Tumor Tissue MIR92a and Plasma MIRs21 and 29a as Predictive Biomarkers Associated with Clinicopathological Features and Surgical Resection in a Prospective Study on Colorectal Cancer Patients
Abstract
:1. Introduction
2. Experimental Section
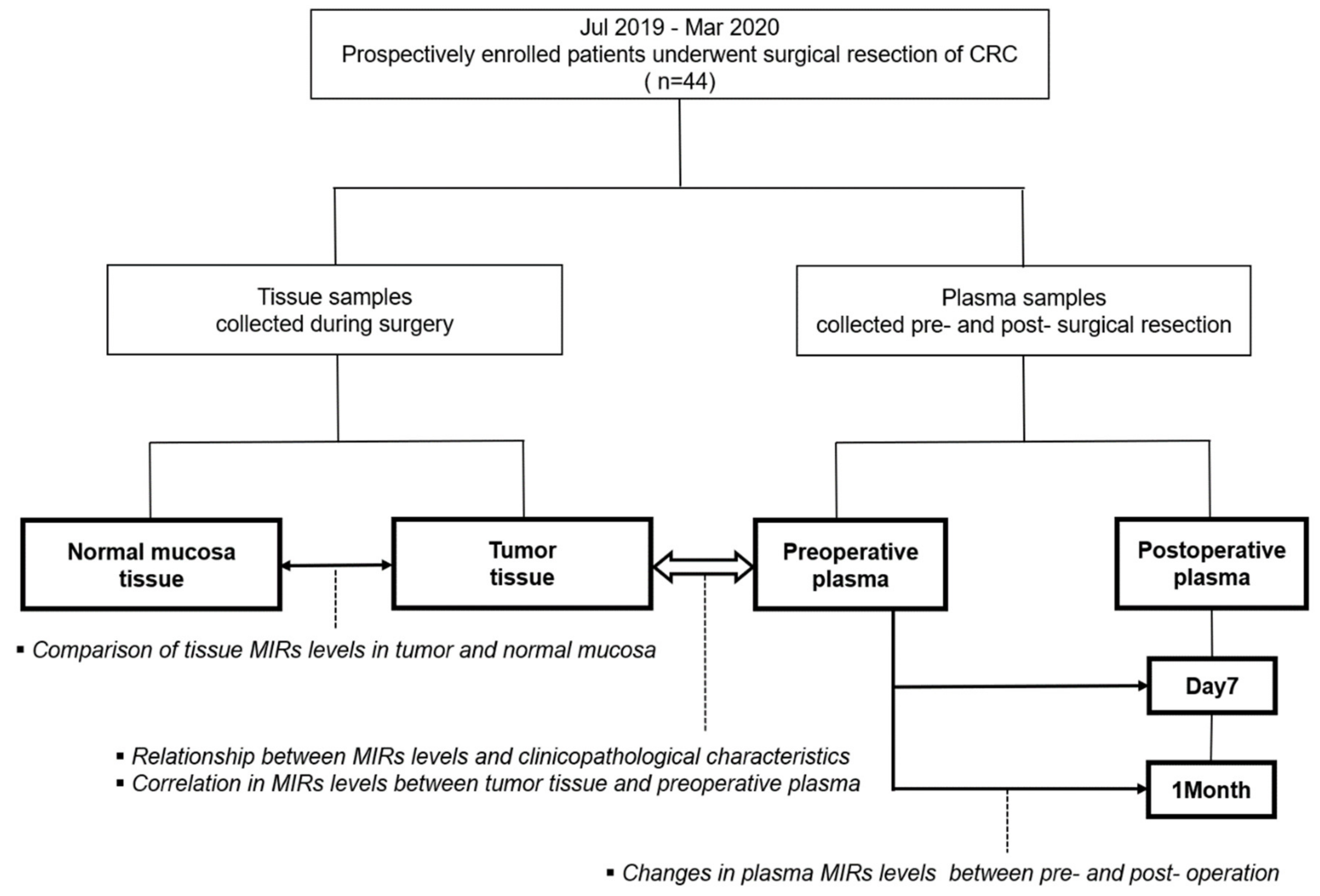
2.1. Study Population
2.2. Sample Processing
2.3. Preparation of Exosome-Enriched Fractions
2.4. Extraction of MiRNA from Tissue and Plasma Samples
2.5. Reverse Transcription-Quantitative Polymerase Chain Reaction Using Real-Time Polymerase Chain Reaction
2.6. Collection of Clinical and Pathological Characteristics
2.7. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Evaluation of the Relative Levels of Exosomal MiRNAs to the Circulating MiRNAs in Plasma
3.3. Evaluation of the Potential Internal Control for Quantification of Tumor Tissue MiRNA
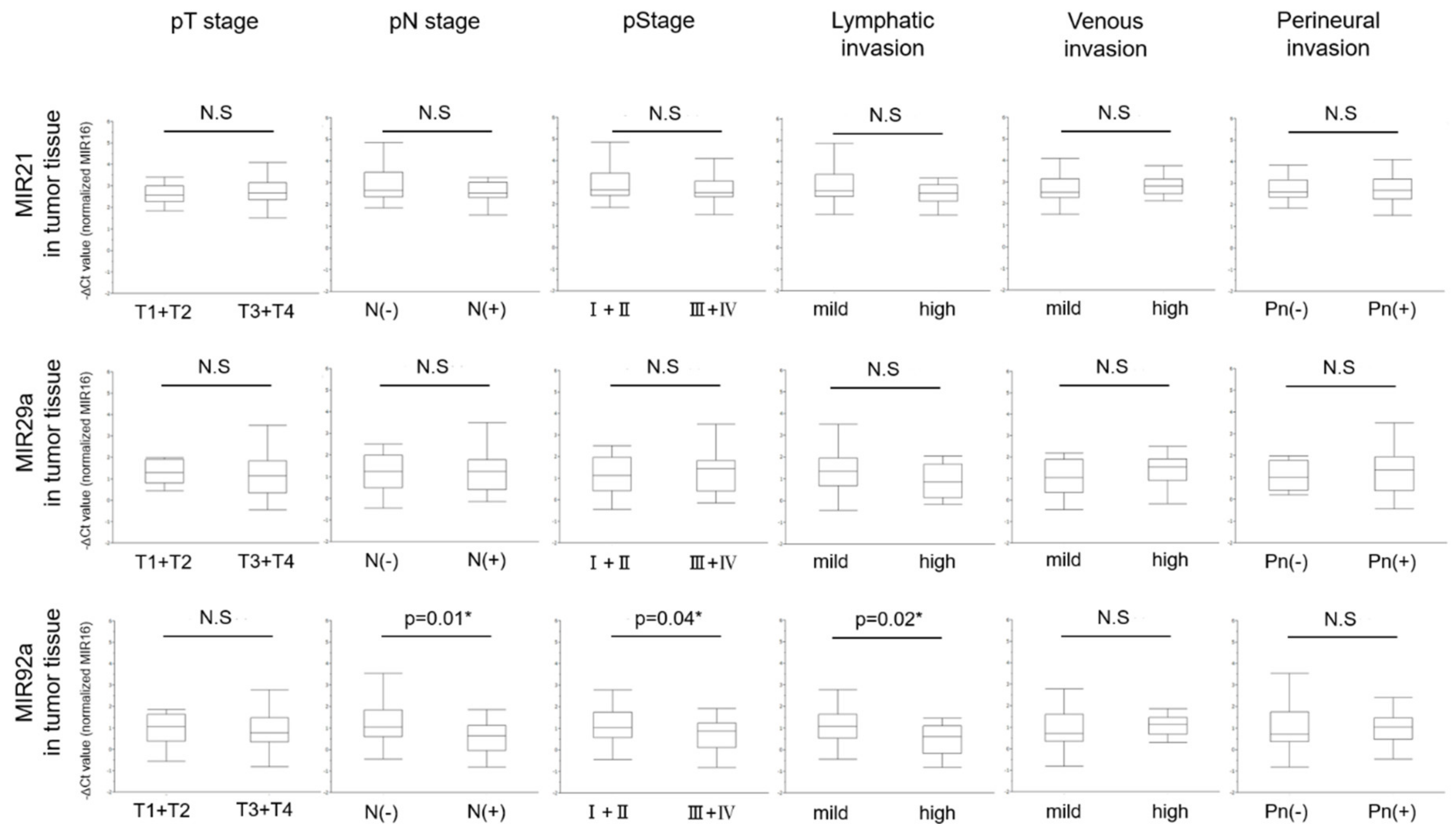
3.4. Levels of MIRs21, 29a, and 92a Evaluated by RT-qPCR in Colorectal Tumor Tissues and the Relationship with Clinicopathological Characteristics
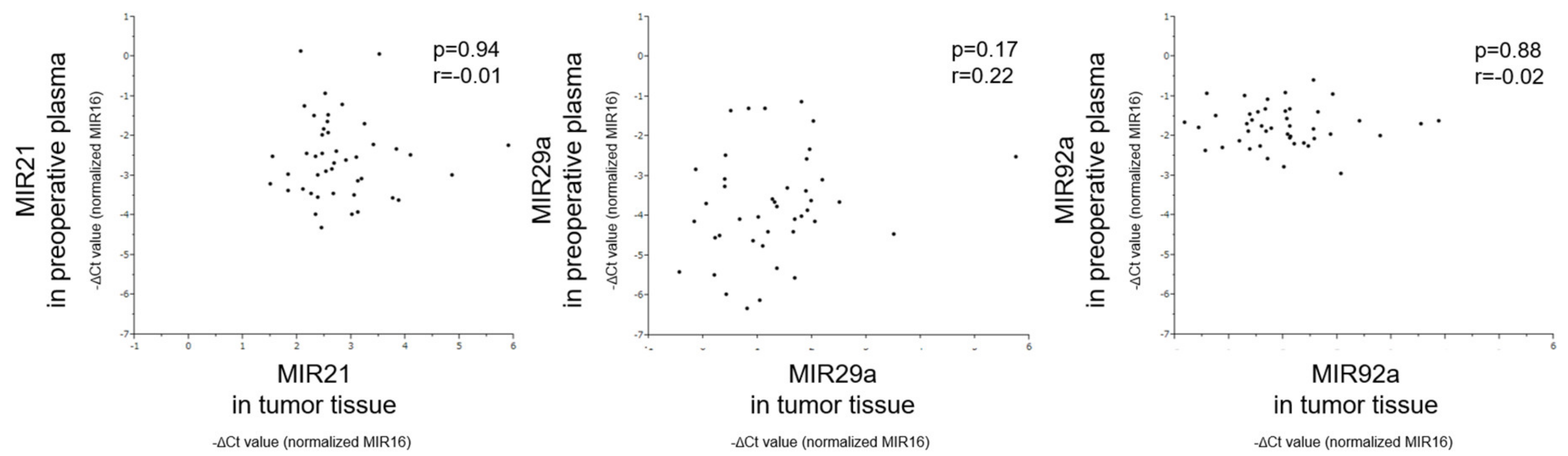
3.5. Correlation of MiRNA Levels Between Tumor Tissue and Preoperative Plasma
3.6. Relationship between Levels of MiRNAs in Preoperative Plasma and Clinicopathological Characteristics
3.7. Multivariate Analysis of MiRNAs and Pathological Characteristics in the CRC with High Lymphatic Invasion
3.8. Changes in Level of Plasma MIR21, 29a, and 92a Between Pre- and Post-Surgical Resection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarzenbach, H.; Nishida, N.; Calin, G.A.; Pantel, K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [Green Version]
- Yáñez-Mó, M.; Siljander, P.R.M.; Andreu, Z.; Bedina Zavec, A.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [Green Version]
- Azmi, A.S.; Bao, B.; Sarkar, F.H. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metastasis Rev. 2013, 32, 623–642. [Google Scholar] [CrossRef] [Green Version]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [Green Version]
- De Jong, O.G.; Verhaar, M.C.; Chen, Y.; Vader, P.; Gremmels, H.; Posthuma, G.; Schiffelers, R.M.; Gucek, M.; van Balkom, B.W.M. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J. Extracell. Vesicles 2012, 1, 18396. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, B.S.; Jorgensen, S.; Fog, J.U.; Sokilde, R.; Christensen, I.J.; Hansen, U.; Brunner, N.; Baker, A.; Moller, S.; Nielsen, H.J. