Evolving Technologies in Gastrointestinal Microbiome Era and Their Potential Clinical Applications
Abstract
:1. Introduction
2. Methods
3. Results and Discussion
3.1. The Human Gut and Its Microbiota
3.2. Technologies in Gastrointestinal Microbiome Study
3.3. 16S rRNA Gene Amplicon Sequencing
3.4. Microfluidics
3.5. High-Throughput Metabolomics
3.6. Metatranscriptomics
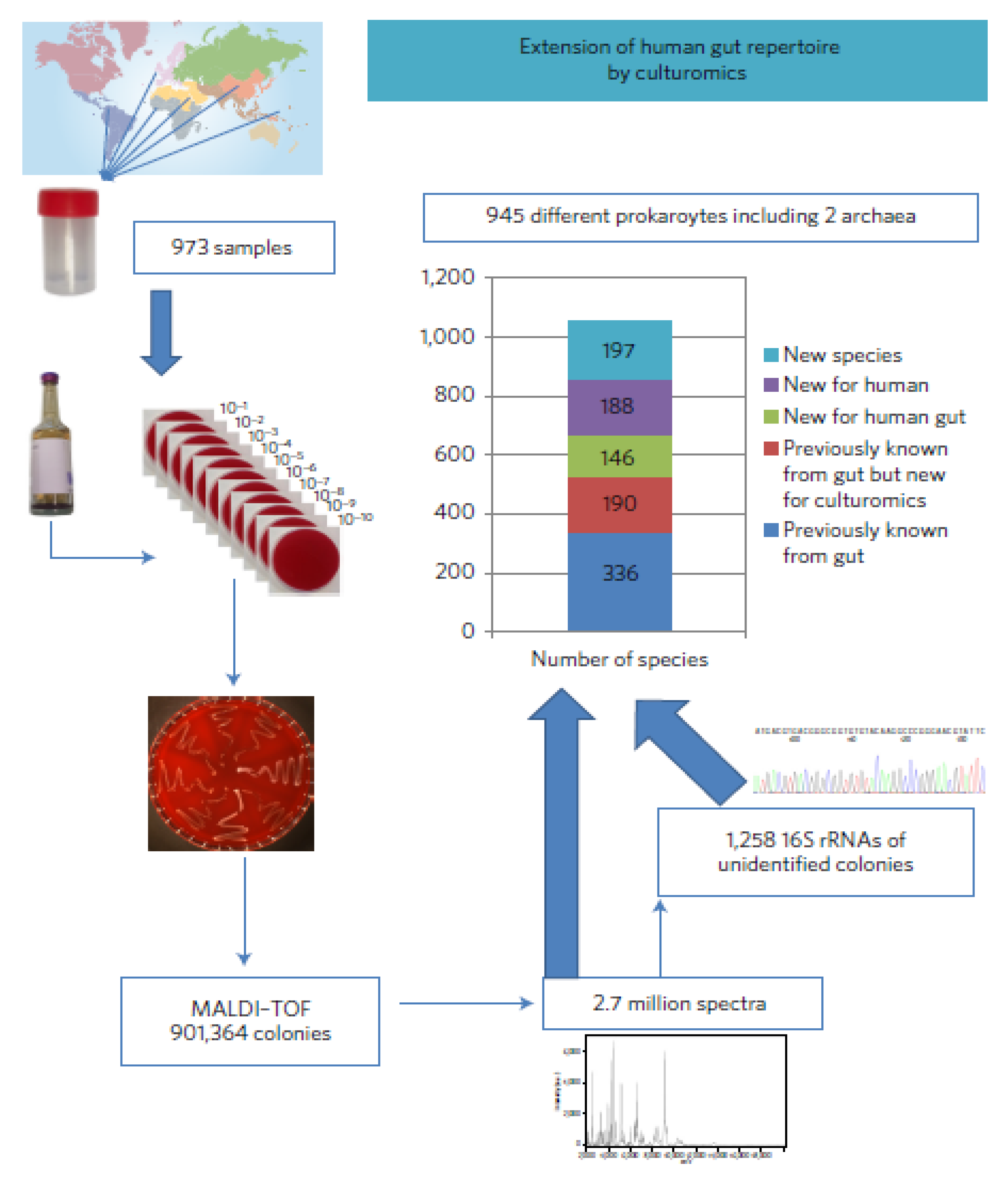
3.7. High-Throughput Culturing
4. Future Perspectives and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Matsuki, T.; Tanaka, R. Function of the human gut microbiota. In The Human Microbiota and Microbiome; CABI Publishing: Cardiff, UK, 2014; pp. 90–106. [Google Scholar]
- Blaut, M. Composition and function of the gut microbiome. In The Gut Microbiome in Health and Disease; Springer Science and Business Media LLC: Berlin, Germany, 2018; pp. 5–30. [Google Scholar]
- Valdes, A.; Walter, J.; Segal, E.; Spector, T. Role of the gut microbiota in nutrition and health. BMJ 2018, 361, k2179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pires, E.S.; Hardoim, C.C.P.; Miranda, K.R.; Secco, D.A.; Lobo, L.A.; de Carvalho, D.P.; Han, J.; Borchers, C.H.; Ferreira, R.B.R.; Salles, J.F.; et al. The gut microbiome and metabolome of two riparian communities in the amazon. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Karkman, A.; Lehtimäki, J.; Ruokolainen, L. The ecology of human microbiota: Dynamics and diversity in health and disease. Ann. N. Y. Acad. Sci. 2017, 1399, 78–92. [Google Scholar] [CrossRef]
- Vila, A.V.; Imhann, F.; Collij, V.; Jankipersadsing, S.A.; Gurry, T.; Mujagic, Z.; Kurilshikov, A.; Bonder, M.J.; Jiang, X.; Tigchelaar, E.F.; et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci. Transl. Med. 2018, 10, eaap8914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Sun, G.; Feng, T.; Zhang, J.; Huang, X.; Wang, T.; Xie, Z.; Chu, X.; Yang, J.; Wang, H.; et al. Sodium oligomannate therapeutically remodels gut microbiota and suppresses gut bacterial amino acids-shaped neuroinflammation to inhibit Alzheimer’s disease progression. Cell Res. 2019, 29, 787–803. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.-W.; Adams, J.B.; Gregory, A.C.; Borody, T.; Chittick, L.; Fasano, A.; Khoruts, A.; Geis, E.; Maldonado, J.; McDonough-Means, S.; et al. Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: An open-label study. Microbiome 2017, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Wu, K.; Pan, W.; Zeng, Y.; Hu, K.; Chen, D.; Huang, X.; Zhang, Q. Intestinal flora alterations in patients with early chronic kidney disease: A case-control study among the Han population in southwestern China. J. Int. Med. Res. 2020, 48, 300060520926033. [Google Scholar] [CrossRef]
