapy for end-stage renal disease: A National Kidney Foundation/Kidney Disease Outcomes Quality Initiative (NKF/KDOQITM) conference. Clin. J. Am. Soc. Nephrol. CJASN 2008, 3, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Pomfret, E.A.; Sung, R.S.; Allan, J.; Kinkhabwala, M.; Melancon, J.K.; Roberts, J.P. Solving the organ shortage crisis: The 7th annual American Society of Transplant Surgeons’ State-of-the-Art Winter Symposium. Am. J. Transplant. 2008, 8, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kriesche, H.U.; Schold, J.D.; Srinivas, T.R.; Kaplan, B. Lack of improvement in renal allograft survival despite a marked decrease in acute rejection rates over the most recent era. Am. J. Transplant. 2004, 4, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Gaston, R.S. Improving Long-Term Outcomes in Kidney Transplantation: Towards a New Paradigm of Post-Transplant Care in the United States. Trans. Am. Clin. Climatol. Assoc. 2016, 127, 350–361. [Google Scholar] [PubMed]
- Nasr, M.; Sigdel, T.; Sarwal, M. Advances in diagnostics for transplant rejection. Expert Rev. Mol. Diagn. 2016, 16, 1121–1132. [Google Scholar] [CrossRef]
- Loupy, A.; Haas, M.; Solez, K.; Racusen, L.; Glotz, D.; Seron, D.; Nankivell, B.J.; Colvin, R.B.; Afrouzian, M.; Akalin, E.; et al. The Banff 2015 Kidney Meeting Report: Current Challenges in Rejection Classification and Prospects for Adopting Molecular Pathology. Am. J. Transplant. 2017, 17, 28–41. [Google Scholar] [CrossRef]
- Filler, G.; Bendrick-Peart, J.; Christians, U. Pharmacokinetics of mycophenolate mofetil and sirolimus in children. Ther. Drug Monit. 2008, 30, 138–142. [Google Scholar] [CrossRef]
- Yang, J.Y.; Sarwal, M.M. Transplant genetics and genomics. Nat. Rev. Genet. 2017, 18, 309–326. [Google Scholar] [CrossRef]
- Reeve, J.; Bohmig, G.A.; Eskandary, F.; Einecke, G.; Lefaucheur, C.; Loupy, A.; Halloran, P.F. The MMDx-Kidney Study Group. Assessing rejection-related disease in kidney transplant biopsies based on archetypal analysis of molecular phenotypes. JCI Insight 2017, 2, 1–14. [Google Scholar] [CrossRef]
- Roedder, S.; Sigdel, T.; Salomonis, N.; Hsieh, S.; Dai, H.; Bestard, O.; Metes, D.; Zeevi, A.; Gritsh, A.; Cheeseman, J.; et al. The kSORT assay to detect renal transplant patients at high risk for acute rejection: Results of the multicenter AART study. PLoS Med. 2014, 11, e1001759. [Google Scholar] [CrossRef]
- Sigdel, T.K.; Salomonis, N.; Nicora, C.D.; Ryu, S.; He, J.; Dinh, V.; Orton, D.J.; Moore, R.J.; Hsieh, S.C.; Dai, H.; et al. The identification of novel potential injury mechanisms and candidate biomarkers in renal allograft rejection by quantitative proteomics. Mol. Cell. Proteom. MCP 2014, 13, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Khatri, P.; Roedder, S.; Kimura, N.; De Vusser, K.; Morgan, A.A.; Gong, Y.; Fischbein, M.P.; Robbins, R.C.; Naesens, M.; Bute, A.J.; et al. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J. Exp. Med. 2013, 210, 2205–2221. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Sigdel, T.K.; Sarwal, M.M. Self-antigens and rejection: A proteomic analysis. Curr. Opin. Organ Transplant. 2016, 21, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Sarwal, M.; Chua, M.S.; Kambham, N.; Hsieh, S.C.; Satterwhite, T.; Masek, M.; Salvatierra, O., Jr. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N. Engl. J. Med. 2003, 349, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Erpicum, P.; Hanssen, O.; Weekers, L.; Lovinfosse, P.; Meunier, P.; Tshibanda, L.; Ktzesinski, J.M.; Hustinx, R.; Jouret, F. Non-invasive approaches in the diagnosis of acute rejection in kidney transplant recipients, part II: Omics analyses of urine and blood samples. Clin. Kidney J. 2017, 10, 106–115. [Google Scholar] [CrossRef][Green Version]