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin. Exp. Metastasis 2011, 28, 27–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.S.; Nam, SK.; Koh, J.; Kim, D.W.; Kang, S.B.; Choe, G.; Kim, W.H.; Lee, H.S. Stromal expression of microRNA-21 in advanced colorectal cancer patients with distant metastases. J. Pathol. Transl. Med. 2016, 50, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Zhang, Y.Y.; Wu, Y.Y.; Wang, X.D.; Wan, L.H.; Li, L.; Zhou, L.M. Correlation of over-expressions of miR-21 and Notch-1 in human colorectal cancer with clinical stages. Life Sci. 2014, 106, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wang, X.; Huang, Z.; Wang, J.; Zhu, W.; Shu, Y.; Liu, P. Prognostic value of miR-21 in various cancers: An updating meta-analysis. PLoS ONE 2014, 9, e102413. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.G.; Gu, J. Serum microRNA-29a is a promising novel markerfor early detection of colorectal liver metastasis. Cancer Epidemiol. 2012, 36, e61–e67. [Google Scholar] [CrossRef]
- Zhi, M.L.; Liu, Z.J.; Yi, X.Y.; Zhang, L.J.; Bao, Y.X. Diagnostic performance of microRNA-29a for colorectal cancer: A meta-analysis. Genet. Mol. Res. 2015, 14, 18018–18025. [Google Scholar] [CrossRef]
- Wu, C.W.; Ng, S.S.; Dong, Y.J.; Ng, S.C.; Leung, W.W.; Lee, C.W.; Wong, Y.N.; Chan, F.K.; Yu, J.; Sung, J.J. Detection of miR-92a and miR-21 in stool samples as potential screening biomarkers for colorectal cancer and polyps. Gut 2012, 61, 739–745. [Google Scholar] [CrossRef]
- Huang, Z.; Huang, D.; Ni, S.; Peng, Z.; Sheng, W.; Du, X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int. J. Cancer 2010, 127, 118–126. [Google Scholar] [CrossRef]
- Heishima, K.; Mori, T.; Ichikawa, Y.; Sakai, H.; Kuranaga, Y.; Nakagawa, T.; Tanaka, Y.; Okamura, Y.; Masuzawa, M.; Sugito, N.; et al. MicroRNA-214 and MicroRNA-126 are potential biomarkers for malignant endothelial proliferative diseases. Int. J. Mol. Sci. 2015, 16, 25377–25391. [Google Scholar] [CrossRef] [Green Version]
- Brierley, J.D.; Gospodarowicz, M.K.; Wittekind, C. International union against cancer. In TNM Classification of Malignant Tumors, 8th ed.; Wiley-Blackwell: Chichester, UK, 2017. [Google Scholar]
- Japanese Society for Cancer of the Colon and Rectum. Japanese classification of colorectal, appendiceal, and anal carcinoma: The 3rd English edition. J. Anus. Rectum. Colon. 2019, 3, 175–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Editorial Board of the Cancer Statistics in Japan. Cancer Statistics in Japan 2019; Foundation of Promotion of Cancer Research: Tokyo, Japan, 2020. [Google Scholar]
- Betge, J.; Pollheimer, M.J.; Lindtner, R.A.; Kornprat, P.; Schlemmer, A.; Rehak, P.; Vieth, M.; Hoefler, G.; Langner, C. Intramural and extramural vascular invasion in colorectal cancer: Prognostic significance and quality of pathology reporting. Cancer 2012, 118, 628–638. [Google Scholar] [CrossRef] [PubMed]