- Lippert, K.; Kedenko, L.; Antonielli, L.; Gemeier, C.; Leitner, M.; Kautzky-Willer, A.; Paulweber, B.; Hackl, E.; Kedenko, I. Gut microbiota dysbiosis associated with glucose metabolism disorders and the metabolic syndrome in older adults. Benef. Microbes 2017, 8, 545–556. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Zhuang, Z.-Q.; Shen, L.-L.; Li, W.-W.; Fu, X.; Zeng, F.; Gui, L.; Lv, Y.; Cai, M.; Zhu, C.; Tan, Y.-L.; et al. Gut Microbiome is Altered in Patients with Alzheimer’s Disease. J. Alzheimer’s Dis. 2018, 63, 1337–1346. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Li, B.; Sun, D.; Chen, S. Gut microbiota: Implications in Alzheimer’s disease. J. Clin. Med. 2020, 9, 2042. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Han, Y.; Dy, A.B.C.; Hagerman, R.J. The Gut Microbiota and Autism Spectrum Disorders. Front. Cell. Neurosci. 2017, 11, 120. [Google Scholar] [CrossRef] [PubMed]
- Jie, Z.; Xia, H.; Zhong, S.-L.; Feng, Q.; Li, S.; Liang, S.; Zhong, H.; Liu, Z.; Gao, Y.; Zhao, H.; et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 2017, 8, 845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feeney, A.; Sleator, R.D. The human gut microbiome: The ghost in the machine. Future Microbiol. 2012, 7, 1235–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khosravi, A.; Mazmanian, S.K. Disruption of the gut microbiome as a risk factor for microbial infections. Curr. Opin. Microbiol. 2013, 16, 221–227. [Google Scholar] [CrossRef] [Green Version]
- Sommer, M.O. Advancing gut microbiome research using cultivation. Curr. Opin. Microbiol. 2015, 27, 127–132. [Google Scholar] [CrossRef]
- Dabke, K.; Hendrick, G.; Devkota, S. The gut microbiome and metabolic syndrome. J. Clin. Investig. 2019, 129, 4050–4057. [Google Scholar] [CrossRef]
- Barko, P.C.; McMichael, M.A.; Swanson, K.S.; Williams, D.A. The gastrointestinal microbiome. J. Vet. Intern. Med. 2018, 32, 9–25. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [Green Version]
- Shao, Y.; Forster, S.C.; Tsaliki, E.; Vervier, K.; Strang, A.; Simpson, N.; Kumar, N.; Stares, M.D.; Rodger, A.; Brocklehurst, P.; et al. Stunted Microbiota and Opportunustic pathogens colonization in caesarean-section birth. Nature 2019, 574, 117–121. [Google Scholar] [CrossRef]
- Satokari, R.; Gronroo, T.; Laitinen, K.; Salminen, S.; Isolauri, E. Bifidobacterium and Lactobacillus DNA in the human placenta. Lett. Appl. Microbiol. 2009, 48, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014, 6, 237ra65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, K.; Rodriguez, M.D.; Paul, Z.; Gordon, E.; Rice, K.; Triplett, W.E.; Keller-Wood, M.; Wood, C.E. Proof of principle: Physiological transfer of small numbers of bacteria from mother to fetus in late-gestation pregnant sheep. PLoS ONE 2019, 14, e0217211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Goffau, M.C.; Lager, S.; Sovio, U.; Gaccioli, F.; Cook, E.; Peacock, S.J.; Parkhill, J.; Charnock-Jones, D.S.; Smith, G.C.S. Human placenta has no microbiome but can contain potential pathogens. Nature 2019, 572, 329–334. [Google Scholar] [CrossRef]
