H2A Histone Family Member X (H2AX) Is Upregulated in Ovarian Cancer and Demonstrates Utility as a Prognostic Biomarker in Terms of Overall Survival
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Clinical Samples
2.2. Cell Culture
2.3. DNA Damage Induction by H2O2 Exposure
2.4. Immunofluorescence—γ-H2AX Assay
2.5. RNA Isolation, cDNA Synthesis and Quantitative RT-PCR
2.6. Immunohistochemistry for Tissue Microarray
2.7. Bioinformatic Analysis
2.8. Population Sample and Genotyping
2.9. Statistical Analysis
3. Results
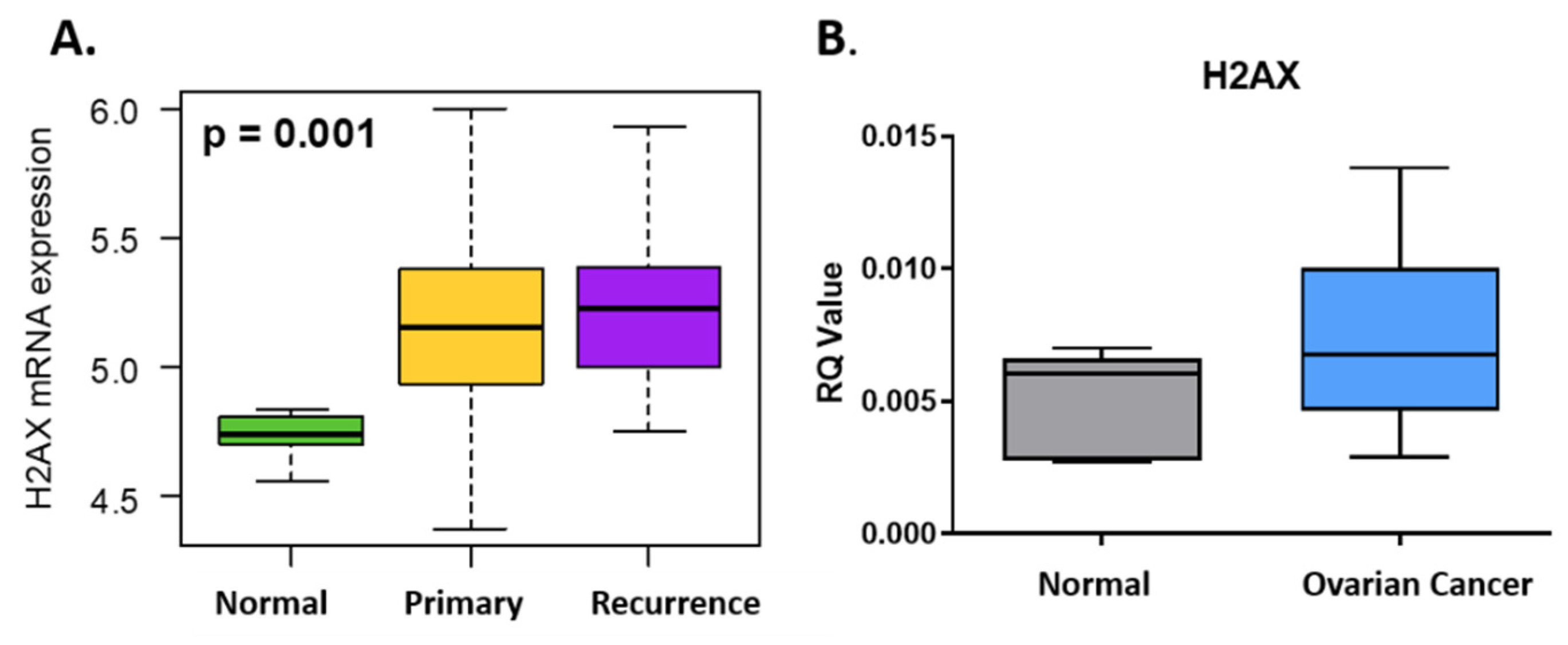
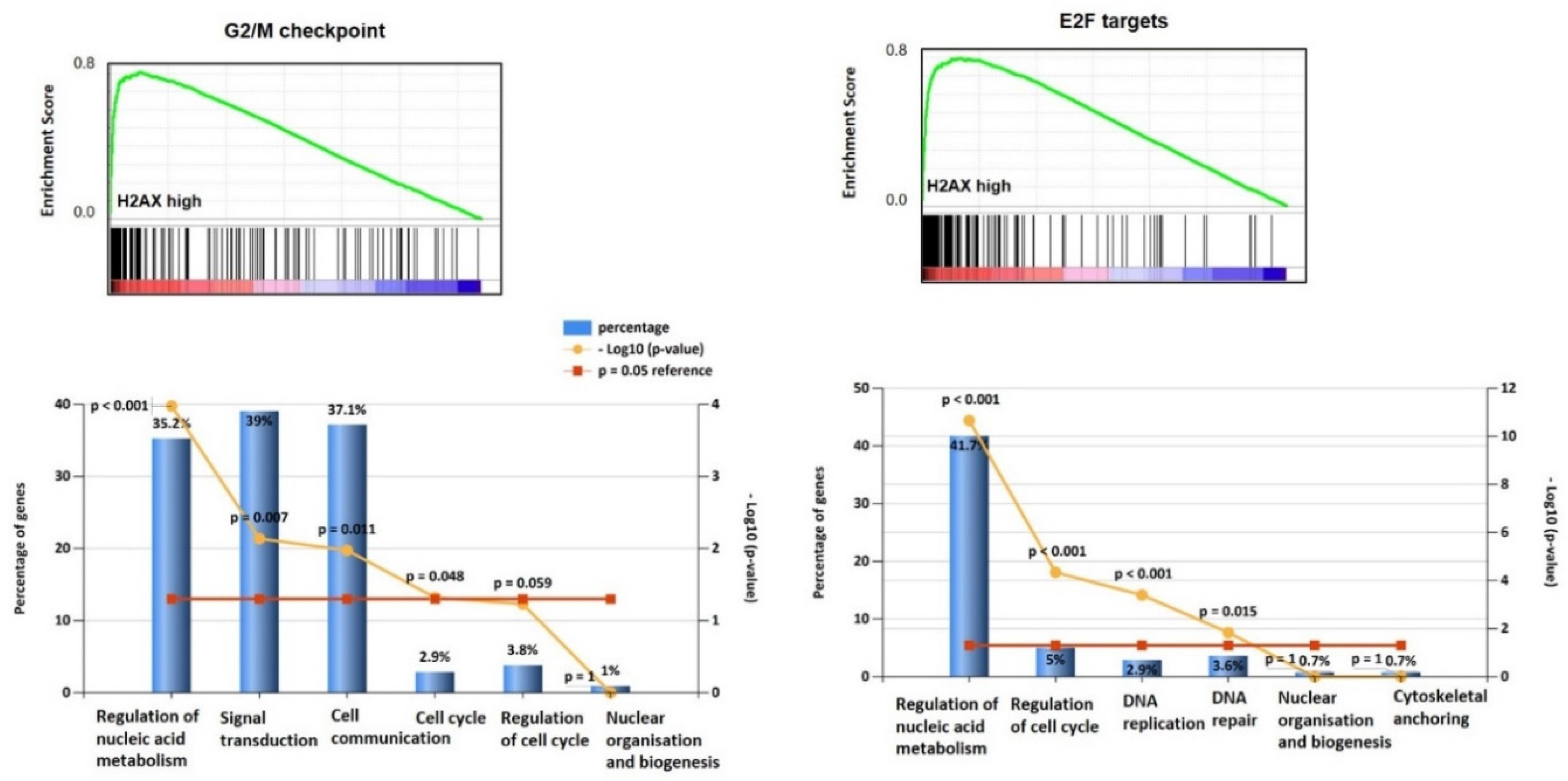
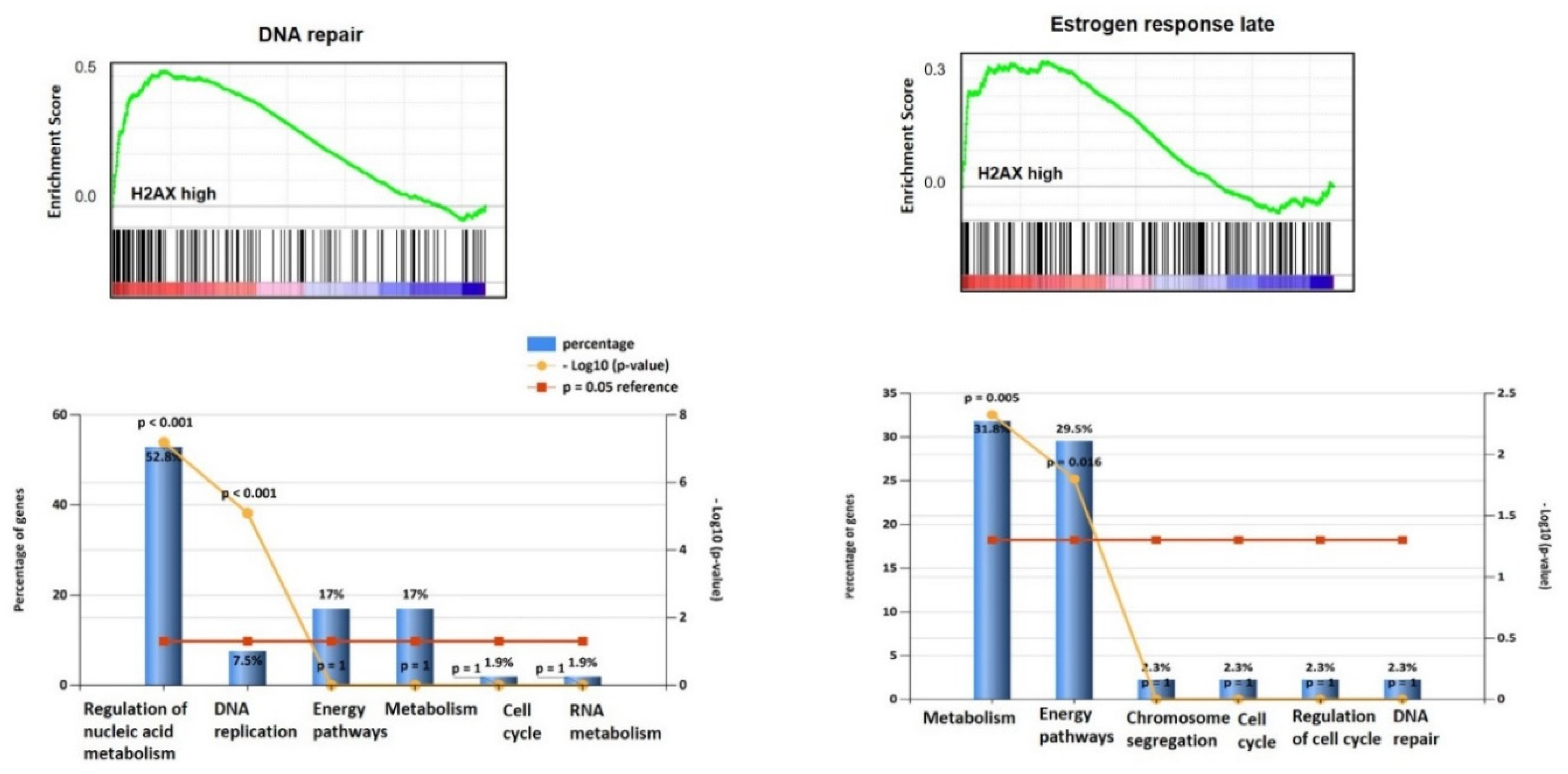
3.1. H2AX Is Over-Expressed in Ovarian Cancer and Predicts Survival
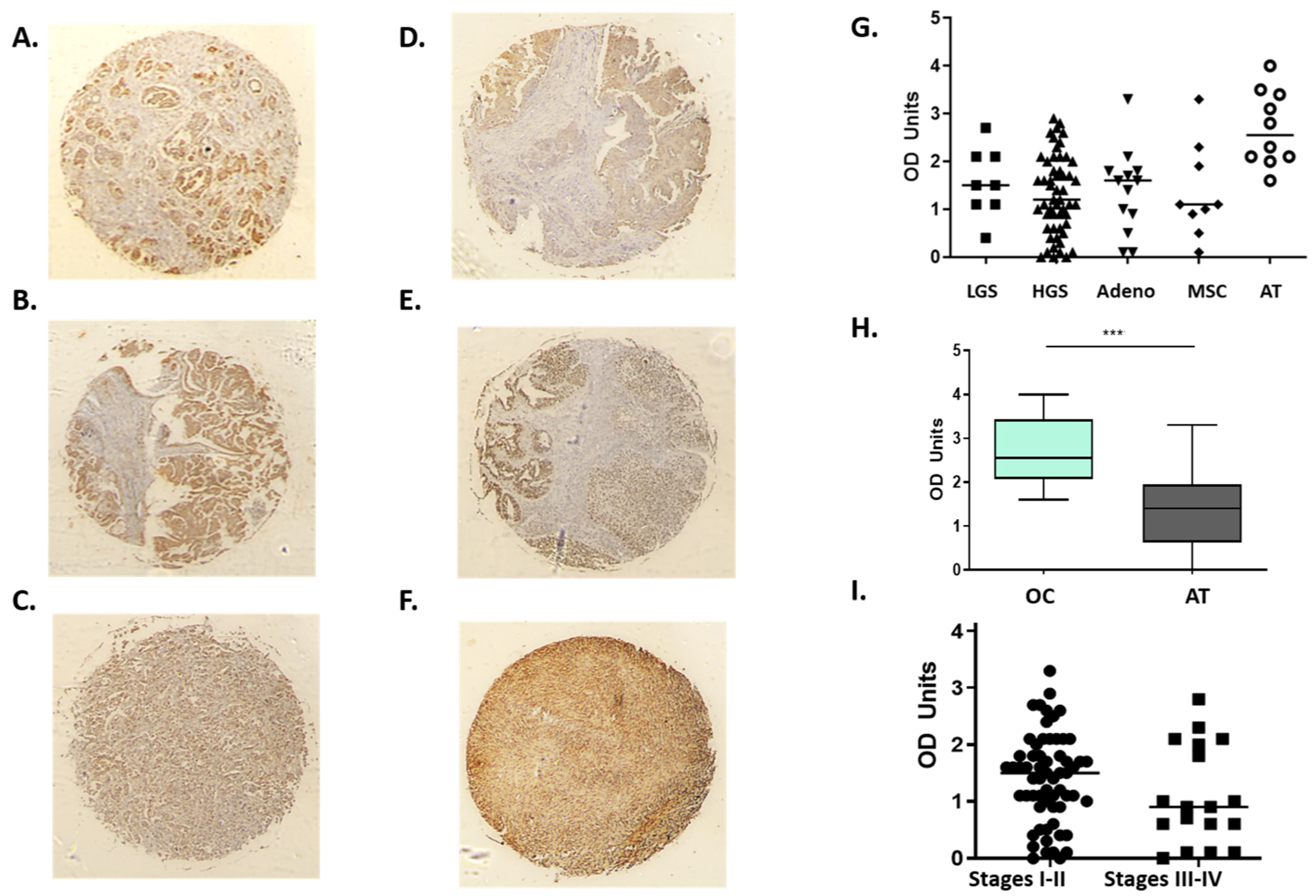
3.2. H2AX Protein Is Abundantly Expressed in Ovarian Cancer Tissues
3.3. Genetic Association Analysis for H2AX
3.4. H2AX Expression Correlates with γ-H2AX Staining In Vitro
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ledermann, J.A.; Drew, Y.; Kristeleit, R.S. Homologous recombination deficiency and ovarian cancer. Eur. J. Cancer 2016, 60, 49–58. [Google Scholar] [CrossRef]
- O’Donovan, P.J.; Livingston, D.M. BRCA1 and BRCA2: Breast/ovarian cancer susceptibility gene products and participants in DNA double-strand break repair. Carcinogenesis 2010, 31, 961–967. [Google Scholar] [CrossRef]
- Hoppe, M.M.; Sundar, R.; Tan, D.S.P.; Jeyasekharan, A.D. Biomarkers for Homologous Recombination Deficiency in Cancer. J. Natl. Cancer Inst. 2018, 110, 704–713. [Google Scholar] [CrossRef]