- Sejben, I.; Bori, R.; Csemi, G. Venous invasion demonstrated by orcein staining of colorectal carcinoma specimens is associated with the development of distant metastasis. J. Clin. Pathol. 2010, 63, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Schoppmann, A.; Tamandl, D.; Herberger, B.; Längle, F.; Birner, P.; Geleff, S.; Grünberger, T.; Schoppmann, S.F. Comparison of lymphangiogenesis between primary colorectal cancer and corresponding liver metastases. Anticancer Res. 2011, 31, 4605–4611. [Google Scholar] [PubMed]
- Akagi, Y.; Adachi, Y.; Ohchi, T.; Kinugasa, T.; Shirouzu, K. Prognostic impact of lymphatic invasion of colorectal cancer: A single-center analysis of 1616 patients over 24 years. Anticancer Res. 2013, 33, 2965–2970. [Google Scholar]
- Hayashita, Y.; Osada, H.; Tatematsu, Y.; Yamada, H.; Yanagisawa, K.; Tomida, S.; Yatabe, Y.; Kawahara, K.; Sekido, Y.; Takahashi, T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005, 65, 9628–9632. [Google Scholar] [CrossRef] [Green Version]
- Shigoka, M.; Tsuchida, A.; Matsudo, T.; Nakagawa, Y. Deregulation of miR-92a expression is implicated in hepatocellular carcinoma development. Pathol. Int. 2010, 60, 351–357. [Google Scholar]
- Chen, ZL.; Zhao, X.H.; Wang, J.W.; Li, B.Z.; Wang, Z.; Sun, J.; Tan, F.W.; Ding, D.P.; Xu, X.H.; Zhou, F.; et al. MicroRNA-92a promotes lymph node metastasis of human esophageal squamous cell carcinoma via E-cadherin. J. Biol. Chem. 2010, 286, 10725–10734. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, M.; Oikawa, K.; Takanashi, M.; Kudo, M.; Ohyashiki, J.; Ohyashiki, K.; Kuroda, M. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS ONE 2009, 4, e5532. [Google Scholar] [CrossRef]
- Diosdado, B.; Van De Wiel, M.A.; Droste, J.T.S.; Mongera, S.; Postma, C.; Meijerink, W.J.H.J.; Carvalho, B.; Meijer, G.A. MiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br. J. Cancer 2009, 101, 707–714. [Google Scholar]
- Connolly, E.; Melegari, M.; Landgraf, P.; Tchaikovskaya, T.; Tennant, B.C.; Slagle, B.L.; Rogler, L.E.; Zavolan, M.; Tuschl, T.; Rogler, C.E. Elevated expression of the miR-17-92 polycistron and miR-21 in hepadnavirusassociated hepatocellular carcinoma contributes to the malignant phenotype. Am. J. Pathol. 2008, 173, 856–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuchida, A.; Ohno, S.; Wu, W.; Borjigin, N.; Fujita, K.; Aoki, T.; Ueda, S.; Takanashi, M.; Kuroda, M. miR-92 is a key oncogenic component of the miR-17-92 cluster in colon cancer. Cancer Sci. 2011, 102, 2264–2271. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Zhang, Z.; Wang, Y.; Li, C.; Gong, W.; Wang, X. MicroRNA-92a promotes colorectal cancer cell growth and migration by inhibiting KLF4. Oncol. Res. 2016, 23, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Yamada, N.; Nakagawa, Y.; Tsujimura, N.; Kumazaki, M.; Noguchi, S.; Mori, T.; Hirata, I.; Maruo, K.; Akao, Y. Role of intracellular and extracellular microRNA-92ain colorectal cancer. Transl. Oncol. 