- Koenig, J.E.; Spor, A.; Scalfone, N.; Fricker, A.D.; Stombaugh, J.; Knight, R.; Angenent, L.T.; Ley, R.E. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA 2011, 108, 4578–4585. [Google Scholar] [CrossRef] [Green Version]
- Differding, M.K.; Benjamin-Neelon, S.E.; Hoyo, C.; Østbye, T.; Mueller, N.T. Timing of complementary feeding is associated with gut microbiota diversity and composition and short chain fatty acid concentrations over the first year of life. BMC Microbiol. 2020, 20, 56. [Google Scholar] [CrossRef] [Green Version]
- Cozzolino, A.; Vergalito, F.; Tremonte, P.; Iorizzo, M.; Lombardi, S.J.; Sorrentino, E.; Luongo, D.; Coppola, R.; Marco, R.D.; Succi, M. Preliminary evaluation of the safety and probiotic potential of Akkermansiamuciniphila OSM 22959 in comparison with Lactobacillus rhamnosus GG. Microorganisms 2020, 8, 189. [Google Scholar] [CrossRef] [Green Version]
- Amabebe, E.; Robert, F.O.; Agbalalah, T.; Orubu, E.S.F. Microbial dysbiosis-induced obesity: Role of gut microbiota in homeostasis of energy metabolism. Br. J. Nutr. 2020, 3, 1–23. [Google Scholar] [CrossRef]
- Barone, M.; Turroni, S.; Rampelli, S.; Soverini, M.; D’Amico, F.; Biagi, E.; Brigidi, P.; Troiani, E.; Candela, M. Gut microbiome response to a modern Paleolithic diet in a western lifestyle context. PLoS ONE 2019, 14, e0220619. [Google Scholar] [CrossRef] [Green Version]
- Caviglia, G.P.; Rosso, C.; Ribaldone, D.G.; Dughera, F.; Fagoonee, S.; Astegiano, M.; Pellicano, R. Physiopathology of intestinal barrier and the role of Zonulin. Minerva Biotecnol. 2019, 31, 83–92. [Google Scholar] [CrossRef]
- Kim, K.O.; Gluck, M. Fecal Microbiota Transplantation: An Update on Clinical Practice. Clin. Endosc. 2019, 52, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Jie, Z.; Cui, B.; Wang, H.; Feng, Q.; Zou, Y.; Zhang, X.; Yang, H.; Wang, J.; Zhang, F.; et al. Faecalmicrobiota transplantation results in bacterial strain displacement in patients with inflammatory bowel diseases. FEBS Open Bio 2019, 10, 41–55. [Google Scholar] [CrossRef] [PubMed]
- Bibbò, S.; Settanni, C.R.; Porcari, S.; Bocchino, E.; Ianiro, G.; Cammarota, G.; Gasbarrin, A. Fecal Microbiota Transplantation: Screening and Selection to Choose the Optimal Donor. J. Clin. Med. 2020, 9, 1757. [Google Scholar] [CrossRef]
- Sbahi, H.; Di Palma, J.A. Faecalmicrobiota transplantation: Applications and limitations in treating gastrointestinal disorders. BMJ Open Gastroenterol. 2016, 3, e000087. [Google Scholar] [CrossRef] [PubMed]
- Antushevich, H. Faecalmicrobita transplantation in disease therapy. Clin. Chim. Acta 2019, 503, 90–98. [Google Scholar] [CrossRef]
- Cammarota, G.; Laniro, G.; Kelly, C.R.; Mullish, B.H.; Allegretti, J.R.; Kassam, Z.; Putignani, L.; Fischer, M.; Keller, J.J.; Castello, S.P.; et al. International consensus conference on stool banking for faecalmicrobiota transplantation in clinical practice. Gut 2019, 68, 2111–2121. [Google Scholar] [CrossRef] [Green Version]
- Belizário, J.E.; Faintuch, J.; Garay-Malpartida, M. Gut Microbiome Dysbiosis and Immunometabolism: New Frontiers for Treatment of Metabolic Diseases. Mediat. Inflamm. 2018, 2037838, 1–12. [Google Scholar] [CrossRef]