- Brandsma, I.; Van Gent, D.C. Pathway choice in DNA double strand break repair: Observations of a balancing act. Genome Integr. 2012, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Kuo, L.J.; Yang, L.X. γ-H2AX- A novel biomaker for DNA double-strand breaks. In Vivo (Brooklyn) 2008, 22, 305–310. [Google Scholar]
- Palla, V.-V.; Karaolanis, G.; Katafigiotis, I.; Anastasiou, I.; Patapis, P.; Dimitroulis, D.; Perrea, D. gamma-H2AX: Can it be established as a classical cancer prognostic factor? Tumor Biol. 2017, 39. [Google Scholar] [CrossRef]
- Bassing, C.H.; Chua, K.F.; Sekiguchi, J.; Suh, H.; Whitlow, S.R.; Fleming, J.C.; Monroe, B.C.; Ciccone, D.N.; Yan, C.; Vlasakova, K.; et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc. Natl. Acad. Sci. USA 2002, 99, 8173–8178. [Google Scholar] [CrossRef] [PubMed]
- Celeste, A.; Difilippantonio, S.; Difilippantonio, M.J.; Fernandez-Capetillo, O.; Pilch, D.R.; Sedelnikova, O.A.; Eckhaus, M.; Ried, T.; Bonner, W.M.; Nussenzweig, A. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell (Cambridge) 2003, 114, 371–383. [Google Scholar] [CrossRef]
- Bassing, C.H.; Suh, H.; Ferguson, D.O.; Chua, K.F.; Manis, J.; Eckersdorff, M.; Gleason, M.; Bronson, R.; Lee, C.; Alt, F.W. Histone H2AX. Cell 2003, 114, 359–370. [Google Scholar] [CrossRef]
- Dickey, J.S.; Redon, C.E.; Nakamura, A.J.; Baird, B.J.; Sedelnikova, O.A.; Bonner, W.M. H2AX: Functional roles and potential applications. Chromosoma 2009, 118, 683–692. [Google Scholar] [CrossRef]