2013, 6, 482–492. [Google Scholar] [CrossRef] [Green Version]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs-microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Liu, Z.L.; Wang, H.; Liu, J.; Wang, Z.X. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol. Cell. Biochem. 2012, 372, 35–45. [Google Scholar] [CrossRef]
- Zhang, B.G.; Li, J.F.; Yu, B.Q.; Zhu, Z.G.; Liu, B.Y.; Yan, M. microRNA-21 promotes tumor proliferation and invasion in gastric cancer by targeting PTEN. Oncol. Rep. 2012, 27, 1019–1026. [Google Scholar] [CrossRef] [Green Version]
- Qin, X.; Yan, L.; Zhao, X.; Li, C.; Fu, Y. microRNA-21 overexpression contributes to cell proliferation by targeting PTEN in endometrioid endometrial cancer. Oncol. Lett. 2012, 4, 1290–1296. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Yu, J.; Yu, S.; Lavker, R.M.; Cai, L.; Liu, W.; Yang, K.; He, X.; Chen, S. MicroRNA-21 acts as an oncomir through multiple targets in human hepatocellular carcinoma. J. Hepatol. 2010, 53, 98–107. [Google Scholar] [CrossRef]
- Lei, B.X.; Liu, Z.H.; Li, ZJ.; Li, C.; Deng, Y.F. miR-21 induces cell proliferation and suppresses the chemosensitivity in glioblastoma cells via downregulation of FOXO1. Int. J. Clin. Exp. Med. 2014, 7, 2060–2066. [Google Scholar]
- Yamamichi, N.; Shimomura, R.; Inada, K.; Sakurai, K.; Haraguchi, T.; Ozaki, Y.; Fujita, S.; Mizutani, T.; Furukawa, C.; Fujishiro, M.; et al. Locked nucleic acid in situ hybridization analysis of miR-21 expression during colorectal cancer development. Clin. Cancer Res. 2009, 15, 4009–4016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Song, Y.; Xiong, Y.; Wang, X.; Xu, K.; Han, B.; Bai, Y.; Li, L.; Zhang, Y.; Zhou, L. MicroRNA-21 (Mir-21) promotes cell growth and invasion by repressing tumor suppressor PTEN in colorectal cancer. Cell. Physiol. Biochem. 2017, 43, 945–958. [Google Scholar] [CrossRef]
- Wei, J.; Liu, L.K.; Gao, W.; Zhu, C.J.; Liu, Y.Q.; Cheng, T.; Shu, Y.Q. Reduction of Plasma MicroRNA-21 is associated with chemotherapeutic response in patients with non-small cell lung cancer. Chin. J. Cancer Res. 2011, 23, 123–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, J.; Tang, W.; Du, P.; Wang, G.; Chen, W.; Li, J.; Zhu, Y.; Gao, J.; Cui, L. Identifying microRNA-mRNA regulatory network in colorectal cancer by a combination of expression profile and bioinformatics analysis. BMC Syst. Biol. 2012, 6, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.; Zhu, Y.; Gao, J.; Fu, J.; Liu, C.; Liu, Y.; Song, C.; Zhu, S.; Leng, Y.; Wang, G.; et al. MicroRNA-29a promotes colorectal cancer metastasis by regulating matrix metalloproteinase 2 and E-cadherin via KLF4. Br. J. Cancer 2014, 110, 450–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, L.L.; Li, L.; Liu, J.N.; Mei, J.; Lei, C.J. Down-regulation of miR-29a facilitates apoptosis of colorectal carcinoma cell SW480 and suppresses its Paclitaxel resistance. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5499–5507. [Google Scholar]