- Wong, A.C.; Levy, M. New approaches to microbiome-based therapies. mSystems 2019, 4, e00122-19. [Google Scholar] [CrossRef] [Green Version]
- Guimaraes, N.; Azevedo, N.F.; Figueiredo, C.; Keevil, C.W.; Vieira, M. Development and application of a novel peptide nucleic acid probe for the specific detection of Helicobacter pylori in gastric biopsy specimen. Clin. Microbiol. 2007, 45, 3089–3094. [Google Scholar] [CrossRef] [Green Version]
- Baysal, A.H. Comparison of conventional culture method and fluorescent in situ hybridization technique from detection of Listeria Spp. In ground beef, turkey and chicken breast fillets in Izmir, Turkey. J. Food Prot. 2014, 77, 2021–2030. [Google Scholar] [CrossRef]
- Becattini, S.; Littmann, E.R.; Carter, R.A.; Kim, S.G.; Morjaria, S.M.; Ling, L.; Gyaltshen, Y.; Fontana, E.; Taur, Y.; Leiner, I.M.; et al. Commensal Microbesprovide First Line Defence Agaimst Listeria monocytogenes Infection. J. Exp. Med. 2017, 214, 1973–1989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prudent, E.; Raoult, D. Fluorescent in situ hybridization, a complementary molecular tool for the clinical diagnosis of infectious diseases by intracellular and fastidious bacteria. FEMS Microbiol. Rev. 2019, 43, 88–107. [Google Scholar] [CrossRef]
- Russmann, H.; Adler, K.; Haas, R.; Gebert, B.; Koletzko, S.; Heesemann, J. Rapid and accurate Determination of genotypic clarithromycin resistance in cultured Helicobacter pylori by fluorescent in situ hybridization. J. Clin. Microbial. 2001, 39, 4142–4144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, J.W.; Roach, J.; Azcarate-Peril, M.A. Emerging technologies for gut microbiome research. Trends Microbiol. 2016, 24, 887–901. [Google Scholar] [CrossRef] [Green Version]
- Borewicz, K.; Gu, F.; Saccenti, E.; Hechler, C.; Beijers, R.; de Weerth, C.; van Leeuwen, S.S.; Schols, H.A.; Smidt, H. The association between breast milk oligosaccharides and faecal microbiota in healthy breastfed infants at two, six, and twelve weeks of age. Sci. Rep. 2020, 10, 4270. [Google Scholar] [CrossRef] [PubMed]
- Backhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva, P.-D.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [Green Version]
- Lau, J.T.; Whelan, F.J.; Herath, I.; Lee, C.H.; Collins, S.M.; Bercik, P.; Surette, M.G. Capturing the diversity of the human gut microbiota through culture-enriched molecular profiling. Genome Med. 2016, 8, 72. [Google Scholar] [CrossRef] [Green Version]
- García-Mantrana, I.; Selma-Royo, M.; González, S.; Parra-Llorcac, A.; Martínez-Costad, C.; Collado, M.C. Distinct maternal microbiota clusters are associated with diet during pregnancy: Impact on neonatal microbiota and infant growth during the first 18 months of life. Gut Microbes 2020, 11, 962–978. [Google Scholar] [CrossRef] [Green Version]