- Wishart, D.S. Metabolomics: A complementary tool in renal transplantation. Contrib. Nephrol. 2008, 160, 76–87. [Google Scholar]
- Klawitter, J.; Klawitter, J.; Kushner, E.; Jonscher, K.; Bendrick-Peart, J.; Leibfritz, D.; Christians, U.; Schmitz, V. Association of immunosuppressant-induced protein changes in the rat kidney with changes in urine metabolite patterns: A proteo-metabonomic study. J. Proteome Res. 2010, 9, 865–875. [Google Scholar] [CrossRef]
- Bouatra, S.; Aziat, F.; Mandal, R.; Guo, A.C.; Wilson, M.R.; Knox, C.; Bjorndahl, T.C.; Krishamurthy, R.; Saleem, F.; Liu, P.; et al. The human urine metabolome. PLoS ONE 2013, 8, e73076. [Google Scholar] [CrossRef]
- Bohra, R.; Klepacki, J.; Klawitter, J.; Klawitter, J.; Thurman, J.; Christians, U. Proteomics and metabolomics in renal transplantation-quo vadis? Transpl. Int. 2013, 26, 225–241. [Google Scholar] [CrossRef]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.I.; Xu, Y.; Correa, E.; Turner, M.L.; Goodacre, R. A tutorial review: Metabolomics and partial least squares-discriminant analysis-a marriage of convenience or a shotgun wedding. Anal. Chim. Acta 2015, 879, 10–23. [Google Scholar] [CrossRef]
- Racusen, L.C.; Halloran, P.F.; Solez, K. Banff 2003 meeting report: New diagnostic insights and standards. Am. J. Transplant. 2004, 4, 1562–1566. [Google Scholar] [CrossRef] [PubMed]
- Racusen, L.C.; Solez, K.; Colvin, R.B.; Bonsib, S.M.; Castro, M.C.; Cavallo, T.; Croker, B.P.; Demetris, A.J.; Drachenberg, C.B.; Fogo, A.B.; et al. The Banff 97 working classification of renal allograft pathology. Kidney Int. 1999, 55, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Solez, K.; Colvin, R.B.; Racusen, L.C.; Haas, M.; Sis, B.; Mengel, M.; Halloran, P.F.; Baldwin, W.; Banfi, G.; Collins, A.B.; et al. Banff 07 classification of renal allograft pathology: Updates and future directions. Am. J. Transplant. 2008, 8, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Nankivell, B.J.; Alexander, S.I. Rejection of the kidney allograft. N. Engl. J. Med. 2010, 363, 1451–1462. [Google Scholar] [CrossRef]
- Fletcher, J.T.; Nankivell, B.J.; Alexander, S.I. Chronic allograft nephropathy. Pediatric Nephrol. 2009, 24, 1465–1471. [Google Scholar] [CrossRef]
- Kind, T.; Wohlgemuth, G.; Lee, D.Y.; Lu, Y.; Palazoglu, M.; Shahbaz, S.; Fiehn, O. FiehnLib: Mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 2009, 81, 10038–10048. [Google Scholar] [CrossRef] [PubMed]