| Patient Total n = 44 | |
|---|---|
| Gender, n (%) | Male 17 (38.6) Female 27 (61.4) |
| Age (years), median (range) | 72 (42–85) |
| BMI, median (range) | 22.1 (15.1–40.3) |
| Primary colorectal tumor location, n (%) | C 3 (6.8)/A 8(18.2)/T 6 (13.6)/D 2 (4.5)/S 8 (18.2)/R 17 (38.6) |
| Colon 27 (61.4)/Rectum 17 (38.6) | |
| Right side 17 (38.6)/Left side 27 (61.4) | |
| Macroscopic types§, n (%) | Type0 4 (9.1)/Type1 4 (9.1)/Type2 29 (65.9)/Type5 7 (15.9) |
| Maximum tumor diameter (mm), median (range) | 45 (15–180) |
| Pathological T Stage§, n (%) | T1 4 (9.1)/T2 7 (15.9)/T3 21 (47.7)/T4 12 (27.3) |
| Pathological N Stage§, n (%) | N0 24 (54.6)/N1 14 (31.8)/N2 6 (13.6) |
| Pathological Stage§, n (%) | I 9 (20.5)/II 13 (29.5)/III 17 (38.6)/IV 5 (11.4) |
| Histological types¶, n (%) | tub1 9 (20.5)/tub2 29 (65.9)/por 1 (2.3)/muc 5 (11.4) |
| Lymphatic invasion¶, n (%) | Ly0 8 (18.2)/Ly1a 23 (52.3)/Ly1b 8(18.2)/Ly1c 5 (11.4) |
| Venous invasion¶, n (%) | V0 12 (27.2)/V1a 17 (38.6)/V1b 8 (18.2)/V1c 7 (15.9) |
| Perineural invasion¶, n (%) | Pn0 19 (43.2)/Pn1 25 (56.8) |
| Preoperative CEA, n (%) | Normal 26 (59.1) Elevated 18 (40.1) |
| Preoperative CA19-9, n (%) | Normal 32 (72.7) Elevated 12 (27.3) |
| Target MicroRNAs | ||||
|---|---|---|---|---|
| MIR16 | MIR21 | MIR29a | MIR92a | |
| −ΔCt value ‡, median (range) | −6.35 (−9.72–−5.10) | −4.81 (−6.86–−3.72) | −5.11 (−7.10–−4.50) | −6.35 (−9.72–−5.1) |
| Relative expression level §, median (range) | 0.014 (0.001–0.029) | 0.038 (0.009–0.076) | 0.029 (0.007–0.12) | 0.036 (0.014–0.044) |
| Target MicroRNAs | ||||||||
|---|---|---|---|---|---|---|---|---|
| MIR21 | MIR29a | MIR92a | ||||||
| −∆Ct value ‡ | p-value | −∆Ct value‡ | p-value | −∆Ct value ‡ | p-value | |||
| Gender | Male | 17 (38.6) | 2.58 (1.51–5.91) | 0.87 | 0.92 (−0.16–3.5) | 0.25 | 1.04 (−0.56–2.07) | 0.41 |
| Female | 27 (61.4) | 2.57 (1.55–4.86) | 1.36 (−0.44–5.75) | 1.01 (−0.82–3.87) | ||||
| Age | ≥75 | 14 (31.8) | 2.30 (1.51–5.91) | 0.66 | 1.42 (−0.44–5.75) | 0.54 | 0.75 (−0.44–3.55) | 0.82 |
| <75 | 30 (68.2) | 2.61 (1.55–4.86) | 1.17 (−0.14–2.50) | 1.04 (−0.82–3.87) | ||||
| BMI | ≥22 | 22 (50.0) | 2.48 (1.55–3.87) | 0.03* | 0.4 (−0.17–2.50) | 0.09† | 0.75 (−0.82–2.41) | 0.28 |
| <22 | 22 (50.0) | 2.96 (1.51–5.91) | 1.62 (−0.44–5.75) | 1.10 (−0.44–3.87) | ||||