- Beaumont, M.; Goodrich, J.K.; Jackson, M.A.; Yet, I.; Davenport, E.R.; Vieira, S.-S.; Debelius, J.; Pallister, T.; Mangino, M.; Raes, J.; et al. Heritable components of the human fecal microbiome are associated with visceral fat. Genome Biol. 2016, 17, 189. [Google Scholar] [CrossRef] [Green Version]
- Chijiwa, R.; Hoskawa, M.; Kogawa, M.; Nishikawa, Y.; Ide, K.; Sakanashi, C.; Takahashi, K.; Takeyama, H. Single-cell genomics of uncultured bacteria reveals dietary fiber responders in the mouse gut microbiota. Microbiome 2020, 8, 5. [Google Scholar] [CrossRef] [Green Version]
- Collado, M.C.; Rautava, S.; Aakko, J.; Isolauri, E.; Salminen, S. Human gut colonization may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016, 6, 23129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albenberg, L.; Kelsen, J. Advances in gut microbiome research and relevance to pediatric diseases. J. Pediatr. 2016, 178, 16–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saulnier, D.M.; Riehle, K.; Mistretta, T.A.; Diaz, M.-A.; Mandal, D.; Raza, S.; Weidler, E.M.; Qin, X.; Coarfa, C.; Milosavljevic, A.; et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 2011, 141, 1782–1791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogtmann, E.; Hua, X.; Zeller, G.; Sunagawa, S.; Voigt, A.Y.; Hercog, R.; Weidler, E.M.; Qin, X.; Coarfa, C.; Milosavljevic, A.; et al. Colorectal cancer and the human gut microbiome: Reproducibility with whole genome shotgun sequencing. PLoS ONE 2016, 11, e0155362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegwald, L.; Caboche, S.; Even, G.; Viscoglisoi, E.; Audebert, C.; Chabe, M. The impact of bioinformatics pipelines on microbiota studies: Does the analytical “Microscope” affect the biological interpretation? Microorganisms 2019, 7, 393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Walther-Antonio, M. Microfluidics: A new tool for microbial single cell analyses in human microbiome studies. Biomicrofluidics 2017, 11, 061501. [Google Scholar] [CrossRef]
- Udayasuryan, B.; Slade, D.J.; Verbridge, S.S. Microfluidics in Microbiome and Cancer Research; Wiley: Hoboken, NJ, USA, 2019; pp. 281–317. [Google Scholar]
- Kasendra, M.; Tovaglieri, A.; Sontheimer, A.-P.; Jalili-Firoozinezhad, S.; Bein, A.; Chalkiadaki, A.; Scholl, W.; Zhang, C.; Rickner, H.; Bein, A.; et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 2018, 8, 2871. [Google Scholar] [CrossRef]
- Cama, J.; Voliotis, M.; Metz, J.; Smith, A.; Iannucci, J.; Keyser, U.F.; Atanasova, K.T.; Pagliara, S. Single-cell microfluidics facilitates the rapid quantification of antibiotic accumulation in Gramnegative bacteria. Lab Chip 2020, 20, 2765–2775. [Google Scholar] [CrossRef]
- Sarangi, A.N.; Goel, A.; Aggarwal, R. Methods for studying gut microbita: A primer for physicians. J. Clin. Exp. Hepatol. 2019, 9, 62–73. [Google Scholar] [CrossRef] [Green Version]