- Bignell, G.R.; Greenman, C.D.; Davies, H.; Butler, A.; Edkins, S.; Andrews, J.M.; Buck, G.; Chen, L.; Beare, D.; Latimer, C.; et al. Europe PMC Funders Group Signatures of mutation and selection in the cancer genome. Nature 2011, 463, 7283. [Google Scholar]
- Beroukhim, R.; Mermel, C.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Yang, Y.; Liu, Y.; Chong, C. The Role of Chromosome Deletions in Human Cancers. In Chromosome Translocation; Zhang, Y., Ed.; Springer: Singapore, 2018; pp. 135–148. [Google Scholar]
- Parikh, A.R.; White, J.S.; Huang, X.; Schoppy, D.W.; Baysal, B.E.; Baskaran, R.; Bakkenist, C.J.; Saunders, W.S.; Hsu, L.; Romkes, M.; et al. Loss of distal 11q is associated with DNA repair deficiency and reduced sensitivity to ionizing radiation in head and neck squamous cell carcinoma. Genes Chrom. Cancer 2007, 46, 761–775. [Google Scholar] [CrossRef] [PubMed]
- Mankarious, A.; Dave, F.; Pados, G.; Tsolakidis, D.; Gidron, Y.; Pang, Y.; Thomas, P.; Hall, M.; Karteris, E. The pro-social neurohormone oxytocin reverses the actions of the stress hormone cortisol in human ovarian carcinoma cells in vitro. Int. J. Oncol. 2016, 48, 1805–1814. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Skanderup, A.J.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK biobank resource with deep phenotyping and genomic data. Nature (Lond.) 2018, 562, 203–209. [Google Scholar] [CrossRef]
- Pruim, R.J.; Welch, R.P.; Sanna, S.; Teslovich, T.M.; Chines, P.S.; Gliedt, T.P.; Boehnke, M.; Abecasis, G.R.; Willer, C.J. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 2010, 26, 2336–2337. [Google Scholar] [CrossRef]