- Kind, T.; Tolstikov, V.; Fiehn, O.; Weiss, R.H. A comprehensive urinary metabolomic approach for identifying kidney cancerr. Anal. Biochem. 2007, 363, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Katajamaa, M.; Miettinen, J.; Oresic, M. MZmine: Toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics 2006, 22, 634–636. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Stekhoven, D.J.; Buhlmann, P. MissForest-non-parametric missing value imputation for mixed-type data. Bioinformatics 2012, 28, 112–118. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Genuer, R.; Poggi, J.M.; Tuleau-Malot, C. VSURF: An R Package for Variable Selection Using Random Forests. R. J. 2015, 7, 19–33. [Google Scholar] [CrossRef]
- Delong, E.R.; Delong, D.M.; Clarkepearson, D.I. Comparing the Areas under 2 or More Correlated Receiver Operating Characteristic Curves-a Nonparametric Approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Nissaisorakarn, V.; Lee, J.R.; Lubetzky, M.; Suthanthiran, M. Urine biomarkers informative of human kidney allograft rejection and tolerance. Hum. Immunol. 2018, 79, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Sigdel, T.K.; Kaushal, A.; Gritsenko, M.; Norbeck, A.D.; Qian, W.J.; Xiao, W.; Camp, D.G.; Smith, R.D.; Sarwal, M.M. Shotgun proteomics identifies proteins specific for acute renal transplant rejection. Proteom. Clin. Appl. 2010, 4, 32–47. [Google Scholar] [CrossRef]
- Sigdel, T.K.; Lee, S.; Sarwal, M.M. Profiling the proteome in renal transplantation. Proteom. Clin. Appl. 2011, 5, 269–280. [Google Scholar] [CrossRef]
- Sigdel, T.K.; Sarwal, M.M. The proteogenomic path towards biomarker discovery. Pediatric Transplant. 2008, 12, 737–747. [Google Scholar] [CrossRef]
- Sigdel, T.K.; Sarwal, M.M. Recent advances in biomarker discovery in solid organ transplant by proteomics. Expert Rev. Proteom. 2011, 8, 705–715. [Google Scholar] [CrossRef]
- Sigdel, T.K.; Gao, Y.; He, J.; Wang, A.; Nicora, C.D.; Fillmore, T.L.; Shi, T.; Webb-Robertson, B.J.; Smith, R.D.; Qian, W.J.; et al. Mining the human urine proteome for monitoring renal transplant injury. Kidney Int. 2016, 89, 1244–1252. [Google Scholar] [CrossRef] [PubMed]
- Sigdel, T.K.; Ng, Y.W.; Lee, S.; Nicora, C.D.; Qian, W.J.; Smith, R.D.; Qian, W.J.; Salvatierra, O.; Camp, D.G.; Sarwal, M.M. Perturbations in the urinary exosome in transplant rejection. Front. Med. 2014, 1, 57. [Google Scholar] [CrossRef] [PubMed]
- Sigdel, T.K.; Vitalone, M.J.; Tran, T.Q.; Dai, H.; Hsieh, S.C.; Salvatierra, O.; Sarwal, M.M. A rapid noninvasive assay for the detection of renal transplant injury. Transplantation 2013, 96, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Suhre, K.; Schwartz, J.E.; Sharma, V.K.; Chen, Q.; Lee, J.R.; Muthukumar, T.; Dadhania, D.M.; Ding, R.; Ikle, D.H.; Bridges, N.D.; et al. Urine Metabolite Profiles Predictive of Human Kidney Allograft Status. J. Am. Soc. Nephrol. JASN 2016, 27, 626–636. [Google Scholar] [CrossRef]
- Fairchild, R.L.; Suthanthiran, M. Urine CXCL10/IP-10 Fingers Ongoing Antibody-Mediated Kidney Graft Rejection. J. Am. Soc. Nephrol. JASN 2015, 26, 2607–2609. [Google Scholar] [CrossRef][Green Version]