- Duscha, A.; Gisevius, B.; Hirschberg, S.; Yissachar, N.; Stangl, G.J.; Eilers, E.; Bader, V.; Haase, S.; Kaisler, J.; David, C.; et al. Propionic acid shapes the multiple sclerosis disease course by an immunomodulatory mechanism. Cell 2020, 180, 1–14. [Google Scholar]
- Murugesan, S.; Nirmalkar, K.; Hoyo-Vadillo, C.; García-Espitia, M.; Ramírez-Sánchez, D.; García-Mena, J. Gut microbiome production of short-chain fatty acids and obesity in children. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 37, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Luan, H.; Wang, X.; Cai, Z. Mass spectrometry-based metabolomics: Targeting the crosstalk between gut microbiota and brain in neurodegenerative disorders. Mass Spectrom. Rev. 2017, 38, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D.; Forse, L.B. Development of Electronic-Nose technologies for early disease detection based on microbial dysbiosis. Proceedings 2019, 4, 32. [Google Scholar] [CrossRef] [Green Version]
- Maier, T.V.; Walker, A.; Heinzmann, S.S.; Forcisi, S.; Martinez, I.; Walter, J.; Kopplin, P.S. Challenges of metabolomics in human gut microbiota research. Int. J. Med. Microbiol. 2016, 306, 266–279. [Google Scholar]
- Bashiardes, S.; Zilberman-Schapira, G.; Elinav, E. Use of Metatranscriptomics in Microbiome Research. Bioinform. Biol. Insights 2016, 10, 19–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gosalbes, M.J.; Durban, A.; Pignatelli, M.; Abellan, J.J.; Jimenez-Hernandez, N.; Perez-Cobas, A.E.; Latorre, A.; Moya, A. Metatranscriptomic Approach to Analyze the Functional Human Gut Microbiota. PLoS ONE 2011, 6, e17447. [Google Scholar] [CrossRef]
- Li, F.; Hitch, T.C.A.; Chen, Y.; Creevey, C.J.; Guan, L.L. Comparative metagenomic and metatranscriptomic analyses reveal the breed effect on the rumen microbiome and its associations with feed efficiency in beef cattle. Microbiome 2019, 7, 6. [Google Scholar] [CrossRef] [Green Version]
- Shakya, M.; Lo, C.C.; Chain, P.S.G. Advances and Challenges in Metatranscriptomic Analysis. Front. Genet. 2019, 10, 904. [Google Scholar] [CrossRef] [Green Version]
- Lagier, J.-C.; Khelaifia, S.; Alou, M.T.; Ndongo, S.; Dione, N.; Hugon, P.; Caputo, A.; Cadoret, F.; Traore, S.I.; Secck, E.H.; et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 2016, 1, 16203. [Google Scholar] [CrossRef]
- Traore, S.I.; Bilen, M.; Cadoret, F.; Khelaifa, S.; Million, M.; Raoult, D.; Lagier, J.C. Study of huma gastrointestinal microbiota by culturomics in Africa. Med. Sante Trop. 2019, 29, 366–370. [Google Scholar]
- Goodman, A.L.; Kallstrom, G.; Faith, J.J.; Reyes, A.; Moore, A.; Dantas, G.; Gordon, J.I. Extensive personal human gut microbiota culture collection characterized and manipulated in gnobiotic mice. Proc. Natl. Acad. Sci. USA 2011, 108, 6252–6257. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Subject | Metabolic Syndrome/Disease | Correlated Clinical Indicator | Indicator Microbe (Healthy Metabolic State) | Indicator Microbe (Dysfunctional Metabolic State) | Reference |
|---|---|---|---|---|---|