- Katsuta, E.; Ouchi, M.; Ouchi, T.; Takabe, K. Abstract 3216: H2AX is a novel prognostic marker of breast cancer. Tumor Biol. 2018, 78, 3216. [Google Scholar] [CrossRef]
- Mei, L.; Hu, Q.; Peng, J.; Ruan, J.; Zou, J.; Huang, Q.; Liu, S.; Wang, H. Phospho-histone H2AX is a diagnostic and prognostic marker for epithelial ovarian cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 5597–5602. [Google Scholar]
- Altomare, D.A.; Wang, H.Q.; Skele, K.L.; De Rienzo, A.; Klein-Szanto, A.J.; Godwin, A.K.; Testa, J.R. AKT and mTOR phosphorylation is frequently detected in ovarian cancer and can be targeted to disrupt ovarian tumor cell growth. Oncogene 2004, 23, 5853–5857. [Google Scholar] [CrossRef]
- Rogers-Broadway, K.; Kumar, J.; Sisu, C.; Wander, G.; Mazey, E.; Jeyaneethi, J.; Pados, G.; Tsolakidis, D.; Klonos, E.; Grunt, T.; et al. Differential expression of mTOR components in endometriosis and ovarian cancer: Effects of rapalogues and dual kinase inhibitors on mTORC1 and mTORC2 stoichiometry. Int. J. Mol. Med. 2018, 43, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Rogers-Broadway, K.-R.; Chudasama, D.; Pados, G.; Tsolakidis, D.; Goumenou, A.; Hall, M.; Karteris, E. Differential effects of rapalogues, dual kinase inhibitors on human ovarian carcinoma cells in vitro. Int. J. Oncol. 2016, 49, 133–143. [Google Scholar] [CrossRef][Green Version]