- Li, B.; Hartono, C.; Ding, R.; Sharma, V.K.; Ramaswamy, R.; Qian, B.; Serur, D.; Mouradian, J.; Schwartz, J.E.; Suthanthiran, M. Noninvasive diagnosis of renal-allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. N. Engl. J. Med. 2001, 344, 947–954. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Guo, L.X.; Li, Y.; Chang, W.Q.; Liu, J.Q.; Liu, L.F.; Xin, G.Z. Dose-response characteristics of Clematis triterpenoid saponins and clematichinenoside AR in rheumatoid arthritis rats by liquid chromatography/mass spectrometry-based serum and urine metabolomics. J. Pharm. Biomed. Anal. 2017, 136, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Schaub, S.; Mayr, M.; Honger, G.; Bestland, J.; Steiger, J.; Regeniter, A.; Mihatsch, M.J.; Wilkins, J.A.; Rush, D.; Nickerson, P. Detection of subclinical tubular injury after renal transplantation: Comparison of urine protein analysis with allograft histopathology. Transplantation 2007, 84, 104–112. [Google Scholar] [CrossRef]
- Schaub, S.; Rush, D.; Wilkins, J.; Gibson, I.W.; Weiler, T.; Sangster, K.; Nicolle, L.; Karpinski, M.; Jeffery, J.; Nickerson, P. Proteomic-based detection of urine proteins associated with acute renal allograft rejection. J. Am. Soc. Nephrol. JASN 2004, 15, 219–227. [Google Scholar] [CrossRef]
- Torng, S.; Rigatto, C.; Rush, D.N.; Nickerson, P.; Jeffery, J.R. The urine protein to creatinine ratio (P/C) as a predictor of 24-hour urine protein excretion in renal transplant patients. Transplantation 2001, 72, 1453–1456. [Google Scholar] [CrossRef]
- Choi, J.Y.; Yoon, Y.J.; Choi, H.J.; Park, S.H.; Kim, C.D.; Kim, I.S.; Kwon, T.H.; Do, J.Y.; Kim, S.H.; Ryu, D.H.; et al. Dialysis modality-dependent changes in serum metabolites: Accumulation of inosine and hypoxanthine in patients on haemodialysis. Nephrol. Dial. Transpl. 2011, 26, 1304–1313. [Google Scholar] [CrossRef]
- Blydt-Hansen, T.D.; Sharma, A.; Gibson, I.W.; Mandal, R.; Wishart, D.S. Urinary metabolomics for noninvasive detection of borderline and acute T cell-mediated rejection in children after kidney transplantation. Am. J. Transplant. 2014, 14, 2339–2349. [Google Scholar] [CrossRef]
- Dieme, B.; Halimi, J.M.; Emond, P.; Buchler, M.; Nadal-Desbarat, L.; Blasco, H.; Guellec, C.L. Assessing the metabolic effects of calcineurin inhibitors in renal transplant recipients by urine metabolic profiling. Transplantation 2014, 98, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.; Sharma, A.; Mandal, R.; Wishart, D.S.; Wiebe, C.; Storsley, L.; Karpinski, M.; Gibson, I.M.; Nickerson, P.W.; Rush, D.N. Detecting Renal Allograft Inflammation Using Quantitative Urine Metabolomics and CXCL10. Transplant. Direct 2016, 2, e78. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, N.E.; Iser, J.H.; Smallwood, R.A. Hepatic bile acid transport: Effect of conjugation and position of hydroxyl groups. Am. J. Physiol 1975, 229, 298–302. [Google Scholar] [CrossRef]
- Chesney, R.W.; Han, X.; Patters, A.B. Taurine and the renal system. J. Biomed. Sci. 2010, 17 (Suppl. 1):S4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Trachtman, H.; Sturman, J.A. Taurine: A therapeutic agent in experimental kidney disease. Amino Acids 1996, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Trachtman, H.; Futterweit, S.; Prenner, J.; Hanon, S. Antioxidants reverse the antiproliferative effect of high glucose and advanced glycosylation end products in cultured rat mesangial cells. Biochem. Biophys. Res. Commun. 1994, 199, 346–352. [Google Scholar] [CrossRef]
- Dantzler, W.H.; Silbernagl, S. Renal tubular reabsorption of taurine, gamma-aminobutyric acid (GABA) and beta-alanine studied by continuous microperfusion. Pflug. Arch. 1976, 367, 123–128. [Google Scholar] [CrossRef]