| Adult (Male & Female) | Prediabetes or type 2 diabetes mellitus | Impaired lipid & glucose metabolism | Clostridia and Rikenellaceae members | Holdernania & Blautia genera | [10] |
| Adult | Obesity | NI | Balance population of Bacteroidetes & Firmicutes | Few Bacteroidetes & more Firmicutes | [11] |
| Adult | Alzheimer’s Disease | Low Mini-Mental State Examination score, APOE ε4 carriers, high Clinical Dementia Rating and Activity of Daily Living scores | Normal gut microbiota population comprising Firmicutes, Proteobacteria, Bacteroidetes and Actinobacteria | Significant decrease in the population of Negativicutes and Bacteroidia | [12,13] |
| Children | Autism | NI | Moderate level of Clostridium histolyticum, normal population of Firmicutes, Bacteroidetes | Lower levels of Prevotella, Coprococcus, Veillonellaceae, Firmicutes and Bifidobacterium and higher levels of Clostridium histolyticum, Desulfovibrio, Lactobacillus, Sarcina Clostridium, Bacteroidetes and Caloramator | [14] |
| Adult | Early chronic kidney disease | abnormal kidney structure, urinary albumin excretion rate ≥30 mg/24 h, glomerular filtration rate, 30–90 mL/minute/1.73 m2 | Abundance of Roseburia and other genera | Abundance of Ruminococcus and other genera | [9] |
| Adult | Atherosclerotic cardiovascular disease | stable angina, unstable angina, or acute myocardial infarction, ≥50% stenosis in single or multiple vessel | Higher population of Bacteroides and Prevotella | Relative reduction in Bacteroides and Prevotella and enrichment in Streptococcus spp. and Escherichia, Klebsiella spp., Enterobacter aerogenes | [15] |
| Subject | Methods Employed | Outcome | Reference |
|---|---|---|---|
| Association between breast milk oligosaccharides and fecal microbiota in healthy breast fed infants | 16S rRNA genes sequencing of V4 region using the Illumina Hiseq 2000 platform, porous graphitized carbon-ultra high-performance liquid chromatography (PGC-UPLC-MS) and bioinformatics (QIIME) | Microbiota composition strongly influenced by infant age, associated mode of delivery and breast milk | [47] |
| Dynamics and stabilization of the human gut microbiome during the first year of life | Metagenomics (DNA extraction from stool samples and preparation of DNA library using Illumina Hiseq2000) and bioinformatics (SOAPdenovo2, GeneMark v2.7, NCBI database) | Nutrition has a far reaching influence on infant microbiota composition and function with halting of breast-feeding other than introduction of solid food | [48] |
| Determining the diversity of human gut microbiota | Culture with enrichment, 16S rRNA gene sequencing of V3 region using the Illumina Miseq platform and bioinformatics (QIIME) | Use of enriched culture method enhanced the culturability of bacteria identified by 16S sequencing of the microbiota of the human gut | [49] |