| Tumor location | Colon | 27 (61.4) | 2.67 (1.51–5.91) | 0.70 | 1.14 (−0.44–3.5) | 0.47 | 0.71 (−0.82–2.79) | 0.25 |
| Rectum | 17 (38.6) | 2.53 (1.55–4.86) | 1.32 (−0.16–5.75) | 1.12 (−0.56–3.87) | ||||
| Right side | 17 (38.6) | 2.52 (1.51–4.1) | 0.16 | 0.84 (−0.44–2.19) | 0.06† | 0.68 (−0.82–2.41) | 0.11 | |
| Left side | 27 (61.4) | 2.65 (1.55–5.91) | 1.36 (−0.16–5.75) | 1.12 (−0.56–3.87) | ||||
| Preoperative CEA | Elevated | 18 (40.1) | 2.57 (1.55–5.91) | 0.87 | 1.21 (−0.44–5.75) | 0.64 | 1.04 (−0.82–3.55) | 0.57 |
| Normal | 26 (59.1) | 2.61 (1.51–4.86) | 1.27 (−0.17–2.5) | 0.86 (−0.56–3.87) | ||||
| Preoperative CA19-9 | Elevated | 12 (27.3) | 2.54 (1.55–3.13) | 0.14 | 1.01 (−0.16–2.05) | 0.21 | 1.10 (−0.82–1.56) | 0.07 † |
| Normal | 32 (72.7) | 2.62 (1.51–5.91) | 1.36 (−0.44–5.75) | 1.08 (−0.56–3.78) | ||||
| Maximum tumor diameter | ≥45 | 22 (50.0) | 2.57 (1.51–5.91) | 0.95 | 1.34 (−0.17–3.5) | 0.97 | 0.75 (−0.56–2.07) | 0.16 |
| <45 | 22 (50.0) | 2.65 (1.84–4.86) | 1.04 (−0.44–5.75) | 1.07 (−0.82–3.87) | ||||
| Pathological T stage § | T3 + T4 | 33 (75.0) | 2.67 (1.51–5.91) | 0.77 | 1.14 (−0.44–5.75) | 0.79 | 0.78 (−0.82–3.55) | 0.51 |
| T1 + T2 | 11 (25.0) | 2.57 (1.84–4.86) | 1.28 (0.43–1.99) | 1.07 (−0.56–3.87) | ||||
| Pathological N stage § | Present | 20 (45.4) | 2.53 (1.51–5.91) | 0.35 | 1.23 (−0.16–3.5) | 0.57 | 0.64 (−0.82–1.87) | 0.01 * |
| Absent | 24 (54.5) | 2.67 (1.84–4.86) | 1.24 (−0.44–5.75) | 1.06 (−0.44–3.87) | ||||
| Pathological Stage § | III + IV | 22 (50.0) | 2.53 (1.51- 5.91) | 0.55 | 1.44 (−0.16–3.50) | 0.85 | 0.86 (−0.82–1.92) | 0.04 * |
| I + II | 22 (50.0) | 2.665 (1.84–4.86) | 1.12 (−0.44–5.75) | 1.03 (−0.44–3.87) | ||||
| Histological differentiation ¶ | Poorly | 6 (13.6) | 2.23 (1.51–2.83) | 0.03* | 0.6 (−0.16–1.92) | 0.16 | 0.13 (−0.56–1.38) | 0.03 * |
| Well and moderately | 38 (86.4) | 2.66 (1.84–5.91) | 1.32 (−0.44–5.75) | 1.06 (−0.82–3.87) | ||||
| Lymphatic invasion ¶ | High | 13 (29.5) | 2.53 (1.51–3.24) | 0.14 | 0.84 (−0.17–2.05) | 0.13 | 0.6 (−0.82–1.46) | 0.02 * |
| Mild | 31 (70.4) | 2.65 (1.55–5.91) | 1.35 (−0.44–5.75) | 1.08 (−0.44–3.87) | ||||
| Venous invasion ¶ | High | 15 (34.1) | 2.83 (2.14–5.91) | 0.29 | 1.55 (−0.17–3.5) | 0.33 | 1.12 (−0.56–1.87) | 0.97 |
| Mild | 29(65.9) | 2.52 (1.51–4.86) | 1.04 (−0.44–5.75) | 0.71 (−0.82–3.87) | ||||
| Perineural invasion ¶ | Positive | 25 (56.8) | 2.68 (1.51–5.91) | 0.97 | 1.35 (−0.44–3.5) | 0.97 | 1.04 (−0.44–2.41) | 0.61 |
| Negative | 19 (43.2) | 2.58 (1.84–4.86) | 1.01 (0.2–5.75) | 0.71(−0.82–3.87) | ||||
| Target MicroRNAs | ||||||||
|---|---|---|---|---|---|---|---|---|
| MIR21 | MIR29a | MIR92a | ||||||
| −∆Ct value ‡ | p-value | −∆Ct value ‡ | p-value | −∆Ct value‡ | p-value | |||