- Aran, D.; Camarda, R.; Odegaard, J.; Paik, H.; Oskotsky, B.; Krings, G.; Goga, A.; Sirota, M.; Butte, A.J. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat. Commun. 2017, 8, 1077. [Google Scholar] [CrossRef] [PubMed]
- Loret, N.; Denys, H.; Tummers, P.; Berx, G. The Role of Epithelial-to-Mesenchymal Plasticity in Ovarian Cancer Progression and Therapy Resistance. Cancers 2019, 11, 838. [Google Scholar] [CrossRef] [PubMed]
- Lal, A.; Pan, Y.; Navarro, F.; Dykxhoorn, D.M.; Moreau, L.; Meire, E.; Bentwich, Z.; Lieberman, J.; Chowdhury, D. miR-24–mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat. Struct. Mol. Biol. 2009, 16, 492–498. [Google Scholar] [CrossRef]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stähler, C.; Meese, E.; et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar] [CrossRef]
- Liu, W.; Wang, S.; Zhou, S.; Yang, F.; Jiang, W.; Zhang, Q.; Wang, L. A systems biology approach to identify microRNAs contributing to cisplatin resistance in human ovarian cancer cells. Mol. BioSyst. 2017, 13, 2268–2276. [Google Scholar] [CrossRef]
- Launonen, V.; Stenbäck, F.; Puistola, U.; Bloigu, R.; Huusko, P.; Kytölä, S.; Kauppila, A.; Winqvist, R. Chromosome 11q22.3-q25 LOH in Ovarian Cancer: Association with a More Aggressive Disease Course and Involved Subregions. Gynecol. Oncol. 1998, 71, 299–304. [Google Scholar] [CrossRef]
- Launonen, V.; Mannermaa, A.; Stenbäck, F.; Kosma, V.; Puistola, U.; Huusko, P.; Anttila, M.; Bloigu, R.; Saarikoski, S.; Kauppila, A.; et al. Loss of heterozigocity at chromosomes 3, 6, 8, 11, 16, and 17 in Ovarian Cancer: Correlation to Clinicopathological Variables. Cancer Genet. Cytogenet. 2000, 122, 49–54. [Google Scholar] [CrossRef]