- Brocker, C.; Thompson, D.C.; Vasiliou, V. The role of hyperosmotic stress in inflammation and disease. Biomol. Concepts 2012, 3, 345–364. [Google Scholar] [CrossRef]
- Yorek, M.A.; Dunlap, J.A.; Lowe, W.L., Jr. Osmotic regulation of the Na+/myo-inositol cotransporter and postinduction normalization. Kidney Int. 1999, 55, 215–224. [Google Scholar] [CrossRef][Green Version]
- Niewczas, M.A.; Sirich, T.L.; Mathew, A.V.; Skupien, J.; Mohney, R.P.; Warram, J.H.; Smiles, A.; Huang, X.; Walker, W.; Byun, J.; et al. Uremic solutes and risk of end-stage renal disease in type 2 diabetes: Metabolomic study. Kidney Int. 2014, 85, 1214–1224. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, H.; Yamauchi, A.; Sugiura, T.; Matsuoka, Y.; Horio, M.; Tohyama, M.; Shimada, S.; Imai, E.; Hori, M. Inhibition of myo-inositol transport causes acute renal failure with selective medullary injury in the rat. Kidney Int. 1998, 53, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Gil, R.B.; Ortiz, A.; Sanchez-Nino, M.D.; Markoska, K.; Schepers, E.; Vanholder, R.; Glorieux, G.; Schmitt-Kopplin, P.; Heinzmann, S.S. Increased urinary osmolyte excretion indicates chronic kidney disease severity and progression rate. Nephrol. Dial. Transplant. 2018, 33, 2156–2164. [Google Scholar] [CrossRef] [PubMed]
- Burg, M.B.; Ferraris, J.D. Intracellular organic osmolytes: Function and regulation. J. Biol. Chem. 2008, 283, 7309–7313. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.C.; Sarwal, R.; Ky, K.; Dong, V.; Stoller, M.; Sarwal, M.; Chi, T. Non-Radiologic Assessment of Kidney Stones by KIT, a Spot Urine Assay. Br. J. Urol. Int. 2020, 125, 732–738. [Google Scholar] [CrossRef]
- Yang, J.Y.C.; Sarwal, R.D.; Fervenza, F.C.; Sarwal, M.M.; Lafayette, R.A. Noninvasive Urinary Monitoring of Progression in IgA Nephropathy. Int. J. Mol. Sci. 2019, 20, 4463. [Google Scholar] [CrossRef]
- Sigdel, T.K.; Yang, J.Y.C.; Bestard, O.; Schroeder, A.; Hsieh, S.-C.; Liberto, J.M.; Qamm, I.; Geraedts, A.C.M.; Sarwal, M.M. A urinary Common Rejection Module (uCRM) score for non-invasive kidney transplant monitoring. PLoS ONE 2019, 7, 1–15. [Google Scholar] [CrossRef]
- Sigdel, T.; Yang, J.; Bestard, O.; Hsieh, S.; Roedder, S.; Damm, I.; Liberto, J.; Nandoe, S.; Sarwal, M. A Non-Invasive Urinary Common Rejection Module (uCRM) Gene Expression Score Quantifies and Differentiates Kidney Transplant Injury. Am. J. Transplant. 2017. [Google Scholar] [CrossRef][Green Version]
- Yang, J.Y.C.; Sarwal, R.D.; Sigdel, T.K.; Damm, I.; Rosenbaum, B.; Liberto, J.M.; Chan-on, C.; Arreola-Guerra, J.M.; Alberu, J.M.; Vincenti, F.; et al. A urine score for noninvasive accurate diagnosis and prediction of kidney transplant rejection. Sci. Transl. Med. 2020, 12, 1–11. [Google Scholar] [CrossRef]
- Bi, H.; Guo, Z.; Jia, X.; Liu, H.; Ma, L.; Xue, L. The key points in the pre-analytical procedures of blood and urine samples in metabolomics studies. Metabolomics 2020, 16, 1–15. [Google Scholar] [CrossRef]
- Moreso, F.; Carrera, M.; Goma, M.; Hueso, M.; Sellares, J.; Martorell, J.; Grinyó, J.M.; Serón, D. Early subclinical rejection as a risk factor for late chronic humoral rejection. Transplantation 2012, 93, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Bassi, R.; Niewczas, M.A.; Biancone, L.; Bussolino, S.; Merugumala, S.; Tezza, S.; D’Addio, F.; Nasr, M.B.; Valderrama-Vasquez, A.; Usuelli, V. Metabolomic profiling in individuals with a failing kidney allograft. PLoS ONE 2017, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]