| Gender | Male | 17 (38.6) | −2.54 [−3.91–0.14) | 0.63 | −3.86 (−5.63–−1.3) | 0.44 | −1.79 (−2.95–−0.59) | 0.98 |
| Female | 27 (61.4) | −2.61 (−4.31–0.07) | −3.76 (−1.14–−6.33) | −1.70 (−2.78–−0.95) | ||||
| Age | ≥75 | 14 (31.8) | −2.74 (−3.91–0.07) | 0.22 | −3.58 (−5.42–−2.32) | 0.74 | −1.66 (−2.36–−0.92) | 0.17 |
| <75 | 30 (68.2) | −2.86 (−4.31–0.14) | −4.03 (−6.33–−1.14) | −1.85 (−2.95–−0.59) | ||||
| BMI | ≥22 | 22 (50.0) | −2.62 (−4.31–0.14) | 0.92 | −4.08 (−6.33–−1.35) | 0.21 | −1.80 (−2.95–−0.59) | 0.34 |
| <22 | 22 (40.0) | −2.57 (−3.98–−0.93) | −3.61 (−5.97–−1.14) | −1.73 (−2.36–−0.90) | ||||
| Tumor location | Colon | 27 (61.4) | −2.52 (−3.91–0.07) | 0.71 | −3.92 (−6.33–−1.14) | 0.67 | −1.81 (−2.95–−0.90) | 0.70 |
| Rectum | 17 (38.6) | −2.61 (−4.31–0.14) | −3.76 (−5.97–−1.35) | −1.75 (−2.57–−0.59) | ||||
| Right side | 17 (38.6) | −2.48 (−3.91–0.07) | 0.17 | −3.38 (−6.13–−1.14) | 0.16 | −1.66 (−2.36–−0.90) | 0.23 | |
| Left side | 27 (61.4) | −2.84 (−4.31–0.14) | −4.01 (−6.33–−1.35) | −1.83 (−2.95–−0.59) | ||||
| Preoperative CEA | Elevated | 18 (40.1) | −2.52 (−4.31–−0.93) | 0.51 | −3.66 (−6.13–−1.14) | 0.51 | −1.78 (−2.57–−0.59) | 0.74 |
| Normal | 26 (59.1) | −2.86 (−3.98–0.14) | −3.86 (−6.33–−1.35) | −1.77 (−2.95–−0.99) | ||||
| Preoperative CA19-9 | Elevated | 12 (27.3) | −2.45 (−3.54–0.14) | 0.07† | −3.66 (−4.76–−1.14) | 0.10 | −1.81 (−2.95–−0.99) | 0.58 |
| Normal | 32 (72.7) | −2.77 (−4.31–0.07) | −3.86 (−6.33–−1.35) | −1.77 (−2.95–−0.95) | ||||
| Maximum tumor diameter | ≥45 | 22 (50.0) | −2.50 (−3.61–0.14) | 0.07† | −3.68 (−5.32–−1.30) | 0.046* | −1.77 (−2.95–−0.59) | 0.30 |
| <45 | 22 (50.0) | −2.99 (−4.31–0.07) | −4.08 (−6.33–−1.35) | −1.70 (−2.78–−1.38) | ||||
| Pathological T stage § | T3 + T4 | 33 (75.0) | −2.52 (−4.31–0.14) | 0.14 | −3.70 (−6.13–−1.14) | 0.19 | −1.75 (−2.95–−0.59) | 0.64 |
| T1 + T2 | 11 (25.0) | −2.99 (−3.98–−1.46) | −4.03 (−6.33–−2.32) | −1.79 (−2.78–−1.39) | ||||
| Pathological N stage § | Present | 20 (45.4) | −2.53 (−3.97–0.14) | 0.22 | −3.70 (−5.56–−1.14) | 0.09† | −1.77 (−2.33–−0.59) | 0.39 |
| Absent | 24 (54.5) | −2.77 (−4.31–0.07) | −4.03 (−6.33–−1.61) | −1.76 (−2.95–−0.95) | ||||
| Pathological Stage § | III + IV | 22 (50.0) | −2.52 (−3.97–0.14) | 0.21 | −3.66 (−5.56–−1.14) | 0.02* | −1.77 (−2.33–−0.59) | 0.24 |
| I + II | 22 (50.0) | −2.90 (−4.31–0.07) | −4.09 (−6.33–−2.32) | −1.76 (−2.95–−1.07) | ||||
| Histological differentiation ¶ | Poorly | 6 (13.6) | −2.70 (−3.21–0.14) | 0.24 | −3.66 (−3.86–−1.30) | 0.22 | −1.98 (−2.33–−0.92) | 0.56 |
| Well and moderately | 38 (86.4) | −2.58 (−4.31–0.07) | −4.03 (−6.33–−1.14) | −1.73 (−2.95–−0.59) | ||||
| Lymphatic invasion ¶ | High | 13 (29.5) | −1.92 (−3.98–0.14) | 0.05† | −3.58 (−5.97–−1.14) | 0.05† | −1.75 (−2.29–−0.9) | 0.61 |