- Staff, S.; Tolonen, T.; Laasanen, S.-L.; Mecklin, J.-P.; Isola, J.; Mäenpää, J. Quantitative Analysis of γ-H2AX and p53 Nuclear Expression Levels in Ovarian and Fallopian Tube Epithelium from Risk-reducing Salpingo-Oophorectomies in BRCA1 and BRCA2 Mutation Carriers. Int. J. Gynecol. Pathol. 2014, 33, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Ly, T.; Endo, A.; Brenes, A.; Gierlinski, M.; Afzal, V.; Pawellek, A.; Lamond, A.I. Proteome-wide analysis of protein abundance and turnover remodelling during oncogenic transformation of human breast epithelial cells. Wellcome Open Res. 2018, 3, 51. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.; Tutt, A.; Ashworth, A. Hallmarks of ’BRCAness’ in sporadic cancers. Nat. Rev. Cancer 2004, 4, 814–819. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.P.; Zhu, Y.-L.; Lo, Y.-C.; Moscarelli, J.; Xiong, A.; Korayem, Y.; Huang, P.H.; Giri, S.; Lorusso, P.; Ratner, E.S. Combination of triapine, olaparib, and cediranib suppresses progression of BRCA-wild type and PARP inhibitor-resistant epithelial ovarian cancer. PLoS ONE 2018, 13, e0207399. [Google Scholar] [CrossRef]
- Beaufort, C.M.; Helmijr, J.C.A.; Piskorz, A.M.; Hoogstraat, M.; Ruigrok-Ritstier, K.; Besselink, N.; Murtaza, M.; Van Ijcken, W.F.J.; Heine, A.A.J.; Smid, M.; et al. Ovarian Cancer Cell Line Panel (OCCP): Clinical Importance of In Vitro Morphological Subtypes. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saravi, S.; Katsuta, E.; Jeyaneethi, J.; Amin, H.A.; Kaspar, M.; Takabe, K.; Pados, G.; Drenos, F.; Hall, M.; Karteris, E. H2A Histone Family Member X (H2AX) Is Upregulated in Ovarian Cancer and Demonstrates Utility as a Prognostic Biomarker in Terms of Overall Survival. J. Clin. Med. 2020, 9, 2844. https://doi.org/10.3390/jcm9092844
Saravi S, Katsuta E, Jeyaneethi J, Amin HA, Kaspar M, Takabe K, Pados G, Drenos F, Hall M, Karteris E. H2A Histone Family Member X (H2AX) Is Upregulated in Ovarian Cancer and Demonstrates Utility as a Prognostic Biomarker in Terms of Overall Survival. Journal of Clinical Medicine. 2020; 9(9):2844. https://doi.org/10.3390/jcm9092844
Chicago/Turabian StyleSaravi, Sayeh, Eriko Katsuta, Jeyarooban Jeyaneethi, Hasnat A. Amin, Matthias Kaspar, Kazuaki Takabe, George Pados, Fotios Drenos, Marcia Hall, and Emmanouil Karteris. 2020. "H2A Histone Family Member X (H2AX) Is Upregulated in Ovarian Cancer and Demonstrates Utility as a Prognostic Biomarker in Terms of Overall Survival" Journal of Clinical Medicine 9, no. 9: 2844. https://doi.org/10.3390/jcm9092844
APA StyleSaravi, S., Katsuta, E., Jeyaneethi, J., Amin, H. A., Kaspar, M., Takabe, K., Pados, G., Drenos, F., Hall, M., & Karteris, E. (2020). H2A Histone Family Member X (H2AX) Is Upregulated in Ovarian Cancer and Demonstrates Utility as a Prognostic Biomarker in Terms of Overall Survival. Journal of Clinical Medicine, 9(9), 2844. https://doi.org/10.3390/jcm9092844