| Impact of diet during pregnancy on maternal microbiota clusters and its influence on neonatal microbiota and infant growth during the first 18 months of life | 16S rRNA gene sequencing of V3-V4 region using Miseq Illumina platform. Bioinformatics (QIIME, LEfSe, Calypso online platform) | Diet is an important perinatal factor in the initial phase of life and have significant impact on neonatal microbiome | [50] |
| Heritable components of the human fecal microbiome are associated with visceral fat | Measuring of body composition by dual-energy X-ray absorptiometry, 16S rRNA gene sequencing of V4 region on Illumina Miseq platform and bioinformatics (QIIME 1.7.0, PICRUSt v1.0.0, STAMP) | There was significant association of adiposity-OTU abundance with host genetic variations indicating possible role of host genes in influencing the link between obesity and fecal microbiome | [51] |
| Succession of microbial consortia in the developing infant gut microbiome | 454-pyrosequencing of 16S rRNA gene, GC-MS analysis of SCFA, quantitative PCR and bioinformatics (QIIME, MG-RAST, NCBI database) | Revealed shifts in microbiome associated with life events | [26] |
| Identification of uncultured bacteria that are metabolic responders in a microbiota | Massively parallel single-cell genome sequencing technique (SAG-gel Platform), 16S rRNA gene sequencing of V3-V4 using Illumina Miseq 2 x 300bp platform and bioinformatics (QIIME2 v.2019.1). Determination of the concentration of SCFA was done by GC-mass spectrophotometry | Functions of uncultured bacteria in the microbiota were elucidated | [52] |
| Study of human gut colonization linked to in utero by microbial communities in the amniotic fluid and placenta | Culture, Gradient Gel Electrophoresis (DGGE), 16S rRNA gene pyrosequencing of V1-V3 region, quantitative PCR and bioinformatics (PICRUSt, QIIME, LEfSe) | The microbiota composition of infant gut at the age of 3-4 days begins to look like that detected in colostrum hence, the presumption that colonization is initiated prenatally by a distinct microbiota in the amniotic fluid and placenta | [53] |
| Technology/Methodology | Advantage | Disadvantage | Potential Clinical Application |
|---|---|---|---|
| Metagenomics (High-through sequencing) |
|
|
|
| High-throughput Metabolomics |
|
|
|
| High-throughput Metatranscriptomics |
|
|
|
| Microfluidics |
|
|
|
| High-throughput Culturing |
|
|
|
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ajayi, A.; Jolaiya, T.; Smith, S. Evolving Technologies in Gastrointestinal Microbiome Era and Their Potential Clinical Applications. J. Clin. Med. 2020, 9, 2565. https://doi.org/10.3390/jcm9082565
Ajayi A, Jolaiya T, Smith S. Evolving Technologies in Gastrointestinal Microbiome Era and Their Potential Clinical Applications. Journal of Clinical Medicine. 2020; 9(8):2565. https://doi.org/10.3390/jcm9082565
Chicago/Turabian StyleAjayi, Abraham, Tolulope Jolaiya, and Stella Smith. 2020. "Evolving Technologies in Gastrointestinal Microbiome Era and Their Potential Clinical Applications" Journal of Clinical Medicine 9, no. 8: 2565. https://doi.org/10.3390/jcm9082565