| Mild | 31 (70.4) | −2.69 (−4.31–0.07) | −4.03 (−6.33–−1.61) | −1.81 (−2.95–−0.59) | ||||
| Venous invasion ¶ | High | 15 (34.1) | −2.43 (−3.98–−1.20) | 0.85 | −4.14 (−5.97–−1.30) | 0.43 | −1.75 (−2.26–−0.59) | 0.10 |
| Mild | 29 (65.9) | −2.84 (−4.31–0.14) | −3.66 (−6.33–−1.14) | −1.81 (−2.95–−0.92) | ||||
| Perineural invasion ¶ | Positive | 25 (56.8) | −2.48 (−3.61–0.14) | 0.01* | −3.66 (−6.13–−1.14) | 0.26 | −1.81 (−2.95–−0.59) | 0.58 |
| Negative | 19 (43.2) | −3.08 (−4.31–−1.49) | −4.03 (−6.33–−1.35) | −1.69 (−2.78–−1.39) | ||||
| Prognostic factors | p-Value | Odds Ratio | 95% Confidence Interval |
|---|---|---|---|
| Pathological T stage § (T3+T4/T1+T2) | 0.66 | 1.84 | 0.12–33.45 |
| Venous invasion ¶ (high/mild) | <0.001 *** | 66.84 | 5.10–2853.98 |
| Perineural invasion ¶ (positive/negative) | 0.53 | 0.46 | 0.03–5.70 |
| Level of MIR92a in tumor tissue (−∆Ct value‡) (high/low) | 0.01 * | 0.08 | 0.003–0.60 |
| Level of MIR21 in preoperative plasma (−∆Ct value ‡) (high/low) | 0.19 | 0.20 | 0.008–2.16 |
| Level of MIR29a in preoperative plasma (−∆Ct value ‡) (high/low) | 0.009 ** | 30.72 | 2.24–1038.11 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fukada, M.; Matsuhashi, N.; Takahashi, T.; Sugito, N.; Heishima, K.; Akao, Y.; Yoshida, K. Tumor Tissue MIR92a and Plasma MIRs21 and 29a as Predictive Biomarkers Associated with Clinicopathological Features and Surgical Resection in a Prospective Study on Colorectal Cancer Patients. J. Clin. Med. 2020, 9, 2509. https://doi.org/10.3390/jcm9082509
Fukada M, Matsuhashi N, Takahashi T, Sugito N, Heishima K, Akao Y, Yoshida K. Tumor Tissue MIR92a and Plasma MIRs21 and 29a as Predictive Biomarkers Associated with Clinicopathological Features and Surgical Resection in a Prospective Study on Colorectal Cancer Patients. Journal of Clinical Medicine. 2020; 9(8):2509. https://doi.org/10.3390/jcm9082509
Chicago/Turabian StyleFukada, Masahiro, Nobuhisa Matsuhashi, Takao Takahashi, Nobuhiko Sugito, Kazuki Heishima, Yukihiro Akao, and Kazuhiro Yoshida. 2020. "Tumor Tissue MIR92a and Plasma MIRs21 and 29a as Predictive Biomarkers Associated with Clinicopathological Features and Surgical Resection in a Prospective Study on Colorectal Cancer Patients" Journal of Clinical Medicine 9, no. 8: 2509. https://doi.org/10.3390/jcm9082509
APA StyleFukada, M., Matsuhashi, N., Takahashi, T., Sugito, N., Heishima, K., Akao, Y., & Yoshida, K. (2020). Tumor Tissue MIR92a and Plasma MIRs21 and 29a as Predictive Biomarkers Associated with Clinicopathological Features and Surgical Resection in a Prospective Study on Colorectal Cancer Patients. Journal of Clinical Medicine, 9(8), 2509. https://doi.org/10.3390/jcm9082509