| Phenotype | AR | STA | IFTA | BKVN | p-Value |
|---|---|---|---|---|---|
| Number of Patients | 106 | 111 | 71 | 22 | |
| Maintenance (% Steroid-free) | 63.2% | 50.5% | 56.3% | 36.4% | 0.078 |
| Recipient Gender (% M) | 64.2% | 58.6% | 67.6% | 59.1% | 0.002 |
| Recipient Age * (years) | 13 ± 5 (14; 2–21) | 14 ± 5 (15; 1–21) | 10 ± 6 (10; 1–20) | 14 ± 5 (17; 1–18) | 0.003 |
| Donor Gender (% M) | 46.2% | 52.3% | 52.1% | 72.7% | 0.123 |
| Donor Age * (years) | 29 ± 11 (29; 4–50) | 30 ± 10 (28; 14–51) | 30 ± 10 (32; 12–50) | 28 ± 10 (29; 16–49) | 0.353 |
| Month post-Tx (mean ± SD) | 71 ± 32 | 15 ± 24 | 23 ± 32 | 8 ± 7 | 0.311 |
| Donor Source (%): | |||||
| 1 = Living Related | 1 = 24.5% | 1 = 37.8% | 1 = 43.7% | 1 = 9.1% | |
| 2 = Living Unrelated | 2 = 40.6% | 2 = 8.1% | 2 = 8.5% | 2 = 31.8% | |
| 3 = Deceased | 3 = 34.0% | 3 = 44.1% | 3 = 47.9% | 3 = 54.5% | |
| Recipient Race (%): | |||||
| 1 = Caucasian | 1 = 42.5% | 1 = 43.2% | 1 = 50.7% | 1 = 27.3% | |
| 2 = Asian | 2 = 5.7% | 2 = 4.5% | 2 = 7.0% | 2 = 0.0% | |
| 3 = African American | 3 = 16.0% | 3 = 18.0% | 3 = 18.3% | 3 = 13.6% | |
| 4 = Hispanic | 4 = 7.5% | 4 = 2.7% | 4 = 5.6% | 4 = 18.2% | |
| 5 = Mixed and Others | 5 = 12.3% | 5 = 16.2% | 5 = 9.9% | 5 = 0.0% | |
| HLA Mismatch | 4.64 ± 1.41 | 4.15 ± 1.35 | 3.62 ± 1.67 | 4.80 ± 1.15 | 0.245 |
| eGFR | 75.3 ± 42.3 | 95.4 ± 28.5 | 104.1 ± 36.7 | N/A # | 0.171 |
| Injury-Specific (n = 9) | AR (n = 11) | BKVN (n = 5) |
|---|---|---|
| Glycine | Glycine | Arabinose |
| N-methylalanine | Glutaric acid | 2-hydroxy-2-methylbutanoic acid |
| Adipic acid | Adipic acid | Hypoxanthine |
| Glutaric acid | Inulobiose | Benzyl alcohol |
| Inulobiose | Threose | N-acetyl-d-mannosamine |
| Threitol | Sulfuric acid | |
| Isothreitol | Taurine | |
| Sorbitol | N-methylalanine | |
| Isothreonic acid | Asparagine | |
| 5-aminovaleric acid lactam | ||
| Myo-inositol |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sigdel, T.K.; Schroeder, A.W.; Yang, J.Y.C.; Sarwal, R.D.; Liberto, J.M.; Sarwal, M.M. Targeted Urine Metabolomics for Monitoring Renal Allograft Injury and Immunosuppression in Pediatric Patients. J. Clin. Med. 2020, 9, 2341. https://doi.org/10.3390/jcm9082341
Sigdel TK, Schroeder AW, Yang JYC, Sarwal RD, Liberto JM, Sarwal MM. Targeted Urine Metabolomics for Monitoring Renal Allograft Injury and Immunosuppression in Pediatric Patients. Journal of Clinical Medicine. 2020; 9(8):2341. https://doi.org/10.3390/jcm9082341
Chicago/Turabian StyleSigdel, Tara K., Andrew W. Schroeder, Joshua Y. C. Yang, Reuben D. Sarwal, Juliane M. Liberto, and Minnie M. Sarwal. 2020. "Targeted Urine Metabolomics for Monitoring Renal Allograft Injury and Immunosuppression in Pediatric Patients" Journal of Clinical Medicine 9, no. 8: 2341. https://doi.org/10.3390/jcm9082341
APA StyleSigdel, T. K., Schroeder, A. W., Yang, J. Y. C., Sarwal, R. D., Liberto, J. M., & Sarwal, M. M. (2020). Targeted Urine Metabolomics for Monitoring Renal Allograft Injury and Immunosuppression in Pediatric Patients. Journal of Clinical Medicine, 9(8), 2341. https://doi.org/10.3390/jcm9082341






